Abstract
Here we review the application of molecular biological approaches to mineral precipitation in modern marine microbialites. The review focuses on the nearly two decades of nucleotide sequencing studies of the microbialites of Shark Bay, Australia; and The Bahamas. Molecular methods have successfully characterized the overall community composition of mats, pinpointed microbes involved in key metabolisms, and revealed patterns in the distributions of microbial groups and functional genes. Molecular tools have become widely accessible, and we can now aim to establish firmer links between microbes and mineralization. Two promising future directions include “zooming in” to assess the roles of specific organisms, microbial groups, and surfaces in carbonate biomineralization and “zooming out” to consider broader spans of space and time. A middle ground between the two can include model systems that contain representatives of important microbial groups, processes, and metabolisms in mats and simplify hypothesis testing. These directions will benefit from expanding reference datasets of marine microbes and enzymes and enrichments of representative microbes from mats. Such applications of molecular tools should improve our ability to interpret ancient and modern microbialites and increase the utility of these rocks as long-term recorders of microbial processes and environmental chemistry.
1. Introduction
Fossilized microbial mats are the earliest record of life on Earth [1,2,3]. Even though the identities of individual organisms in fossilized mats remain unknown, the macroscopic sizes of lithified microbial mats—microbialites—tell us that a myriad organisms must have come together to bind sediments, drape rock surfaces, form cohesive layers, and promote mineral precipitation (Figure 1). Most microbialites were preserved by carbonate minerals, but exceptional preservation of textures, organic matter, and microbial fossils in some Archean and many Proterozoic laminated microbialites and stromatolites also required more localized precipitation of silica (Figure 1). Studies of modern microbial mats and their fossilized counterparts in the 20th and the 21st century revealed the dependence of macroscopic microbialite morphologies, as well as the microscopic textures and microbial fossils in carbonate microbialites on environmental physics and chemistry; the distribution and nature of different primary producers, such as coccoidal vs. filamentous cyanobacteria; the types of microbial surfaces present; and the small-scale gradients in microbial activity (see References [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24]).
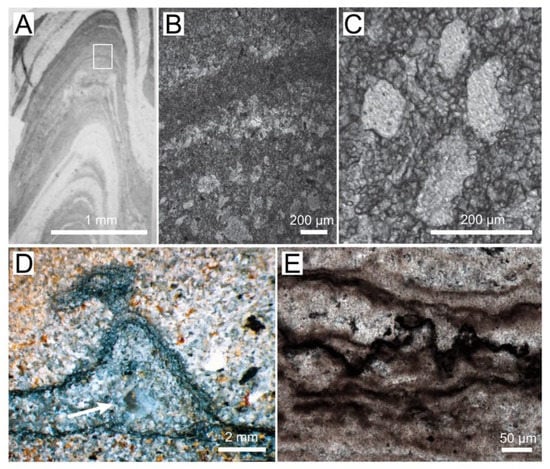
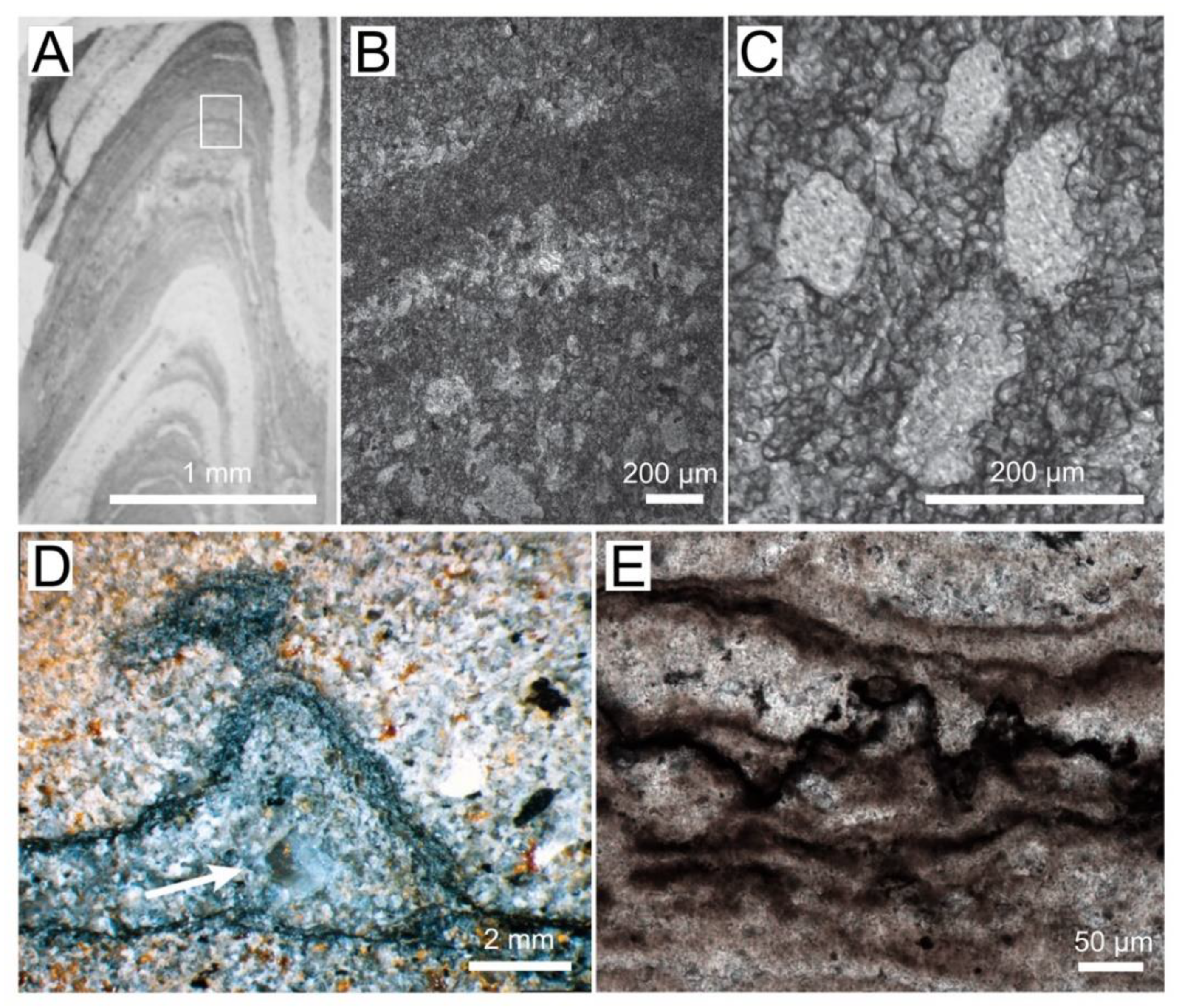
Figure 1.
Textures in Archaean and Proterozoic microbial mats and stromatolites. (A) Photograph of a thin section of conical stromatolite from the 3 Ga Chobeni Formation, South Africa. The light areas contain silica, and the dark laminae contain dolomite. White rectangle outlines the area enlarged in panel (B). (B) Photomicrograph of laminae preserved by microcrystalline and microsparitic dolomite (dark). The lighter laminae contain pores filled by silica. (C) Enlarged view of silica-filled pores surrounded by 10–100 mm–thick dolomite laminae. (D) Photograph of a polished slab showing a tufted silicified mat in a sandstone from the Archean Moodies Group, South Africa [25]. The arrow points to a silica-filled void. (Reprinted with permission from ref. [25]. Copyright 2015 Elsevier) (E) Photomicrograph of the dark, organic-rich layers in a pustular silicified mat from the ~1600 Ma Proterozoic Balbirini Dolomite, Australia.
