Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Birds, Experimental Design and Sampling
2.2. Gene Expression Analysis
2.3. T-RFLP Analysis of Bacterial Community
2.4. Statistical Methods
3. Results
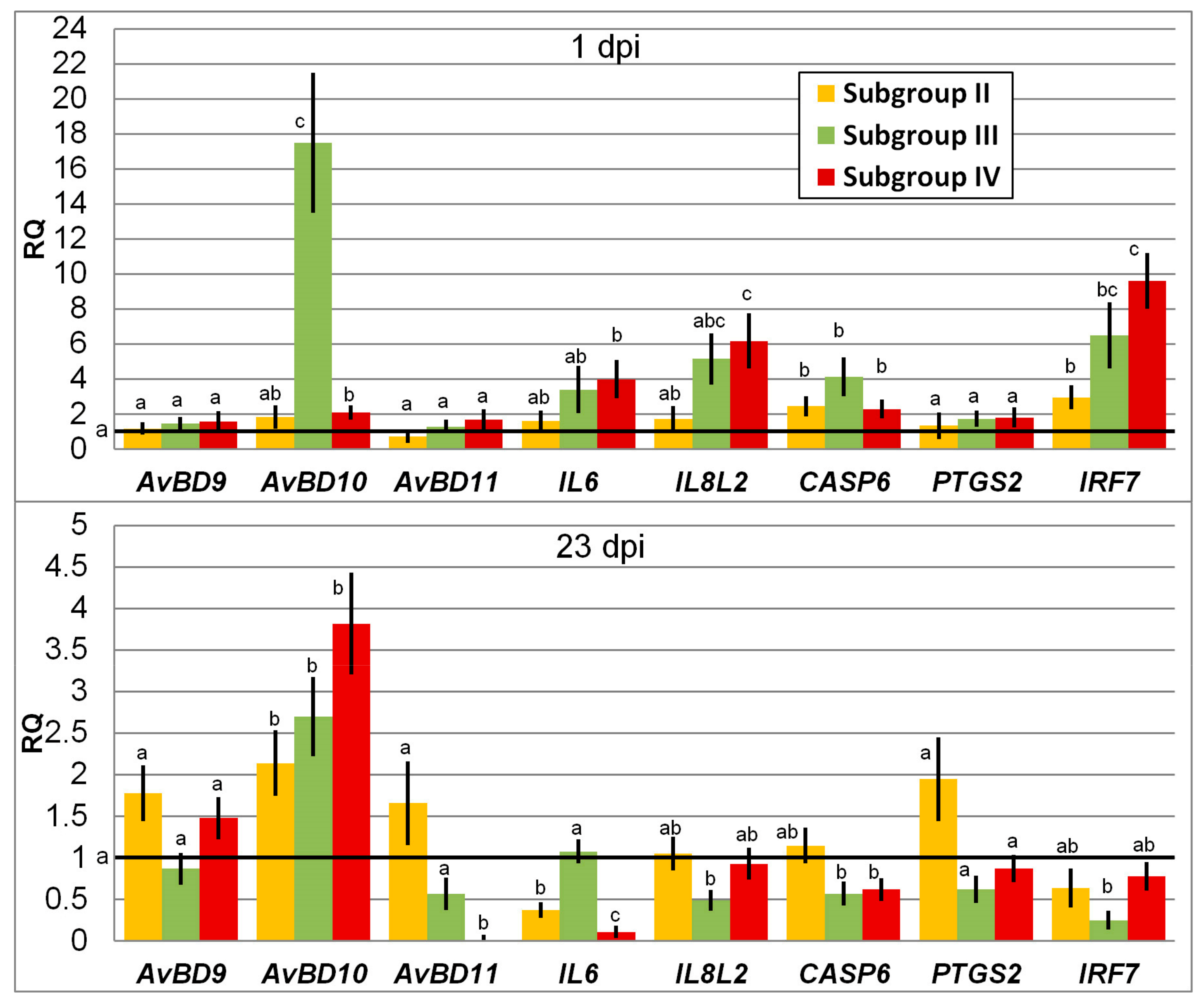
3.1. Change in Expression of Genes Involved in Immune Response
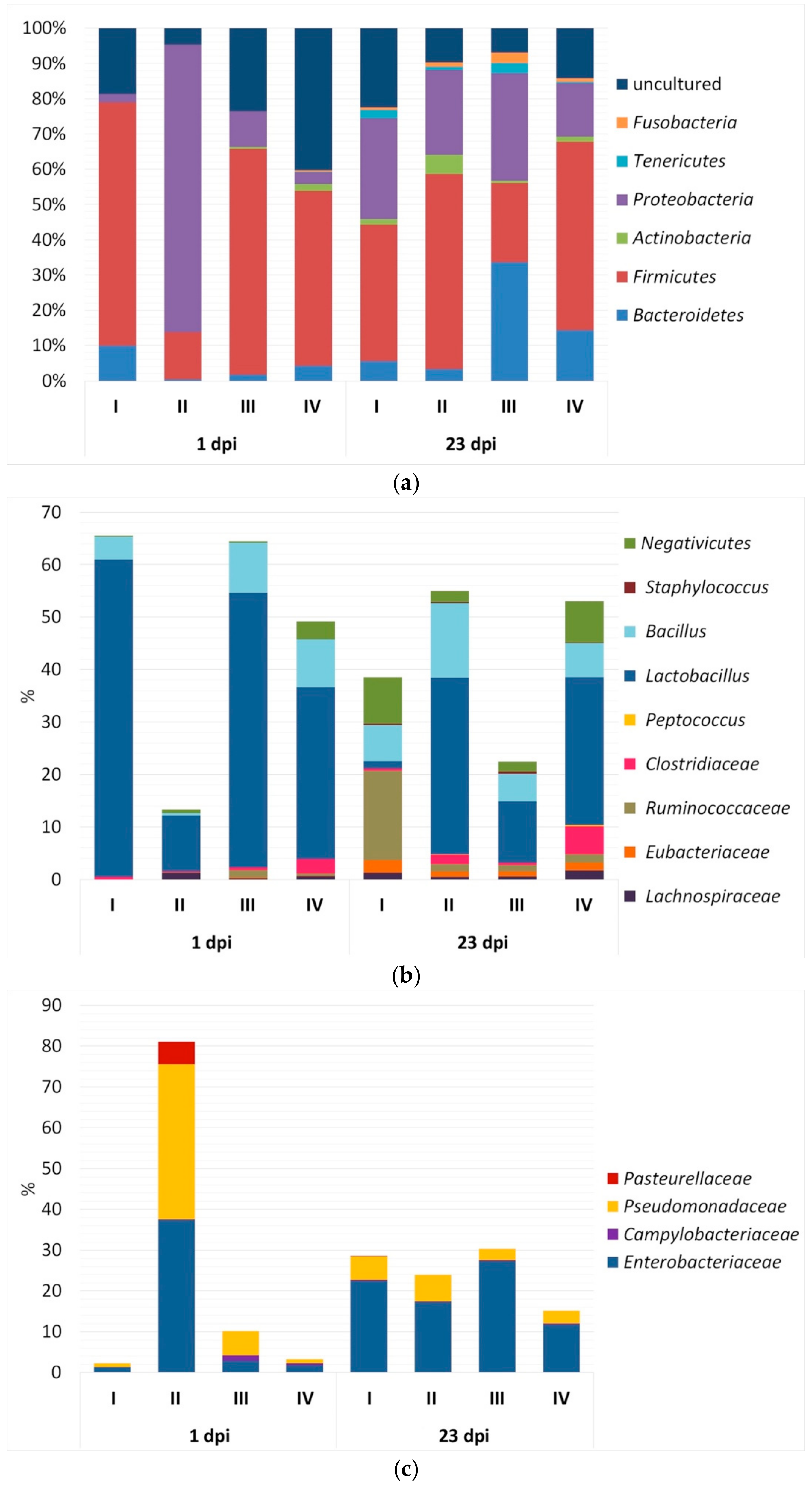
3.2. Bacterial Community Composition in Caecal Contents
3.3. Differences in Performance of Growing Chickens
4. Discussion
4.1. Immunity Gene Expression in Response to SE and Phytobiotic
4.2. Microbial Community Composition Due to Infection and Phytobiotic
4.3. Observed Effects on Performance
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fisinin, V.I.; Surai, P. Gut immunity in birds: Facts and reflections (review). Sel’skokhozyaistvennaya Biol. 2013, 4, 3–25. [Google Scholar] [CrossRef]
- Klasing, K.C. Nutrition and the immune system. Br. Poult. Sci. 2007, 48, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Verstegen, M.; Tamminga, S.; Williams, B. The role of the commensal gut microbial community in broiler chickens. World’s Poult. Sci. J. 2005, 61, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Adegunloye, D.V. Microorganism associated with poultry faeces. J. Food Agric. Environ. 2006, 4, 41–42. [Google Scholar]
- McDougald, L.R. Protozoal Infections. In Diseases of Poultry, 11th rev. ed.; Saif, Y.M., Ed.; Iowa State University Press: Ames, IA, USA, 2003; pp. 973–974. [Google Scholar]
- Reynolds, D.L. Multicausal Enteric Diseases. In Diseases of Poultry, 11th rev. ed.; Saif, Y.M., Ed.; Iowa State University Press: Ames, IA, USA, 2003; pp. 1169–1171. ISBN 978-0-8138-0423-1. [Google Scholar]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Grozina, A.A. Influence of the Ration Composition on the Microbiological Indices in the Gastrointestinal Tract of Broiler Chickens When Feeding Stafac 110 and Cellobacterin-T Preparations. Candidate of Biological Sciences Dissertation, Federal State Budget Scientific Institution ‘All-Russian Poultry Research and Technological Institute’, Sergiyev Posad, Russia, 2015. [Google Scholar]
- Nikonov, I.N.; Kochish, I.I.; Ilina, L.A.; Romanov, M.N.; Shevkhuzhev, A.F. Microbiota in the intestines of cross chick Lohmann Brown in ontogeny. Res. J. Pharm. Biol. Chem. Sci. 2017, 8, 645–654. [Google Scholar]
- Tlaskalová-Hogenová, H.; Stepánková, R.; Hudcovic, T.; Tucková, L.; Cukrowska, B.; Lodinová-Zádníková, R.; Kozáková, H.; Rossmann, P.; Bártová, J.; Sokol, D.; et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 2004, 93, 97–108. [Google Scholar] [CrossRef]
- Esmaeilipour, O.; Moravej, H.; Shivazad, M.; Rezaian, M.; Aminzadeh, S.; Van Krimpen, M. Effects of diet acidification and xylanase supplementation on performance, nutrient digestibility, duodenal histology and gut microflora of broilers fed wheat based diet. Br. Poult. Sci. 2012, 53, 235–244. [Google Scholar] [CrossRef]
- Spiridonov, A.N.; Petrova, O.N.; Irza, V.N.; Karaulov, A.K.; Nikiforov, V.V. Epizootic situation on infectious avian diseases based on analysis of data from veterinary reports. Vet. Segodnia 2015, 4, 18–28. [Google Scholar]
- Mughini-Gras, L.; Enserink, R.; Friesema, I.; Heck, M.; Van Duynhoven, Y.; Van Pelt, W. Risk factors for human salmonellosis originating from pigs, cattle, broiler chickens and egg laying hens: A combined case-control and source attribution analysis. PLoS ONE 2014, 9, e87933. [Google Scholar] [CrossRef]
- Barrow, P.A. Further observations on the effect of feeding diets containing avoparcin on the excretion of salmonellas by experimentally infected chickens. Epidemiol. Infect. 1989, 102, 239–252. [Google Scholar] [CrossRef] [Green Version]
- Sekirov, I.; Tam, N.M.; Jogova, M.; Robertson, M.L.; Li, Y.; Lupp, C.; Finlay, B.B. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008, 76, 4726–4736. [Google Scholar] [CrossRef]
- Croswell, A.; Amir, E.; Teggatz, P.; Barman, M.; Salzman, N.H. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect. Immun. 2009, 77, 2741–2753. [Google Scholar] [CrossRef]
- Wilson, J.L.; Buhr, R.J.; Cosby, D.E.; Cox, N.A.; Harrison, M.A.; Fedorka-Cray, P.J. Salmonella and antimicrobial resistance in broilers: A review. J. Appl. Poult. Res. 2015, 24, 408–426. [Google Scholar]
- Nair, V.T.D.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 2018, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Berndt, A.; Wilhelm, A.; Jugert, C.; Pieper, J.; Sachse, K.; Methner, U. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 2007, 75, 5993–6007. [Google Scholar] [CrossRef]