Most of the microbial constituents in marine carbonate-precipitating mats are not readily identifiable by eye [7,26,27], but Cyanobacteria, the primary producers in modern microbialites, excrete copious extracellular polymeric substances (EPS) that bind sediments (Figure 2) [7,24]. Sediment-trapping alone cannot build stromatolites—carbonate minerals precipitated in situ cement the microbialites and create some of their textures (Figure 2C,D), and processes within mats could potentially influence the growth or dissolution of trapped grains, as well. Microchemical measurements and visualization techniques combined with isotope labeling and microscopy have linked the activity of sulfate reducing bacteria and cyanobacteria to carbonate precipitation and cementation in modern microbialites and carbonate grains (see References [7,8,28,29,30,31,32,33,34,35,36,37,38,39]). However, these approaches alone cannot explain the mind-boggling diversity of microbes in modern mats [40,41,42,43,44,45], confirm specific microbial interactions that effect mineralization, or predict which textures and microbialite morphologies will form. Consequently, the incomplete understanding of modern microbialites impacts what we can learn from Archean and Proterozoic stromatolites, which are characterized by a notably greater diversity of stromatolite textures compared to modern stromatolites (Figure 1).
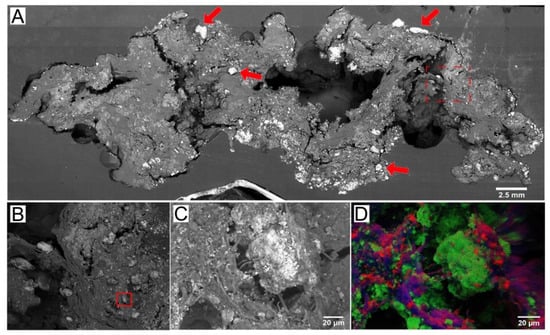
Figure 2.
SEM and EDS images of microbial–carbonate interactions in a microbial mat. (A) SEM of a peritidal pustular mat from Shark Bay, Australia. Arrows point to the aragonite grains trapped by the mat. Red box (dashed) shows the area enlarged in panel (B). (B) Enlarged view of the area outlined by the red box in panel (A). Red box shows the area enlarged in panel (C). (C) Microcrystalline aragonite that precipitates in EPS, some cyanobacterial filaments are also visible. (D) EDS map of carbon (red), calcium (green), and sulfur (blue) of the region shown in panel (C).
The tools of molecular biology can address biomineralization mechanisms at very fine scales to reveal processes by which specific organisms, genes, biomolecules, and microbial interactions influence the precipitation of carbonates and other minerals. Broadly defined molecular techniques encompass all methods that elucidate the structure and function of biomolecules (namely nucleic acids, proteins, lipids, or carbohydrates) or harness the properties of biomolecules to observe biological processes. Over the past three decades, molecular tools—specifically those based on nucleic acid sequencing—have revealed substantial microbial, gene, metabolic, and organic diversity in microbial mats.
Here, we review the application of molecular biological approaches to the study of mineral precipitation in modern microbialites. Although the diversity of biomineralizing microbial ecosystems and biomolecules is vast, this review specifically focuses on the use of nucleic acid–based methods to investigate processes in modern marine carbonate precipitating microbialites in Shark Bay, Australia; and Highbourne Cay, The Bahamas, as two extensively studied modern sites. We primarily consider nucleic acid sequencing techniques because they are at once powerful and accessible, and because they have been applied commonly in some of the most studied modern microbial carbonate systems.
2. Microbial Influences on Mineral Precipitation in Microbialites
Molecular techniques are one powerful means of observing and characterizing the biological factors at play in mineralization (Figure 3). These biological factors fall broadly into two categories: metabolic activities that change the chemical environment to promote or inhibit mineralization; and organic compounds that influence mineral nucleation, growth, ordering and shape. The precipitation and dissolution of carbonate minerals depend on the degree to which carbonate minerals are oversaturated or undersaturated in solution, which itself depends on carbonate alkalinity [4,31,46]. Microbial communities locally influence the alkalinity and the saturation state of carbonate minerals via metabolic activities that act as direct sources or sinks for carbonate ions or induce changes in the pH. An understanding of community structure and the effects of various metabolic guilds enables predictions of the net effect of this alkalinity engine (Figure 4).
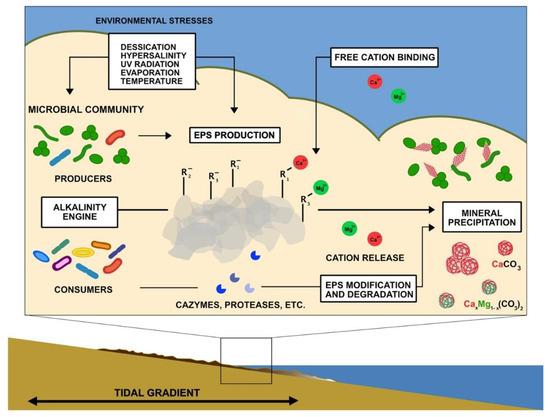
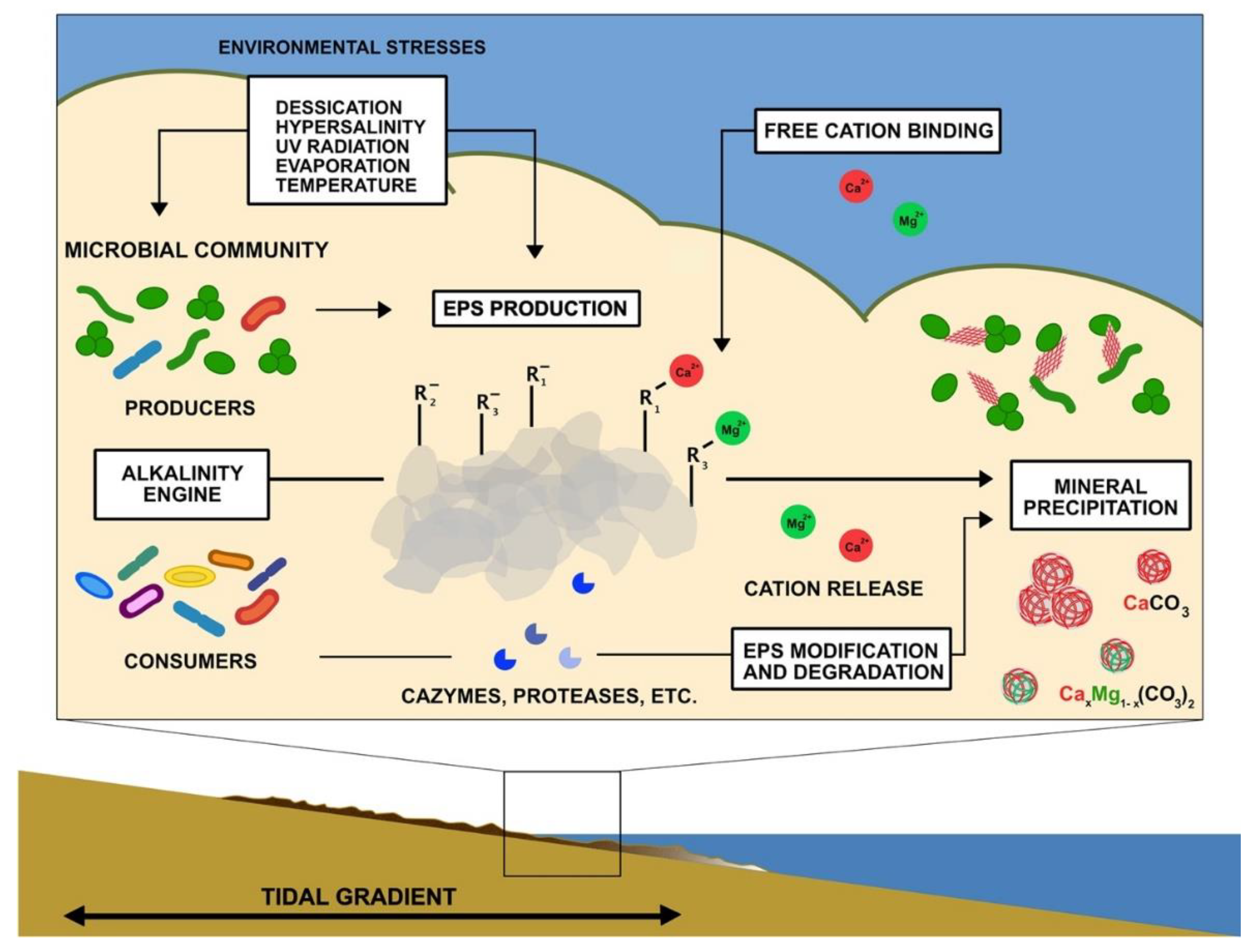
Figure 3.
Summary schematic of potential and known factors contributing to mineralization in microbial mats. Environmental stresses enhance EPS production by photosynthetic microbes and other organisms. Negatively charged functional groups trap and bind free cations on the surface of EPS. Microbial metabolic processes that contribute to EPS production and degradation (alkalinity engine) can influence the concentrations of carbonate ions, release cations, and modify organic surfaces or produce organic compounds that influence mineral nucleation and growth.
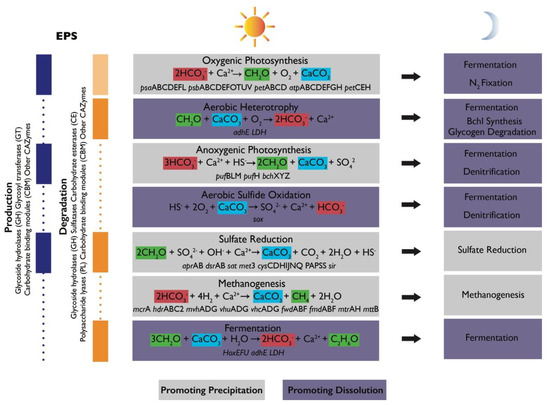
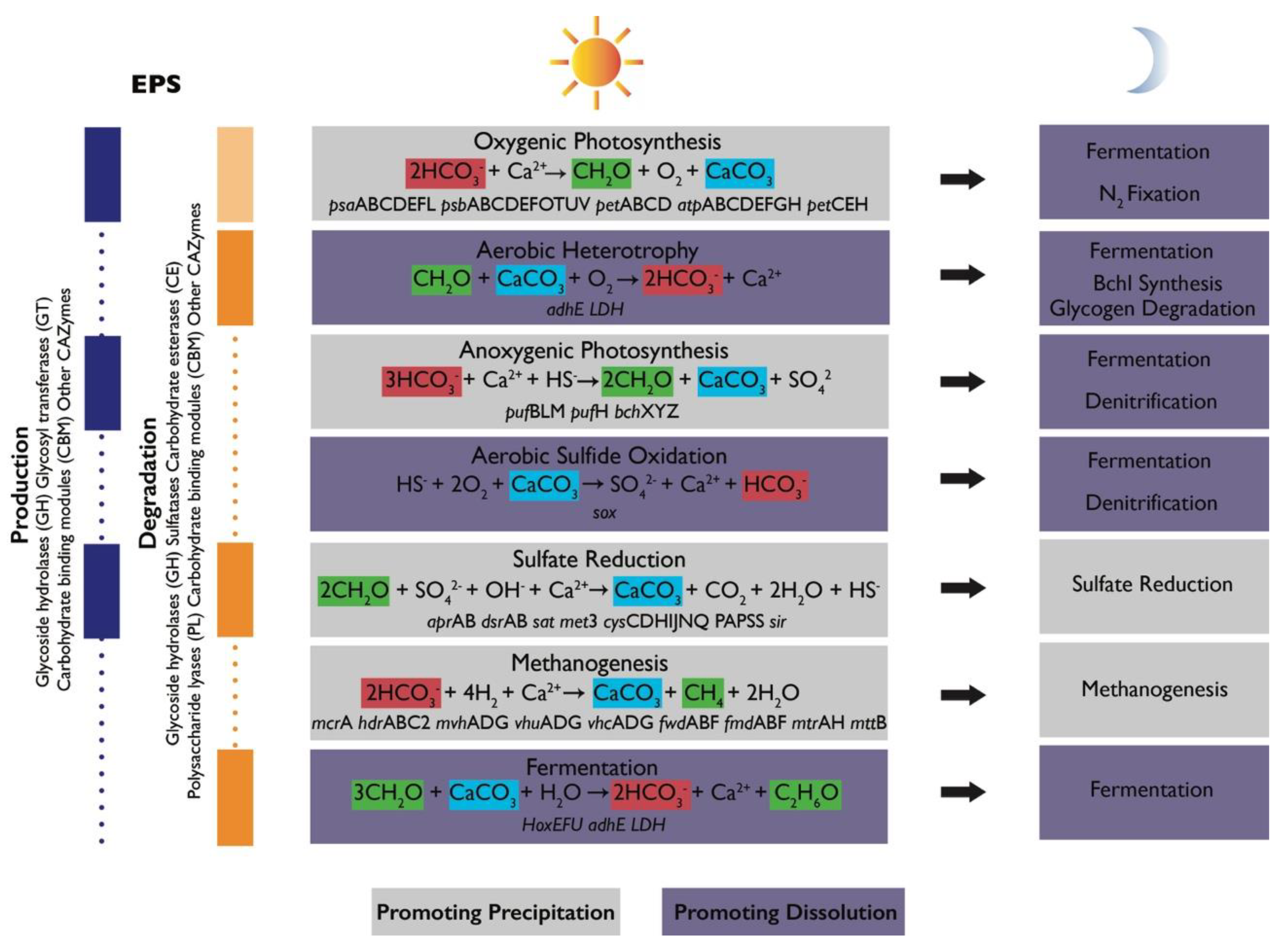
Figure 4.
Schematic of metabolic guilds commonly present in microbial mat communities and their contributions to the alkalinity engine (adapted from Reference [4]). Each group includes a simplified equation representing its major metabolism as it relates to the precipitation of carbonate minerals, alkalinity, and the production or degradation of organic matter [46]. Relevant functional genes associated with some of these metabolisms were detected in Shark Bay mats [47,48,49,50]. Some metabolisms promote carbonate precipitation (gray boxes) or dissolution (purple boxes). Shifts in metabolic activity from day to night can alter the net effects of metabolisms. Left column shows the contribution of each group to EPS cycling and lists some gene families associated with production (blue) and degradation (orange). Light orange box corresponding to oxygenic photosynthesis indicates that oxygenic phototrophs (Cyanobacteria) are known to recycle some of their own EPS but are generally considered net producers of EPS. Vertical purple and orange boxes shown known contributions of some microbial groups to EPS cycling. Dotted lines show that there is still much to learn about EPS cycling in mats and the contribution of various groups of organisms.