- Crhanova, M.; Hradecka, H.; Faldynova, M.; Matulova, M.; Havlickova, H.; Sisak, F.; Rychlik, I. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar Enteritidis infection. Infect. Immun. 2011, 79, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, F.; Giardi, M.F.; Botti, D. Avian cytokines—An overview. Curr. Pharm. Des. 2006, 12, 3083–3099. [Google Scholar] [CrossRef] [PubMed]
- Akbari, M.R.; Haghighi, H.R.; Chambers, J.R.; Brisbin, J.; Read, L.R.; Sharif, S. Expression of antimicrobial peptides in cecal tonsils of chickens treated with probiotics and infected with Salmonella enterica serovar Typhimurium. Clin. Vaccine Immunol. 2008, 15, 1689–1693. [Google Scholar] [CrossRef]
- Wigley, P. Salmonella enterica in the chicken: How it has helped our understanding of immunology in a non-biomedical model species. Front. Immunol. 2014, 5, 482. [Google Scholar] [CrossRef]
- Lee, M.O.; Romanov, M.N.; Plemyashov, K.V.; Dementieva, N.V.; Barkova, O.Y.; Womack, J.E.; Mitrofanova, O.V. Haplotype structure and copy number polymorphism of the beta-defensin 7 genes in diverse chicken breeds. Anim. Genet. 2017, 48, 490–492. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Li, S.; Zhang, L.; Liu, X.; Li, D.; Zhao, X.; Liu, Y. Expression of β-defensins in intestines of chickens injected with vitamin D3 and lipopolysaccharide. Genet. Mol. Res. 2015, 14, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, L.; Li, S.; Ouyang, L.; Robinson, K.; Tang, Y.; Zhu, Q.; Hu, Y.; Liu, Y. 1,25-Dihydroxyvitamin-D3 induces avian β-defensin gene expression in chickens. PLoS ONE 2016, 11, 0154546. [Google Scholar] [CrossRef] [PubMed]
- Menendez, A.; Finlay, B.B. Defensins in the immunology of bacterial infections. Curr. Opin. Immunol. 2007, 19, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Matulova, M.; Rajova, J.; Vlasatikova, L.; Volf, J.; Stepanova, H.; Havlickova, H.; Sisak, F.; Rychlik, I. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. PLoS ONE 2012, 7, e48101. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Higgs, R.; Lloyd, A.T.; O’Farrelly, C.; Hervé-Grépinet, V.; Nys, Y.; Brinkman, F.S.; Yu, P.L.; Soulier, A.; Kaiser, P.; et al. Avian beta-defensin nomenclature: A community proposed update. Immunol. Lett. 2007, 110, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Barkova, O.Y.; Laptev, G.Y.; Kochish, I.I.; Romanov, M.N.; Shevkhuzhev, A.F. Overview of genes associated with egg productivity and resistance of domestic hen. Res. J. Pharm. Biol. Chem. Sci. 2017, 8, 638–644. [Google Scholar]
- Li, X.; Swaggerty, C.L.; Kogut, M.H.; Chiang, H.I.; Wang, Y.; Genovese, K.J.; He, H.; Zhou, H. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS ONE 2010, 5, e11827. [Google Scholar] [CrossRef]
- Yang, D.; Biragyn, A.; Kwak, L.W.; Oppenheim, J.J. Mammalian defensins in immunity: More than just microbicidal. Trends Immunol. 2002, 23, 291–296. [Google Scholar] [CrossRef]
- Feng, Y.; Yang, X.J.; Wang, Y.B.; Li, W.L.; Liu, Y.; Yin, R.Q.; Yao, J.H. Effects of immune stress on performance parameters, intestinal enzyme activity and mRNA expression of intestinal transporters in broiler chickens. Asian Australas. J. Anim. Sci. 2012, 25, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pimenova, V.; Laishevtcev, A.; Pimenov, N. The main directions of health improvement measures for salmonellosis of birds: The principles and disadvantages of antibiotic treatment. Russ. J. Agric. Socio Econ. Sci. 2017, 71, 496–510. [Google Scholar] [CrossRef]
- Manuzon, M.; Lehman, M.; Wan, K.; Luo, H.; Yousef, A.; Wang, H.H.; Wittum, T.E.; Bakaletz, L.O. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 2006, 255, 328. [Google Scholar]
- Yang, C.; Chowdhury, M.A.K.; Hou, Y.; Gong, J. Phytogenic compounds as alternatives to in-feed antibiotics: Potentials and challenges in application. Pathogens 2015, 4, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, V.S.; Glebova, I.V. Antibacterial effect of essential oils of medicinal plants (review). Probl. Biol. Produktivn. Zhivotn. 2017, 3, 5–25. [Google Scholar]
- Jamroz, D.; Wiliczkiewicz, A.; Wertelecki, T.; Orda, J.; Skorupińska, J. Use of active substances of plant origin in chicken diets based on maize and locally grown cereals. Br. Poult. Sci. 2005, 46, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.; Ko, Y.; Kang, S.; Lee, C. Effect of a commercial essential oil on growth performance, digestive enzyme activity and intestinal microflora population in broiler chickens. Anim. Feed. Sci. Technol. 2007, 134, 304–315. [Google Scholar] [CrossRef]
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Idaomar, M. Biological effects of essential oils–A review. Food Chem. Toxicol. 2008, 46, 446–475. [Google Scholar] [CrossRef] [PubMed]
- Adil, S. Impact and manipulation of gut microflora in poultry: A review. J. Anim. Veter Adv. 2012, 11, 873–877. [Google Scholar] [CrossRef]
- Adaszyńska-Skwirzyńska, M.; Szczerbińska, D. Use of essential oils in broiler chicken production—A review. Ann. Anim. Sci. 2017, 17, 317–335. [Google Scholar] [CrossRef]
- Hammer, K.A.; Carson, C.F.; Riley, T.V.; Carson, C.; Hammer, K. Antimicrobial activity of essential oils and other plant extracts. J. Appl. Microbiol. 1999, 86, 985–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulánková, R.; Bořilová, G. In vitro combined effect of oregano essential oil and caprylic acid against Salmonella serovars, Escherichia coli O157:H7, Staphylococcus aureus and Listeria monocytogenes. Acta Veter. Brno 2011, 80, 343–348. [Google Scholar] [CrossRef]
- Roofchaee, A.; Mehradad, I.; Ebrahimzadeeh, M.A.; Akbari, M.R. Effect of dietary oregano (Origanum vulgare L.) essential oil on growth performance, cecal microflora and serum antioxidant activity of broiler chickens. Afr. J. Biotechnol. 2011, 10, 6177–6183. [Google Scholar]
- Zengin, H.; Baysal, A.H. Antibacterial and antioxidant activity of essential oil terpenes against pathogenic and spoilage-forming bacteria and cell structure-activity relationships evaluated by SEM microscopy. Molecules 2014, 19, 17773–17798. [Google Scholar] [CrossRef] [PubMed]
- Windisch, W.; Schedle, K.; Plitzner, C.; Kroismayr, A. Use of phytogenic products as feed additives for swine and poultry. J. Anim. Sci. 2008, 86, E140–E148. [Google Scholar] [CrossRef] [PubMed]
- Brenes, A.; Roura, E. Essential oils in poultry nutrition: Main effects and modes of action. Anim. Feed. Sci. Technol. 2010, 158, 1–14. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, S.; Wang, H.; Piao, X. Essential oil and aromatic plants as feed additives in non-ruminant nutrition: A review. J. Anim. Sci. Biotechnol. 2015, 6, 7. [Google Scholar] [CrossRef]
- Bagno, O.A.; Prokhorov, O.N.; Shevchenko, S.A.; Shevchenko, A.I.; Dyadichkina, T.V. Use of phytobiotics in farm animal feeding (review). Sel’skokhozyaistvennaya Biol. 2018, 53, 687–697. [Google Scholar]
- Hood, J.R.; Burton, D.M.; Wilkinson, J.M.; Cavanagh, H.M. The effect of Leptospermum petersonii essential oil on Candida albicans and Aspergillus fumigatus. Med. Mycol. 2010, 48, 922–931. [Google Scholar] [CrossRef]
- Solórzano-Santos, F.; Miranda-Novales, M.G. Essential oils from aromatic herbs as antimicrobial agents. Curr. Opin. Biotechnol. 2012, 23, 136–141. [Google Scholar] [CrossRef]
- Bharti, P.; Bai, S.; Seasotiya, L.; Malik, A.; Dalal, S. Antibacterial activity and chemical composition of essential oils of ten aromatic plants against selected bacteria. Int. J. Drug Develop. Res. 2012, 4, 342–351. [Google Scholar]
- Mahboubi, M.; Kazempour, N.; Valian, M. Antimicrobial activity of natural Respitol-B and its main components against poultry microorganisms. Pak. J. Biol. Sci. 2013, 16, 1065–1068. [Google Scholar] [CrossRef] [PubMed]
- Krishan, G.; Narang, A. Use of essential oils in poultry nutrition: A new approach. J. Adv. Veter Anim. Res. 2014, 1, 156. [Google Scholar] [CrossRef]
- Ouwehand, A.; Tiihonen, K.; Kettunen, H.; Peuranen, S.; Schulze, H.; Rautonen, N. In vitro effects of essential oils on potential pathogens and beneficial members of the normal microbiota. Veterinární Med. 2010, 55, 71–78. [Google Scholar] [CrossRef]
- Jerzsele, A.; Szeker, K.; Csizinszky, R.; Gere, E.; Jakab, C.; Mallo, J.J.; Gálfi, P. Efficacy of protected sodium butyrate, a protected blend of essential oils, their combination, and Bacillus amyloliquefaciens spore suspension against artificially induced necrotic enteritis in broilers. Poult. Sci. 2012, 91, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Edris, A.E. Pharmaceutical and therapeutic potentials of essential oils and their individual volatile constituents: A review. Phytother. Res. 2007, 21, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Esper, R.H.; Gonçalez, E.; Marques, M.O.M.; Felicio, R.C.; Felício, J.D. Potential of essential oils for protection of grains contaminated by aflatoxin produced by Aspergillus flavus. Front. Microbiol. 2014, 5, 269. [Google Scholar] [CrossRef]
- de Rapper, S.; Kamatou, G.; Viljoen, A.; Van Vuuren, S. The in vitro antimicrobial activity of Lavandula angustifolia essential oil in combination with other aroma-therapeutic oils. Evid. Based Complement. Altern. Med. 2013, 2013, 1–10. [Google Scholar] [CrossRef]
- Sienkiewicz, M.; Łysakowska, M.; Denys, P.; Kowalczyk, E. The antimicrobial activity of thyme essential oil against multidrug resistant clinical bacterial strains. Microb. Drug Resist. 2012, 18, 137–148. [Google Scholar] [CrossRef]