Phototrophic CO2 fixation favors the net precipitation of carbonate minerals. Oxygenic photosynthesis takes in HCO3- and releases OH− ions to increase carbonate alkalinity and pH. Anoxygenic photosynthetic oxidation of HS− decreases alkalinity; however, simultaneous increases in pH maintain favorable conditions for precipitation. These processes are restricted to the upper illuminated parts of the mat and are offset by aerobic respiration, which favors carbonate dissolution by increasing pCO2. The effects of oxygenic photosynthesis and aerobic respiration are balanced in time. At night, oxygenic and anoxygenic phototrophs can ferment, fix N2, and contribute to denitrification— all processes that produce protons and favor dissolution. For these reasons, carbonate minerals in modern microbialites precipitate more extensively in the zones of microbial sulfate reduction and extensive organic degradation [8,32,51] and typically do not preserve the finely laminated photosynthetic textures, such as those shown in Figure 1. The activity of sulfate-reducing bacteria (SRB) is less constrained in time and space. The diversity of organic substrates, the completeness of substrate oxidation, environmental buffering, and the extent of sulfate reduction determine the degree to which sulfate reduction increases alkalinity [52,53]. Some sulfide produced by SRB can feed back into anoxygenic photosynthesis and light-independent sulfide oxidation, with the latter process particularly favoring carbonate dissolution (see References [45,54,55]). Other processes, such as iron reduction or the release of ammonia due to protein degradation, increase alkalinity and may contribute to mineral precipitation (see References [56,57]). Deeper in the mat, methanogenesis and fermentation favor carbonate precipitation and dissolution, respectively (see References [37,58,59]).
The initial nucleation of carbonate mineral phases often imposes the largest kinetic barrier to the precipitation of these minerals from oversaturated solutions. Organic molecules appear to drastically reduce this barrier and control the mineral phase and ordering (see References [60,61,62,63,64,65,66,67]). Incipient carbonate precipitates in modern microbial systems occur as nanometer-sized grains within the EPS and on cell surfaces [21,22,23,32,68,69] (Figure 2). Different cell or viral surfaces and microbial EPS can influence mineral grain sizes, shapes, and crystal ordering [64,70,71,72,73,74]. Thus, organic surfaces within microbial mats are just as critical to mineral nucleation, maturation, and diagenesis in microbial mats, as are major metabolisms, such as photosynthesis and sulfate reducers. These surfaces include both cells themselves, which can have different membrane or cell-wall characteristics (e.g., Gram-negative vs. Gram-positive) relevant to mineralization [75], and their extracellular secretions.
Microorganisms that live in benthic environments, and especially cyanobacteria, secrete copious amounts of EPS. The negatively charged functional groups on organic surfaces (e.g., carboxyl, acetyl, hydroxyl, succinyl, and others) can bind Ca2+ and Mg2+ and are thought to be the most important mineral nucleation sites [61,74]. These functional groups—in particular, carboxylic acids, phosphates, and amines—are typically assumed to be present in EPS by the models used to characterize EPS using titration data [76]. Such models are yet to incorporate the reports of abundant sulfate groups produced by sulfate-reducing bacteria [77], pustule-forming cyanobacteria [50,78,79], and other cyanobacteria [80]. Sulfate-rich EPS produced by pustule-forming cyanobacteria binds magnesium to promote microbial silicification in solutions that contain more silica compared to modern-day seawater [78,79]. In fact, the spatial proximity of fossiliferous cation-rich chert and texture-preserving dolomite has generated testable hypotheses about the contribution of Mg-binding microbial surfaces in the formation of both chert and Mg-rich carbonates (see Figure 1; see References [78,80].
EPS degradation changes the chemical structure of the extracellular matrix. This process modifies and removes functional groups [50], changes the acidity of the surfaces, renders smaller fragments of macromolecules, and releases multivalent cations back into the environment, where they can form minerals [19,20,29,30,35,74,77,81]. The downstream degradation of smaller insoluble and soluble products of EPS degradation may further promote this precipitation by fueling sulfate reduction and the alkalinity engine [34]. Accordingly, extensive carbonate mineral precipitation in modern microbialites occurs primarily in the zones of organic degradation, in voids, and outside of the active photosynthetic layer. Therefore, characterizing, tracking, and quantifying the cycling of EPS in different regions of microbial mats is critical for our understanding of biomineralization in microbial systems.
3. Using Molecular Tools to Link Microbes to Mineralization in Microbialites from Shark Bay, Australia; and Highbourne Cay, Bahamas
The stromatolites and mats from hypersaline Shark Bay, Australia (Figure 5), and marine stromatolites and thrombolites in The Bahamas are among the world’s most prominent and well-studied microbialites [42,43]. While a comprehensive summary of past sequencing-based studies of the many extant microbial carbonate systems is beyond the scope of this review, these two microbialite systems are excellent case studies of how molecular techniques—principally SSU 16/18S rRNA amplicon sequencing, metagenomics, and transcriptomics—have been applied to investigate mechanisms that result in different mat morphologies and textures and influence carbonate alkalinity in marine microbial mats. To date, these studies have described the compositions of microbial communities in mats of different morphological types, at different geographical locations, and within different mat layers, and characterized the diversity of Cyanobacteria and sulfate reducers—the two functional clades with the most explicitly established links to the alkalinity engine and carbonate precipitation. More recent studies have expanded on this by measuring the expression of genes associated with photosynthesis, sulfate reduction, and EPS production and degradation in time and space.