- M’Hir, S.; Sifi, S.; Chammem, N.; Sifaoui, I.; Mejri, A.; Hamdi, M.; Abderrabba, M. Antioxidant effect of essential oils of Thymus, Salvia and Rosemarinus on the stability to auxidation of refined oils. Ann. Biol. Res. 2012, 3, 4259–4263. [Google Scholar]
- Amorati, R.; Foti, M.C.; Valgimigli, L. Antioxidant activity of essential oils. J. Agric. Food Chem. 2013, 61, 10835–10847. [Google Scholar] [CrossRef] [PubMed]
- Awaad, M.H.H.; Abdel-Alim, G.A.; Sayed, K.S.S.; Kawkab, A.A.; Nada, A.A.; Metwalii, A.S.Z.; Alkhalaf, A.N. Immunostimulant effects of essential oils of peppermint and eucalyptus in chickens. Pak. Vet. J. 2010, 30, 61–66. [Google Scholar]
- Faramarzi, S.; Bozorgmehrifard, M.H.; Khaki, A.; Moomivand, H.; Ezati, M.S.; Rasoulinezhad, S.; Bahnamiri, A.J.; Dizaji, B.R. Study on the effect of Thymus vulgaris essential oil on humoral immunity and performance of broiler chickens after La Sota vaccination. Ann. Biol. Res. 2013, 4, 290–294. [Google Scholar]
- Kilkenny, C.; Browne, W.; Cuthill, I.; Emerson, M.; Altman, D. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. Osteoarthr. Cartil. 2012, 20, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Fisinin, V.I.; Egorov, I.A.; Draganov, I.F. Feeding of Poultry: A Textbook; GEOTAR-Media: Moscow, Russia, 2011; ISBN 978-5-9704-1996-0. [Google Scholar]
- Titov, V.Y.; Vertiprakhov, V.G.; Kosenko, O.V.; Fisinin, V.I.; Dmitrieva, M.E.; Novikova, O.B.; Petrov, V.A. Concentrations of nitrite and non-thiol nitroso compounds in tissues as a high sensitive marker of leukocyte activity. Russ. Agric. Sci. 2017, 4, 58–61. [Google Scholar]
- Itoh, N.; Kikuchi, N.; Hiramune, T. Biochemical changes in fowl serum during infection with Salmonella Typhimurium. J. Veter Med. Sci. 1996, 58, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.H.; Berchieri, Â., Jr.; Montassier, H.J.; Fernandes, A.C. Assessment of serological response of chickens to Salmonella Gallinarum and Salmonella Pullorum by Elisa. Rev. Bras. Cienc. Avic. 2004, 6, 111–115. [Google Scholar] [CrossRef]
- Oliveira, G.H.; Berchieri, Â., Jr.; Montassier, H.J. Chicken serologic response to Salmonella enterica serotype Typhimurium assessed by Elisa. Rev. Bras. Cienc. Avic. 2006, 8, 51–54. [Google Scholar] [CrossRef]
- Simon, R.; Tennant, S.M.; Galen, J.E.; Levine, M.M. Mouse models to assess the efficacy of non-typhoidal Salmonella vaccines: Revisiting the role of host innate susceptibility and routes of challenge. Vaccine 2011, 29, 5094–5106. [Google Scholar] [CrossRef]
- Groves, P.J.; Sharpe, S.M.; Cox, J.M. Response of layer and broiler strain chickens to parenteral administration of a live Salmonella Typhimurium vaccine. Poult. Sci. 2015, 94, 1512–1520. [Google Scholar] [CrossRef]
- Vanimmerseel, F. The effect of vaccination with a Salmonella Enteritidis aroA mutant on early cellular responses in caecal lamina propria of newly-hatched chickens. Vaccine 2002, 20, 3034–3041. [Google Scholar] [CrossRef]
- Ribeiro, S.; Berchieri, A., Jr.; Orsi, M.; Mendonça, A.; Ferrati, A. Experimental infection by Salmonella enterica subsp enterica serovar Kottbus in day-old broiler chickens. Rev. Bras. Cienc. Avic. 2005, 7, 107–112. [Google Scholar] [CrossRef]
- Yang, Y.; Tellez, G.; Latorre, J.D.; Ray, P.M.; Hernandez, X.; Hargis, B.M.; Ricke, S.C.; Kwon, Y.M. Salmonella excludes Salmonella in poultry: Confirming an old paradigm using conventional and barcode-tagging approaches. Front. Veter. Sci. 2018, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Titov, V.I.; Ivanova, A.V.; Agapov, A.M.; Petrov, A.V. The content of nitrite and N-nitroso compounds of plasma as a diagnostic test of nonspecific inflammation. Klin. Lab. Diagn. 2011, 56, 13–19. [Google Scholar]
- Ministry of Health of the USSR, Chief Veterinary Directorate at the State Commission for Food and Procurement of the Council of Ministers of the USSR. Laboratory Diagnostics of Human and Animal Salmonellosis, Detection of Salmonella in Feed, Food and Environmental Objects (Methodological Recommendations); Central Research Institute of Epidemiology of the Ministry of Health of the USSR: Moscow, Russia, 1990. [Google Scholar]
- Nääs, I.D.A.; Romanini, C.E.B.; Neves, D.P.; Nascimento, G.R.D.; Vercellino, R.D.A. Broiler surface temperature distribution of 42 days old chickens. Sci. Agric. 2010, 67, 497–502. [Google Scholar] [CrossRef]
- Bohutsky, M.I. Salmonella infection. Ž. Grodn. Gos. Med. Univ. 2011, 1, 7–11. [Google Scholar]
- Garcia, K.; Berchieri, A., Jr.; Santana, A.; Alarcon, M.; Freitas Neto, O.; Fagliari, J. Experimental infection of commercial layers with wild or attenuated Salmonella Gallinarum mutant strains: Anatomic pathology, total blood cell count and serum protein levels. Rev. Bras. Cienc. Avic. 2013, 15, 91–104. [Google Scholar] [CrossRef]
- Kupryś-Caruk, M.; Michalczuk, M.; Chabłowska, B.; Stefańska, I.; Kotyrba, D.; Parzeniecka-Jaworska, M. Efficacy and safety assessment of microbiological feed additive for chicken broilers in tolerance studies. J. Veter Res. 2018, 62, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.R.; McIntyre, D.R.; Pavlidis, H.O.; Archer, G.S. Reducing stress susceptibility of broiler chickens by supplementing a yeast fermentation product in the feed or drinking water. Animals 2018, 8, E173. [Google Scholar] [CrossRef]
- State Commission of the Council of Ministers of the USSR on Food and Procurement, All-Union Production and Research Association ‘Soyuzptitseprom’, Scientific Production Association of the Poultry Processing Industry ‘Complex’. Instructions for the Sanitary and Microbiological Control of Carcasses, Poultry Meat, Poultry Products, Eggs and Egg Products at Poultry and Processing Enterprises; State Commission of the Council of Ministers of the USSR on Food and Procurement, All-Union Production and Research Association ‘Soyuzptitseprom’, Scientific Production Association of the Poultry Processing Industry ‘Complex’: Moscow, Russia, 1990. [Google Scholar]
- Zeka, F.; Vanderheyden, K.; De Smet, E.; Cuvelier, C.A.; Mestdagh, P.; Vandesompele, J. Straightforward and sensitive RT-qPCR based gene expression analysis of FFPE samples. Sci. Rep. 2016, 6, 21418. [Google Scholar] [CrossRef] [Green Version]
- El Khoury, R.; Atoui, A.; Verheecke, C.; Maroun, R.; El Khoury, A.; Mathieu, F. Essential oils modulate gene expression and Ochratoxin A production in Aspergillus carbonarius. Toxins 2016, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.I.; Swaggerty, C.L.; Kogut, M.H.; Dowd, E.S.; Li, X.; Pevzner, I.Y.; Zhou, H. Gene expression profiling in chicken heterophils with Salmonella enteritidis stimulation using a chicken 44 K Agilent microarray. BMC Genom. 2008, 9, 526. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Methods of Genetic Engineering. Molecular Cloning; Mir: Moscow, Russia, 1984. [Google Scholar]
- Li, F.; Hullar, M.A.; Lampe, J.W. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J. Microbiol. Methods 2007, 68, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Ushakova, N.A.; Nekrasov, R.V.; Meleshko, N.A.; Laptev, G.I.; Il’ina, L.A.; Kozlova, A.A.; Nifatov, A.V. Bacillus subtilis influence on rumen microbial consortium and host digestion. Mikrobiologiia 2013, 82, 456–563. [Google Scholar] [PubMed]
- Ilina, L.A.; Yildirim, E.A.; Nikonov, I.N.; Filippova, V.A.; Laptev, G.Y.; Novikova, N.I.; Grozina, A.A.; Lenkova, T.N.; Manukyan, V.A.; Egorov, I.A.; et al. Metagenomic bacterial community profiles of chicken embryo gastrointestinal tract by using T-RFLP analysis. Dokl. Biochem. Biophys. 2016, 466, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Witzig, M.; Da Silva, A.C.; Green-Engert, R.; Hoelzle, K.; Zeller, E.; Seifert, J.; Rodehutscord, M. Spatial variation of the gut microbiota in broiler chickens as affected by dietary available phosphorus and assessed by T-RFLP analysis and 454 pyrosequencing. PLoS ONE 2015, 10, 0143442. [Google Scholar]
- Lindström, S.; Rowe, O.; Timonen, S.; Sundström, L.; Johansson, H. Trends in bacterial and fungal communities in ant nests observed with Terminal-Restriction Fragment Length Polymorphism (T-RFLP) and Next Generation Sequencing (NGS) techniques—validity and compatibility in ecological studies. Peer J. 2018, 6, e5289. [Google Scholar] [CrossRef]
- Gong, J.; Forster, R.J.; Chambers, J.R.; Sabour, P.M.; Wheatcroft, R.; Chen, S.; Yu, H. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 2002, 208, 1–7. [Google Scholar] [CrossRef]
- Hongoh, Y.; Ohkuma, M.; Kudo, T. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 2003, 44, 231–242. [Google Scholar] [CrossRef]
- Amiranashvili, L.L.; Gagelidze, N.A.; Varsimashvili, K.I.; Tinikashvili, L.M.; Tolordava, L.L.; Gamkrelidze, M.D.; Amashukeli, N.V.; Makaradze, L.A. Antimicrobial susceptibility and antibiotic resistance profiles of cultivable lactic acid bacteria from intestinal tract of domestic chickens collected in Adjara. Ann. Agrar. Sci. 2016, 14, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Ma, C.; Sun, Z.; Wang, L.; Huang, S.; Su, X.; Xu, J.; Zhang, H. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 2017, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Al-Gamal, M.S.; Ibrahim, G.A.; Sharaf, O.M.; Radwan, A.A.; Dabiza, N.M.; Youssef, A.M.; El-Ssayad, M.F. The protective potential of selected lactic acid bacteria against the most common contaminants in various types of cheese in Egypt. Heliyon 2019, 5, e01362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryukhanov, A.L.; Rybak, K.V.; Netrusov, A.I. Molecular Microbiology; Izd-vo MGU: Moscow, Russia, 2012; ISBN 978-5-211-05486-8. [Google Scholar]