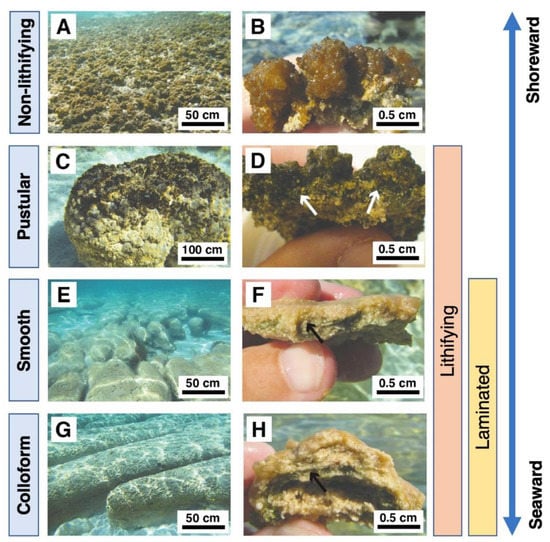
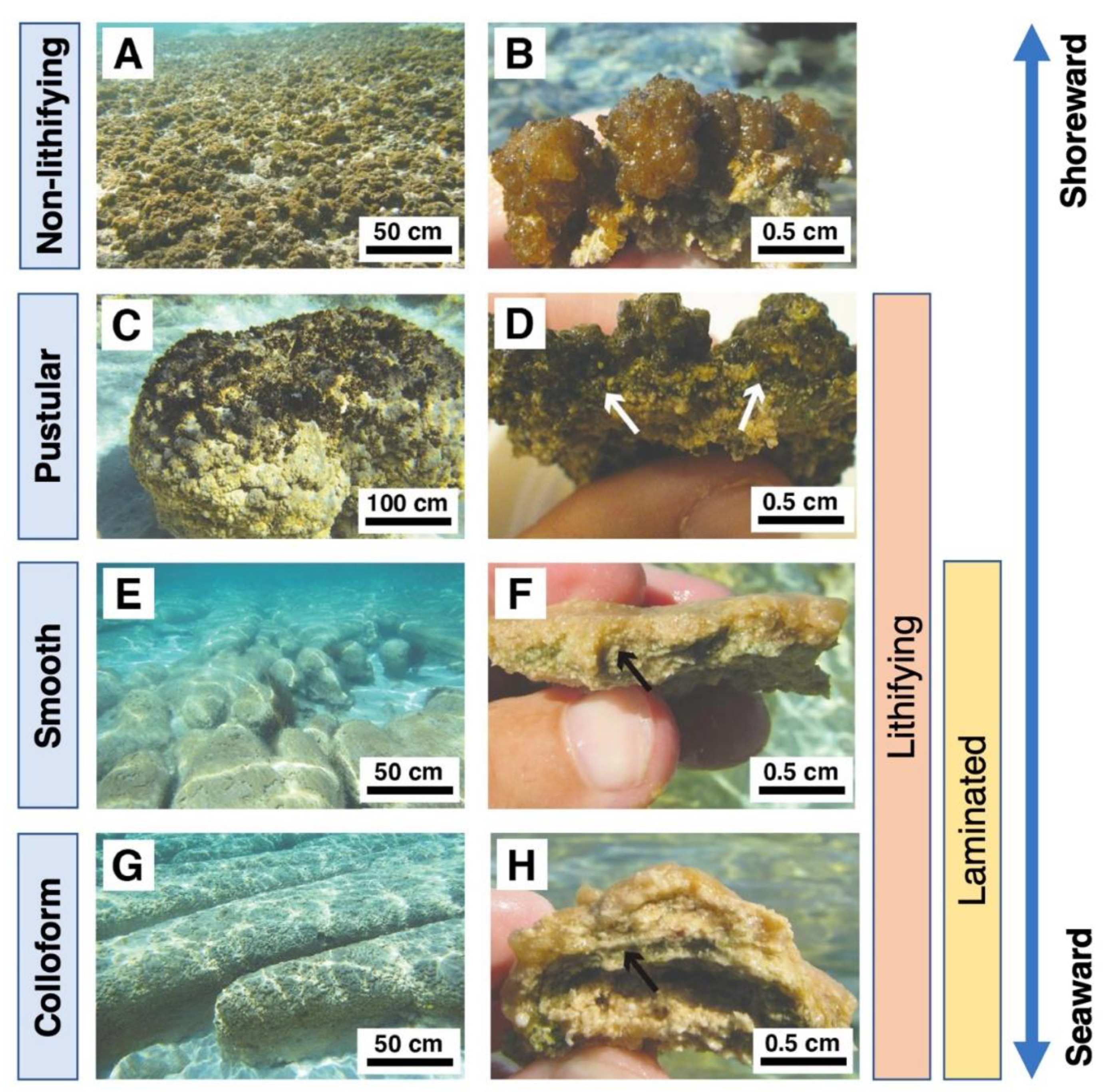
Figure 5.
Modern mat types in Hamelin Pool, Shark Bay, Australia. Adapted from Babilonia, Conesa, Casaburi, Pereira, Louyakis, Reid, and Foster [47], under the license CC-BY 4.0. (A) Sheet of EPS-rich non-lithifying pustular mats in the intertidal zone. (B) Cross-section of EPS-rich non-lithifying pustular mat. (C) Pustular stromatolite-forming mat. (D) Cross-section of lithified pustular mat. (E) Lithified smooth mats (F) Cross-section of smooth mat showing laminations. (G) Lithified colloform mats have a bumpier surface texture and less fine laminations than smooth mats. (H) Cross-section of a colloform mat domal structure. Arrows in panels (D,F,H) indicate the cyanobacterial layers.
The Shark Bay mats have been the subject of molecular ecology studies since the early 2000s [82,83]. Burns, Goh, Allen, and Neilan [83] created the first 16S rDNA clone libraries of Shark Bay stromatolites, and within one year, Papineau et al. [84] had provided the first quantitative description of Shark Bay microbial community structure based on 16S rRNA amplicon data. Molecular studies began a few years later in The Bahamas, where a robust collection of the literature on the biogeochemical and microscopic observations had already established strong links between certain types of microbial activity—primarily photosynthesis, sulfate reduction, and EPS cycling—and mat lithification [7,8,35,40]. The first amplicon-based studies of Bahamian mats characterized the diversity of Cyanobacteria [85,86,87,88] and sulfate reducers [36]. Laboratory studies of whole mat samples and isolates of mat organisms from The Bahamas used nucleic acid sequencing to assess microbial diversity [89,90,91]. Within one decade of the earliest molecular studies, the microbial community compositions of Shark Bay [42,48,92,93] and Bahamian [94,95,96,97,98,99] mats and microbialites had been extensively described, using small-subunit rRNA sequencing. Broadly speaking, these studies found abundant (>10%) Cyanobacteria and Proteobacteria in the Bahamian mats regardless of mat type. In contrast, Cyanobacteria typically accounted for only ~5% of the mat community in Shark Bay, where Proteobacteria, Bacteroidetes, Planctomycetes, and Firmicutes were more abundant. The reasons behind these differences remain unclear and invite questions about the functional roles of organisms from all these groups in different microbial mats.
Even before metagenomics and transcriptomics opened their respective windows on functional potential and gene expression in mats, researchers began attempting to associate the molecular ecology of mats with functions, mat textures, and mineralization potential. Early fluorescence in situ microscopy targeting the 16S rRNA of sulfate-reducing bacteria showed that these presumed anaerobes were actually active in oxygenated mat layers and in close contact with oxygen-producing cyanobacteria [36]. The greater microbial diversity of the more lithified Bahamian mat types was tentatively attributed to a greater metabolic diversity and biogeochemical conditions that would favor mineralization [96,97]. Similarly, the differences in the abundances and types of Cyanobacteria and other community components across the major classes of Bahamian stromatolites and thrombolites were hypothesized to either drive or reflect some morphological and biogeochemical differences [94,96,97]. Low eukaryotic diversity in five thrombolitic mats was interpreted as evidence against thrombolites being simply “bioturbated stromatolites” [94]. A combination of phylogenetic analysis and culture experiments testing for antagonistic interactions between microbes isolated from Bahamian mats suggested that such interactions between heterotrophs, including Gammaproteobacteria, Firmicutes, Bacillus, Halobacillus, or Exigunobacterium, in Bahamian mats could drive lamination [100].
Niche differentiation refers to the spatial distribution of microbes within mats based on amplicon-based microbial community composition. This concept is often used to understand mineralization in Shark Bay and some other environments [101,102]. In arguably the most comprehensive study addressing niche differentiation in Shark Bay, Wong, Smith, Visscher, and Burns [42] combined depth-resolved 16S rRNA amplicon sequencing with biogeochemical data including oxygen depth profiles and measurements of sulfate reduction rates in two Shark Bay mat types. This study hypothesized that the differences in lithification between the lithifying smooth mats and the non-lithifying pustular mats depended on the lower abundances of Cyanobacteria and Deltaproteobacteria the latter [42]. The same study also used the co-occurrence of certain taxonomic groups to motivate ecological-functional hypotheses. Specifically, the detection of Bacteroidetes, Proteobacteria, and Cyanobacteria in the upper layers of mats was interpreted as possible evidence for the “phototrophic consortia” that drive primary production and motivated an early argument for Bacteroidetes as degraders of EPS in mats [42].
As sequencing became less expensive, metagenomic studies enabled researchers to move beyond arguments based on niche differentiation and taxonomy and attempt to connect mineralization to the functional potential of mat microbes more directly [38,41,47,48,86,103,104,105,106]. Building on previous metagenomic work in other mats [10,11], the first metagenomic studies of lithifying mats in the Bahamas [86,103] and Shark Bay [38] confirmed many of the core findings of earlier 16S/18S rRNA-based community profiles and the presence of genes encoding for photosynthesis and sulfate reduction in the mat metagenome.