- Sciarini, S.M. tRFLP Fragment Sorter. The Ohio State University, OARDC. 2005. Available online: https://web.archive.org/web/20100708234941/http://www.oardc.ohio-state.edu/trflpfragsort/index.php (accessed on 8 July 2010).
- Seviour, R.; Nielsen, P.H. Microbial Ecology of Activated Sludge; IWA Publishing: London, UK, 2010; ISBN 978-1-8433-9032-9. [Google Scholar]
- Lakin, G.F. Biometrics; Vysshaya shkola: Moscow, Russia, 1990; ISBN 5-06-000471-6. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R. Version 1.1.453; RStudio: Boston, MA, USA, 2018. [Google Scholar]
- RDocumentation. TukeyHSD. Available online: https://www.rdocumentation.org/packages/stats/versions/3.6.1/topics/TukeyHSD (accessed on 25 July 2019).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9, art. 4. [Google Scholar]
- Hammer, Ø. Past 3.x–the Past of the Future. Natural History Museum, University of Oslo. Available online: http://folk.uio.no/ohammer/past/ (accessed on 25 July 2019).
- Hammer, Ø. PAST: PAleontological STatistics. Version 3.22: Reference manual. Natural History Museum University of Oslo, 1999–2018. Available online: http://folk.uio.no/ohammer/past/past3manual.pdf (accessed on 25 July 2019).
- Hirano, T. Interleukin 6. In The Cytokine Handbook, 3rd ed.; Thomson, A.E., Ed.; Academic Press: San Diego, CA, USA, 1998; pp. 197–227. [Google Scholar]
- Lynagh, G.R.; Bailey, M.; Kaiser, P. Interleukin-6 is produced during both murine and avian Eimeria infections. Veter. Immunol. Immunopathol. 2000, 76, 89–102. [Google Scholar] [CrossRef]
- Kaiser, P.; Wigley, P.; Burnside, J.; Barrow, P.A.; Galyov, E.E.; Rothwell, L. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 2000, 146, 3217–3226. [Google Scholar] [CrossRef]
- Zhao, C.; Nguyen, T.; Liu, L.; Sacco, R.E.; Brogden, K.A.; Lehrer, R.I. Gallinacin-3, an inducible epithelial β-defensin in the chicken. Infect. Immun. 2001, 69, 2684–2691. [Google Scholar] [CrossRef]
- van Dijk, A.; Veldhuizen, E.J.; Kalkhove, S.I.; Tjeerdsma-van Bokhoven, J.L.; Romijn, R.A.; Haagsman, H.P. The β-defensin gallinacin-6 is expressed in the chicken digestive tract and has antimicrobial activity against food-borne pathogens. Antimicrob. Agents Chemother. 2007, 51, 912–922. [Google Scholar] [CrossRef]
- Hasenstein, J.; Lamont, S.J. Chicken Gallinacin Gene Cluster Associated with Salmonella Colonisation in Two Advanced Intercross Lines; Anim. Ind. Report; Iowa State University: Ames, IA, USA, 2007. [Google Scholar]
- Mukhopadhyay, C.S.; Kumar, R.; Brah, G.S. Gallinacin and fowlicidin: Two promising antimicrobial peptides in chickens—A review. Vet. World 2010, 3, 297–300. [Google Scholar]
- Lee, M.O.; Jang, H.J.; Rengaraj, D.; Yang, S.Y.; Han, J.Y.; Lamont, S.J.; Womack, J.E. Tissue expression and antibacterial activity of host defence peptides in chicken. BMC Vet. Res. 2016, 12, 231. [Google Scholar] [CrossRef]
- Higgs, R.; Lynn, D.J.; Gaines, S.; McMahon, J.; Tierney, J.; James, T.; Lloyd, A.T.; Mulcahy, G.; O’Farrelly, C. The synthetic form of a novel chicken beta-defensin identified in silico is predominantly active against intestinal pathogens. Immunogenetics 2005, 57, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Mowbray, C.A.; Niranji, S.S.; Cadwell, K.; Bailey, R.; Watson, K.A.; Hall, J. Gene expression of AvBD6-10 in broiler chickens is independent of AvBD6, 9, and 10 peptide potency. Veter. Immunol. Immunopathol. 2018, 202, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S. Pathogenicity of Salmonella enteritidis in poultry. Int. J. Food Microbiol. 1994, 21, 89–105. [Google Scholar] [CrossRef]
- Beal, R.; Wigley, P.; Powers, C.; Hulme, S.; Barrow, P.; Smith, A. Age at primary infection with Salmonella enterica serovar Typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Veter. Immunol. Immunopathol. 2004, 100, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Dalia, A.M.; Loh, T.C.; Sazili, A.Q.; Jahromi, M.F.; Samsudin, A.A. Effects of vitamin E, inorganic selenium, bacterial organic selenium, and their combinations on immunity response in broiler chickens. BMC Veter. Res. 2018, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Saelao, P.; Wang, Y.; Gallardo, R.A.; Lamont, S.J.; Dekkers, J.M.; Kelly, T.; Zhou, H. Novel insights into the host immune response of chicken Harderian gland tissue during Newcastle disease virus infection and heat treatment. BMC Veter. Res. 2018, 14, 280. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, J.; Finlay, B.B. Type III effector-mediated processes in Salmonella infection. Future Microbiol. 2012, 7, 685–703. [Google Scholar] [CrossRef]