Some studies also proposed new hypotheses that linked the mat microbial community to carbonate precipitation. Khodadad and Foster [103] attributed the primary difference in functional potential between non-lithifying and lithifying stromatolitic mats in the Bahamas to the enrichment of carbohydrate-processing genes in lithifying mats. They interpreted this finding as a potential indication of enhanced EPS degradation in lithifying mats combined with the faster consumption of a wider variety of organic and inorganic sulfur-containing substrates (e.g., sulfate, thiosulfate, etc.) [103]. Sulfate commonly modifies the EPS of pustular mats from Shark Bay, Western Australia, where a combination of culturing and metagenomic analyses identified Cyanobacteria as the main producers of sulfated polysaccharides [50]. The same study proposed a connection among the ecology, chemical properties and biogeochemical cycles in lithifying pustular mats by detecting sulfatases, enzymes required to degrade sulfated polysaccharides, in a number of metagenome-assembled genomes, quantifying sulfatase activity and associating carbonate precipitates with areas with fewer cyanobacteria and less sulfate-rich EPS.
Comparisons among the metagenomes of different mat types in Shark Bay and The Bahamas motivated qualitative hypotheses regarding the relative importance of photosynthesis and heterotrophy in driving the carbonate alkalinity engine. A combination of metagenomic and biogeochemical data from Bahamian thrombolites pointed to photosynthesis as the most important driver of mineralization in thrombolites [105]. In contrast, Ruvindy, White, Neilan, and Burns [38] argued that the greater prevalence of predicted photosynthesis genes in the metagenomes of Bahamian mats compared to Shark Bay mats could indicate that Shark Bay biomineralization is driven more by heterotrophy than by photosynthesis. Analyses of functional genes and microscopy in a gradient of stabilized to high-energy mobilized oolitic sands were used to argue for the roles of diverse microbial metabolisms, including the degradation of EPS, in the precipitation of carbonate ooids [107,108]. Stable isotope analyses of the mat and ooid carbonate provided additional support for photosynthesis, rather than heterotrophy, as the primary driver of mineralization in Bahamian mats [38] and isotopic imprints of photosynthetic processes in marine ooids [39]. Because the relative abundance of genes associated with photosynthesis was greater in pustular lithifying mats from shallower waters in Shark Bay compared to the smooth and colloform mats at greater depths, Babilonia and co-authors [47] argued that lithification in shallower mats might be controlled more by photosynthesis than heterotrophy, and vice versa for deeper mats. Different abundances of genes from different functional pathways in the metagenomes from adjacent stromatolites and thrombolites in Highbourne Cay and Shark Bay were also interpreted as evidence that some as-yet opaque aspect of microbial community metabolism could underpin their differing mineral fabrics [109].
Most recently, transcriptomics has allowed researchers to track gene expression in mats, particularly those associated with photosynthesis and heterotrophy [49,105,109,110]. Depth-resolved metatranscriptomics of Bahamian thrombolites characterized gene expression at midday and, unsurprisingly, found more gene transcripts in pathways related to photosynthesis relative to anaerobic respiration [109]. Most of the photosynthesis gene transcripts were associated with cyanobacterial genera closely related to Dichotrix sp. (order Nostocales), the dominant cyanobacterial clade in Bahamian mats [109]. A subsequent year-long transcriptomic survey identified the filamentous Rivulaceae and coccoidal Xenococcaceae as the most active members of the community in the Bahamian thrombolites [104].
The metatranscriptomic analyses of the microbial activity over seasonal and diel cycles and in multiple mat types in Shark Bay that included cyclone-derived materials (EPS-rich cobbles and sludge) found abundant and active Bacteroidetes and sulfate-reducing bacteria in the EPS-rich cobbles [49]. This study argued that heterotrophic degradation of EPS by Bacteroidetes coupled to sulfate reduction could explain the greater amount of carbonate found in cobbles relative to sludge. This may be the first time that an explicit link between carbonate alkalinity and the activity of Bacteroidetes—an abundant phylum in Shark Bay and Highbourne Cay—has been proposed. Elevated transcription of sialic acid and aTMP–rhamnose synthesis pathways in the cobbles was also interpreted as an indication that there could be two different kinds of EPS present in cobbles, each contributing to the matrix’s resilience against degradation or cohesive properties in different ways that could potentially impact the shape and preservation of cobbles [110]. The high functional potential of Bacteroidetes, Planctomycetes, Verrucomicrobia, Chloroflexi, Myxococcota, and a few other microbial groups from pustular mats in Shark Bay for the degradation of sulfated EPS supports inferences from the transcriptomic data [50]. Molecular tools can now proceed to identify organisms involved in these pathways and their connections to the activities of Cyanobacteria, sulfate-reducing bacteria, and the many other microbial groups in the mats.
4. What Next?
After nearly two decades of molecular studies, the community compositions of the carbonate-precipitating mats from Shark Bay and The Bahamas have been described thoroughly, with studies based on different methods and performed by different researchers yielding broadly consistent findings. Likewise, numerous metagenomic studies and some transcriptomic studies have described the presence and taxonomic distribution of genes and gene transcripts from broadly defined functional categories (e.g., elemental cycling pathways, “carbohydrate metabolism”, etc.) in mats. Even so, we still do not understand why microbial mats host many different microbial groups, genes, and metabolisms and how the diverse microbes participate in building mats, shield the community from environmental stresses, or influence mineral precipitation. The puzzle of relating mineral textures and morphologies preserved in fossil microbialites to biological processes that can be observed in extant microbialites is more confounding still. Ancient mats preserved in fossil microbialites have undergone extensive information loss, from the decay of highly diagnostic biomolecules, such as nucleic acids, proteins, and lipids, to the alteration of original textures by diagenesis and metamorphism [5]. Future studies of modern microbialites should solidify evidence for the connections between molecular data and biogeochemical and microscopic observations of carbonate precipitation, identify testable hypotheses about mechanisms that drive these connections, and then test them in situ or in simplified model systems.
Molecular tools yield bewildering amounts of data. Today, a metagenomic dataset is within reach of any researcher with a few thousand dollars to spare on nucleotide sequencing. Data analyses—and good hypotheses to explore—have displaced technical complexities and cost as the limiting resources. More data and more powerful techniques will surely come. Given the granularity of molecular data, which can track the presence of individual species and their genes in space and time, how do we ensure that we do not miss the mat for the genes? Molecular studies are also only as good as the bioinformatic tools, reference databases, and biochemical studies that inform them. Given the large existing molecular datasets that describe many modern microbial carbonates, one promising avenue of future molecular research is to “zoom in”, narrowing analyses to simple testable hypotheses about the contributions of specific organisms, groups of organisms, and microbial interactions to carbonate precipitation and the formation of microbial textures. This hypothesis-driven approach is necessary to understand how modern marine microbial carbonates form, how different microbes interact in zones of carbonate precipitation, and how these interactions depend on environmental stresses or chemistry (Figure 3). A process-oriented understanding of carbonate precipitating microbial systems under a range of environmental chemical conditions relevant to the past is necessary to understand the textural differences between modern and Archean or Proterozoic microbialites (see Figure 1; see Reference [6]).
The precipitation of dolomite is an example of a geological puzzle that will require consideration of the roles of specific strains, genes, microbial surfaces, and metabolisms to solve. This mineral preserved exquisite microbial textures in rocks of Archean and Proterozoic ages (see Figure 1; see References [5,111,112,113]), but is uncommon or not fabric-retentive in more recent rocks [114,115,116]. The abiotic formation of dolomite is kinetically limited [117,118], so it has been proposed that microbes and/or organic compounds could mediate its formation. Indeed, culture studies and molecular analyses linked various bacteria to the precipitation of protodolomite, including Desulfovibrio brasiliensis [119,120], Virgibacillus sp. [68,76], and Desulfobulbus mediterraneus [67]. The phases precipitated with microbial or organic compounds tend to be disordered (proto)dolomite [60,65,76,121], with only minor amounts of dolomite present [60]. Thus, the formation of ordered dolomite may require additional factors, such as manganese cations [64]. In the future, molecular methods could be used to understand the roles of other microbes in dolomite precipitation, as well as the role of different microbial EPSs and their degradation in the nucleation of nanocrystalline dolomite. Anoxygenic phototrophs may require special attention. Recent 16S rRNA amplicon sequencing revealed a seasonal shift in the hypersaline sabkhas of Qatar from oxygenic phototrophs to anoxygenic phototrophs. Because this shift was accompanied by a change in EPS functional groups, anoxygenic phototrophs were hypothesized to be responsible for dolomite precipitation [122]. Studies of microbial biofilms in Archean-analog solutions also documented the formation of ordered dolomite on the surfaces of the green sulfur bacterium Chlorobium limicola and nanocrystalline dolomite on EPS in Chlorobium-containing biofilms [64,123]. Cyanobacteria, too, have been shown to influence the crystallinity and morphology of magnesium carbonates. They are associated with the globular precipitates of hydromagnesite in lake Salda, Turkey; and dypingite in the alkaline wetlands of Atlin, British Columbia [124,125]. It is possible that these precipitates coalesce to form the larger-scale clotted textures that are common in these environments [73,124], but more targeted molecular studies are needed to elucidate the contribution of specific microbes to mineral textures in these and other magnesium-rich carbonate structures.
Molecular tools, field observations, and experiments could also illuminate the understudied contributions of viruses to mat lithification. Viruses could potentially impact mineralization by providing mineral nucleation sites, modulating microbial community composition via lysis and transferring auxiliary metabolic genes that alter host metabolism [126]. To date, the molecular diversity of viruses in carbonate-precipitating systems has received the most attention [127,128], whereas viral contributions to lithification have been explored less frequently [129,130]. In one such study, Pacton et al. [131] suggested that mineral precipitation can occur directly on viral surfaces and on cell debris created from cell lysis. The inferred viral particles in this study were first permineralized by amorphous magnesium silicates that were subsequently converted to magnesium carbonate nanospheres [131]. A recent study [129] reported potential viruses embedded in high-Mg calcite grains in calcifying microbial mats from the hypersaline lake La Salada de Chiprana, Spain. Molecular methods for virus characterization in lithifying systems combined with mineral growth experiments could evaluate the potential of viruses to nucleate Mg-rich carbonate minerals and distinguish viruses from other biomineralizing surfaces [132,133], such as membrane vesicles that can become coated by dolomite [63,121].
The narrowed scope of “zoomed in” future studies could also explore specific functions or metabolisms, such as EPS degradation. A number of studies of bulk mat biogeochemistry and local EPS properties described above identified EPS cycling as key to biomineralization, but molecular techniques have barely begun to investigate EPS cycling and the role of this cycling in carbonate precipitation [5,12,13,50]. Thus far, molecular tools have identified Bacteroidetes, Alpha- and Gammaproteobacteria, and Planctomycetes as some of the most abundant taxa across many microbial mats [42,48,49,102]. The reasons behind these abundances and the connections of these taxa to EPS cycling, the modification of organic surfaces, and biomineralization remain to be established. This knowledge gap persists due to the difficulties associated with characterizing the chemical composition of EPS and narrowing down the molecular data to a certain function or metabolism. To address these challenges, future studies could tractably focus on microbes that possess or express large numbers of diverse carbohydrate-active enzymes (CAZymes); connect their activities to EPS modification [50] and degradation; track the flow of carbon from these to other mat organisms; and establish stronger links between the diversity of organisms, genes, and compounds present in the zones of active carbonate nucleation. Proteomic tools could also characterize the protein fraction of EPS to reveal any matrix-bound enzymes involved in the degradation of EPS. Given that such enzymes are “available” for use not only by the organism that excreted them but also by its neighbors—which, in many cases, are also bound to the matrix—these enzymes may be key to understanding the relationships among organisms involved in EPS cycling. The organisms whose genomes encode EPS-degrading enzymes may serve as libraries of polysaccharide and protein-degrading potential for the entire community, enabling organisms without their own extracellular CAZymes to play important roles in downstream EPS degradation. Future studies that focus on this question will benefit from the improvement and further development of databases and annotation tools for carbohydrate-active enzymes and sulfatases, such as the CAZy database [134], the dbCAN CAZyme-annotation web server [135,136], and the SulfAtlas sulfatase database [137]. Likewise, obtaining more reference genomes for isolated and characterized marine polysaccharide degraders and more references sequences for isolated and characterized proteins that degrade marine polysaccharides, of which there are very few, will improve our ability to interpret molecular data related to EPS degradation and tie genes to function.
In parallel, future studies could “zoom out” to explore microbial influences on carbonate precipitation over broader spans of time and space. For example, metatranscriptomic analyses of calcifying cobbles in Shark Bay found a greater abundance of genes associated with heterotrophic and anaerobic metabolisms after the cyclone and elevated transcription of genes associated with the production and protection of EPS [110]. Another study of seasonal changes related the growth of subtidal microbial mats during the summer season to the preferential stabilization of sediments in Shark Bay [15], with consequences yet to be determined for lithification. These studies highlight the necessity of frequent sampling across environmental gradients and multiple seasons to better relate lithification to the microbial response to changes in their environment.
Molecular techniques beyond the sequencing-based methods that have already been widely applied to microbial carbonate systems offer yet more opportunities to explore biological processes linked to mineralization. Proteomics [138] and lipidomics [139], as well as the determination of the structures of EPS polysaccharides [140], will be vital for understanding the biochemical environment in mats and how this environment ultimately shapes mineral textures and fabrics. Likewise, a targeted analysis of mat composition using spatially resolved methods such as Mass Spec Imaging (MSI) and desorption electrospray ionization (DESI) could enable spatially resolved, real-time observation of biomolecules in mats [141,142]. Nucleic acid sequencing information will be essential in identifying and tracking the sources and sinks of the biomolecules detectable by these other techniques.
Representative cultures of relevant organisms can help circumvent at least some issues that arise from the inherent complexity of natural EPS and tackle numerous hypotheses that arise from -omic information. Defined or less diverse cultures and laboratory experiments can focus directly onto the roles of specific organisms, metabolic or biosynthetic pathways and related genes, and partnerships among all of these in carbonate precipitation. Laboratory experiments can also establish environments that might not be represented naturally on Earth today but are analogs of conditions in the past and on other planets. Because mineral nucleation and growth in mats begins at scales smaller than or comparable to individual cells, studies of lithification in microbial systems will benefit from molecular techniques that can track gene expression and the synthesis of proteins and polysaccharides in defined systems [143,144,145]. These methods, as well as spatially resolved transcriptomics [146,147], FISH, and NanoSIMS with isotope or other types of labels can then characterize the spatial distributions of the known, as well as the currently understudied, organisms [36,148,149,150,151] and visualize microbial activity, localization, metabolisms, and matrix properties in complex biofilms and microbe–mineral systems [152,153,154,155,156,157].
Although many of these studies focus on individual microbes, genes, and processes, they can eventually be scaled up to multi-microbe model systems to better quantify the effects of microbial interactions on biomineralization and texture formation, starting with model systems, expanding insights to natural mats, engineering and decarbonization applications [158]. Integrating the insights gained from molecular studies in modern microbialites and laboratory model systems with other approaches will be critical for connecting studies of modern mats to mineral textures and mat morphologies in ancient microbialites. Through these multi-pronged approaches, we can start to develop predictions of carbonate microbialites and microbial textures that integrate processes controlled by different genes, organic surfaces, and microbial metabolisms and superimpose them onto the background of environmental evolution to reconstruct the rich record of microbial textures preserved in carbonate microbialites from the Archean Eon onward.