- Li, J.; Hao, H.; Cheng, G.; Liu, C.; Ahmed, S.; Shabbir, M.A.B.; Hussain, H.I.; Dai, M.; Yuan, Z. Microbial shifts in the intestinal microbiota of Salmonella infected chickens in response to enrofloxacin. Front. Microbiol. 2017, 8, 1711. [Google Scholar] [CrossRef]
- Fisinin, V.; All-Russian Research and Technological Poultry Institute, Federal Agency of Scientific Organizations; Laptev, G.; Nikonov, I.; Il’Ina, L.; Yildirim, E.; Filippova, V.; Novikova, N.; Grozina, A.; Egorova, T.; et al. Poultry gastrointestinal microbiome changes during ontogenesis. Sel’skokhozyaistvennaya Biol. 2016, 51, 883–890. [Google Scholar]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef]
- Józefiak, D.; Rutkowski, A.; Kaczmarek, S.; Jensen, B.; Engberg, R.; Højberg, O. Effect of β-glucanase and xylanase supplementation of barley and rye-based diets on caecal microbiota of broiler chickens. Br. Poult. Sci. 2010, 51, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.M.; Shah, T.; Deshpande, S.; Jakhesara, S.J.; Koringa, P.G.; Rank, D.N.; Joshi, C.G.; Jakhesara, S.; Joshi, C. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol. Biol. Rep. 2012, 39, 10595–10602. [Google Scholar] [CrossRef] [PubMed]
- Svetoch, E.A.; Eruslanov, B.V.; Levchuk, V.P.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Stepanshin, J.; Dyatlov, I.; Seal, B.S.; Stern, N.J. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl. Environ. Microbiol. 2011, 77, 2749–2754. [Google Scholar] [CrossRef] [PubMed]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hinton, A.; Corrier, D.E.; Deloach, J.R. In vitro inhibition of Salmonella typhimurium and Escherichia coli 0157:H7 by an anaerobic gram-positive coccus isolated from the cecal contents of adult chickens. J. Food Prot. 1992, 55, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Murry, A., Jr.; Hinton, A.; Morrison, H. Inhibition of growth of Escherichia coli, Salmonella typhimurium, and Clostridia perfringens on chicken feed media by Lactobacillus salivarius and Lactobacillus plantarum. Int. J. Poult. Sci. 2004, 3, 603–607. [Google Scholar]
- Teo, A.Y.L.; Tan, H.M. Inhibition of Clostridium perfringens by a novel strain of Bacillus subtilis isolated from the gastrointestinal tracts of healthy chickens. Appl. Environ. Microbiol. 2005, 71, 4185–4190. [Google Scholar] [CrossRef]
- Stern, N.J.; Svetoch, E.A.; Eruslanov, B.V.; Perelygin, V.V.; Mitsevich, E.V.; Mitsevich, I.P.; Pokhilenko, V.D.; Levchuk, V.P.; Svetoch, O.E.; Seal, B.S. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 2006, 50, 3111–3116. [Google Scholar] [CrossRef]
- Messaoudi, S.; Kergourlay, G.; Dalgalarrondo, M.; Choiset, Y.; Ferchichi, M.; Prévost, H.; Pilet, M.F.; Chobert, J.M.; Manai, M.; Dousset, X. Purification and characterization of a new bacteriocin active against Campylobacter produced by Lactobacillus salivarius SMXD51. Food Microbiol. 2012, 32, 129–134. [Google Scholar] [CrossRef]
- Shin, M.; Han, S.; Ji, A.; Kim, K.; Lee, W. Isolation and characterization of bacteriocin-producing bacteria from the gastrointestinal tract of broiler chickens for probiotic use. J. Appl. Microbiol. 2008, 105, 2203–2212. [Google Scholar] [CrossRef]
- Lawley, T.D.; Walker, A.W. Intestinal colonisation resistance. Immunology 2013, 138, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, I.; Lessire, M.; Mallet, S.; Guillot, J. Microflora of the digestive tract: Critical factors and consequences for poultry. World’s Poult. Sci. J. 2006, 62, 499–511. [Google Scholar]
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010, 467, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Videnska, P.; Sisak, F.; Havlickova, H.; Faldynova, M.; Rychlik, I. Influence of Salmonella enterica serovar Enteritidis infection on the composition of chicken cecal microbiota. BMC Veter Res. 2013, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Xin, H.; Chen, S.; Duan, Y. Effects of a protected inclusion of organic acids and essential oils as antibiotic growth promoter alternative on growth performance, intestinal morphology and gut microflora in broilers. Anim. Sci. J. 2017, 2, 18–1424. [Google Scholar] [CrossRef] [PubMed]
- Betancourt, L.; Phandanouvong, V.; Ariza-Nieto, C.; Hume, M.; Van Kley, A.M.; Nalian, A.; Rodriguez, F.; Nisbet, D.; Afanador-Téllez, G. Effect of Origanum chemotypes on broiler intestinal bacteria. Poult. Sci. 2014, 93, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, M.L.; Quessy, S.; Letellier, A.; Arsenault, J.; Boulianne, M. Impact of a drug-free program on broiler chicken growth performances, gut health, Clostridium perfringens and Campylobacter jejuni occurrences at the farm level. Poult. Sci. 2015, 94, 1791–1801. [Google Scholar] [CrossRef]
- Kelly, C.; Gundogdu, O.; Pircalabioru, G.; Cean, A.; Scates, P.; Linton, M.; Pinkerton, L.; Magowan, E.; Stef, L.; Simiz, E.; et al. The in vitro and in vivo effect of carvacrol in preventing Campylobacter infection, colonisation and in improving productivity of chicken broilers. Foodborne Pathog. Dis. 2017, 14, 341–349. [Google Scholar] [CrossRef]
- Guo, X.; Li, D.; Lu, W.; Piao, X.; Chen, X. Screening of Bacillus strains as potential probiotics and subsequent confirmation of the in vivo effectiveness of Bacillus subtilis MA139 in pigs. Antonie Van Leeuwenhoek 2006, 90, 139–146. [Google Scholar] [CrossRef]
- Egorova, T.; All-Russian Research and Technological Poultry Institute, Federal Agency of Scientific Organizations; Lenkova, T.; Il’Ina, L.; Yildirim, E.; Nikonov, I.; Filippova, V.; Laptev, G.; Novikova, N.; Grozina, A.; et al. The Saccharomyces sp. and Bacillus subtilis based probiotics influence on chicken broiler productivity and caecum microbiome community. Sel’skokhozyaistvennaya Biol. 2016, 51, 891–902. [Google Scholar]
- Tarakanov, B.V. Methods for Studying the Microflora of the Digestive Tract of Farm Animals and Poultry; Nauchnyi mir: Moscow, Russia, 2006; ISBN 5-89176-386-9. [Google Scholar]

| Gene Symbol | Gene/Protein Name | Accession No. | Primer Sequence (5′–3′) 1 | PCR Product Size (bp) | Reference |
|---|---|---|---|---|---|
| AvBD9 | avian β-defensins 9, 10 and 11 (gallinacins) | NM_001001611.2 | F: AACACCGTCAGGCATCTTCACA R: CGTCTTCTTGGCTGTAAGCTGGA | 131 | [25] |
| AvBD10 | CR388516 | F: GCTCTTCGCTGTTCTCCTCT R: CCCAGAGATGGTGAAGGTG | 67 | [32] | |
| AvBD11 | NM_001001779.1 | F: AGTCTGCAATTCGTTAGAGGCG R: GGATGTGGTTTCCAAGGGTTTA | 180 | [26] | |
| IL6 | interleukins 6 and 8-like 2 (cytokines) | AJ309540 | F: AGGACGAGATGTGCAAGAAGTTC R: TTGGGCAGGTTGAGGTTGTT | 78 | [61] |
| IL8L2 | M16199 | F: GGAAGAGAGGTGTGCTTGGA R: TAACATGAGGCACCGATGTG | 102 | [32] | |
| CASP6 | caspase 6 (cysteine protease) | AF082329 | F: CAGAGGAGACAAGTGCCAGA R: CCAGGAGCCGTTTACAGTTT | 250 | [88] |
| PTGS2 | prostaglandin-endoperoxide synthase 2 (cyclooxygenase 2) | M64990 | F: TCGAGATCACACTTGATTGACA R: TTTGTGCCTTGTGGGTCAG | 230 | [88] |
| IRF7 | interferon regulatory factor 7 | U20338 | F: ATCCCTTGGAAGCACAACGCC R: CTGAGGCAACCGCGTAGACCTT | 223 | [88] |
| ACTB | β-actin | NM_205518 | F: ATTGTCCACCGCAAATGCTTC R: AAATAAAGCCATGCCAATCTCGTC | 86 | [34] |
| Indices | Subgroups 1 | |||
|---|---|---|---|---|
| I | II | III | IV | |
| 1 dpi 2 | ||||
| No. of phylotypes | 45.00 ± 2.90 | 19.30 ± 0.90 3 | 52.00 ± 2.10 | 75.00 ± 3.40 3 |
| Fisher’s alpha | 33.20 ± 1.65 | 7.50 ± 0.41 3 | 59.00 ± 3.10 3 | 266.70 ± 15.60 3 |
| 23 dpi 2 | ||||
| No. of phylotypes | 69.00 ± 3.30 | 72.30 ± 3.80 | 49.30 ± 2.20 3 | 73.30 ± 3.90 |
| Fisher’s alpha | 43.10 ± 2.19 | 117.50 ± 7.30 3 | 8.30 ± 0.49 3 | 203.90 ± 12.40 3 |
| Groups 1 | At Day-Old | At 14 Day-Old | Subgroups 2 | 1 dpi 3 | 23 dpi 3 |
|---|---|---|---|---|---|
| I (n = 60) | 38.1 ± 1.9 | 321.82 ± 32.1 * | I (n = 30) | 650.4 ± 104.0 | 2294.0 ± 184.0 |
| II (n = 30) | 656.0 ± 53.0 | 2004.6 ± 233.0 | |||
| II (n = 60) | 38.2 ± 2.4 | 360.02 ± 45.3 * | III (n = 30) | 743.8 ± 54.0 | 2403.2 ± 231.0 |
| IV (n = 30) | 720.3 ± 81.0 | 1936.6 ± 155.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laptev, G.Y.; Filippova, V.A.; Kochish, I.I.; Yildirim, E.A.; Ilina, L.A.; Dubrovin, A.V.; Brazhnik, E.A.; Novikova, N.I.; Novikova, O.B.; Dmitrieva, M.E.; et al. Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic. Animals 2019, 9, 615. https://doi.org/10.3390/ani9090615
Laptev GY, Filippova VA, Kochish II, Yildirim EA, Ilina LA, Dubrovin AV, Brazhnik EA, Novikova NI, Novikova OB, Dmitrieva ME, et al. Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic. Animals. 2019; 9(9):615. https://doi.org/10.3390/ani9090615
Chicago/Turabian StyleLaptev, Georgi Yu., Valentina A. Filippova, Ivan I. Kochish, Elena A. Yildirim, Larisa A. Ilina, Andrei V. Dubrovin, Evgeni A. Brazhnik, Natalia I. Novikova, Oksana B. Novikova, Margarita E. Dmitrieva, and et al. 2019. "Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic" Animals 9, no. 9: 615. https://doi.org/10.3390/ani9090615
APA StyleLaptev, G. Y., Filippova, V. A., Kochish, I. I., Yildirim, E. A., Ilina, L. A., Dubrovin, A. V., Brazhnik, E. A., Novikova, N. I., Novikova, O. B., Dmitrieva, M. E., Smolensky, V. I., Surai, P. F., Griffin, D. K., & Romanov, M. N. (2019). Examination of the Expression of Immunity Genes and Bacterial Profiles in the Caecum of Growing Chickens Infected with Salmonella Enteritidis and Fed a Phytobiotic. Animals, 9(9), 615. https://doi.org/10.3390/ani9090615