Author Contributions
Conceptualization, E.M.C. and T.B.; writing—original draft preparation, E.M.C., M.J.B., J.G., J.H., K.R.M., E.J.S. and T.B.; writing—review and editing, E.M.C. and T.B.; visualization, E.M.C., E.J.S., M.J.B., J.G. and K.R.M.; supervision, T.B.; project administration, E.M.C. and T.B.; funding acquisition, T.B. and E.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Simons Foundation Collaboration on the Origins of Life (SCOL) grant number 327126 to TB and the NASA 80NSSC20K0234 grant to TB. The APC was funded by the John V. Jarve MIT Internal Award to TB. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1745302 to EC. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge the Center for Nanoscale Systems at Harvard University for providing facilities to perform the SEM/EDS analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Allwood, A.C.; Walter, M.R.; Kamber, B.S.; Marshall, C.P.; Burch, I.W. Stromatolite reef from the Early Archaean era of Australia. Nature 2006, 441, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Tice, M.M.; Lowe, D.R. Photosynthetic microbial mats in the 3416-Myr-old ocean. Nature 2004, 431, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Homann, M. Earliest life on earth: Evidence from the Barberton Greenstone Belt, South Africa. Earth-Sci. Rev. 2019, 196, 102888. [Google Scholar] [CrossRef]
- Dupraz, C.; Reid, R.P.; Braissant, O.; Decho, A.W.; Norman, R.S.; Visscher, P.T. Processes of carbonate precipitation in modern microbial mats. Earth-Sci. Rev. 2009, 96, 141–162. [Google Scholar] [CrossRef]
- Bosak, T.; Knoll, A.H.; Petroff, A.P. The meaning of stromatolites. Annu. Rev. Earth Planet. Sci. 2013, 41, 21–44. [Google Scholar] [CrossRef]
- Grotzinger, J.P.; Knoll, A.H. Stromatolites in Precambrian carbonates: Evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Planet. Sci. 1999, 27, 313–358. [Google Scholar] [CrossRef]
- Reid, R.P.; Visscher, P.T.; Decho, A.W.; Stolz, J.F.; Bebout, B.M.; Dupraz, C.; Macintyre, L.G.; Paerl, H.W.; Pinckney, J.L.; Prufert-Bebout, L.; et al. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 2000, 406, 989–992. [Google Scholar] [CrossRef]
- Visscher, P.T.; Reid, R.P.; Bebout, B.M. Microscale observations of sulfate reduction: Correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology 2000, 28, 919–922. [Google Scholar] [CrossRef]
- Petroff, A.; Beukes, N.; Rothman, D.; Bosak, T. Biofilm growth and fossil form. Phys. Rev. X 2013, 3, 041012. [Google Scholar] [CrossRef]
- Petroff, A.P.; Sim, M.S.; Maslov, A.; Krupenin, M.; Rothman, D.H.; Bosak, T. Biophysical basis for the geometry of conical stromatolites. Proc. Natl. Acad. Sci. USA 2010, 107, 9956–9961. [Google Scholar] [CrossRef]
- Grotzinger, J.P.; Rothman, D.H. An abiotic model for stromatolite morphogenesis. Nature 1996, 383, 423–425. [Google Scholar] [CrossRef]
- Walter, M.R.; Bauld, J.; Brock, T.D. Microbiology and morphogenesis of columnar stromatolites (Conophyton, Vacerrilla) from hot springs in Yellowstone National Park. In Stromatolites; Walter, M.R., Ed.; Developments in Sedimentology; Elsevier: Amsterdam, The Netherlands, 1976; Volume 20, pp. 273–310. [Google Scholar]
- Hoffman, P. Environmental diversity of Middle Precambrian stromatolites. In Stromatolites; Walter, M.R., Ed.; Elsevier Scientific Publishing Company: Amsterdam, The Netherlands, 1976; Volume 20, pp. 599–612. [Google Scholar]
- Hoffman, P.F. Stromatolite morphogenesis in Shark Bay, Western Australia. In Stromatolites; Walter, M.R., Ed.; Developments in Sedimentology; Elsevier: Amsterdam, The Netherlands, 1976; Volume 20, pp. 261–272. [Google Scholar]
- Murshid, S.; Mariotti, G.; Pruss, S.B.; Bosak, T.; Suosaari, E.P. Seasonal changes in sediment erodibility in a sandy carbonate environment detected from turbidity time series. Mar. Geol. 2021, 439, 106570. [Google Scholar] [CrossRef]
- Suosaari, E.P.; Reid, R.P.; Araujo, T.A.A.; Playford, P.E.; Holley, D.K.; McNamara, K.J.; Eberli, G.P. Environmental pressures influencing living stromatolites in Hamelin Pool, Shark Bay, Western Australia. Palaios 2016, 31, 483–496. [Google Scholar] [CrossRef]
- Suosaari, E.; Reid, R.; Playford, P.; Foster, J.; Stolz, J.; Casaburi, G.; Hagan, P.; Chirayath, V.; Macintyre, I.; Planavsky, N. New multi-scale perspectives on the stromatolites of Shark Bay, Western Australia. Sci. Rep. 2016, 6, 20557. [Google Scholar] [CrossRef] [PubMed]
- Altermann, W. Accretion, trapping and binding of sediment in Archean stromatolites—Morphological expression of the antiquity of life. Space Sci. Rev. 2008, 135, 55–79. [Google Scholar] [CrossRef]
- Arp, G.; Reimer, A.; Reitner, J. Calcification in cyanobacterial biofilms of alkaline salt lakes. Eur. J. Phycol. 1999, 34, 393–403. [Google Scholar] [CrossRef]
- Arp, G.; Thiel, V.; Reimer, A.; Michaelis, W.; Reitner, J. Biofilm exopolymers control microbialite formation at thermal springs discharging into the alkaline Pyramid Lake, Nevada, USA. Sediment. Geol. 1999, 126, 159–176. [Google Scholar] [CrossRef]
- Couradeau, E.; Benzerara, K.; Gérard, E.; Estève, I.; Moreira, D.; Tavera, R.; López-García, P. Cyanobacterial calcification in modern microbialites at the submicrometer scale. Biogeosciences 2013, 10, 5255–5266. [Google Scholar] [CrossRef]
- Gautret, P.; Camoin, G.; Golubic, S.; Sprachta, S. Biochemical control of calcium carbonate precipitation in modern lagoonal microbialites, Tikehau Atoll, French Polynesia. J. Sediment. Res. 2004, 74, 462–478. [Google Scholar] [CrossRef]
- Sprachta, S.; Camoin, G.; Golubic, S.; Le Campion, T. Microbialites in a modern lagoonal environment: Nature and distribution, Tikehau atoll (French Polynesia). Palaeogeogr. Palaeoclimatol. Palaeoecol. 2001, 175, 103–124. [Google Scholar] [CrossRef]
- Neumann, A.C.; Gebelein, C.D.; Scoffin, T.P. The composition, structure and erodability of subtidal mats, Abaco, Bahamas. J. Sediment. Petrol. 1970, 40, 274–297. [Google Scholar]
- Homann, M.; Heubeck, C.; Airo, A.; Tice, M.M. Morphological adaptations of 3.22 Ga-old tufted microbial mats to Archean coastal habitats (Moodies Group, Barberton Greenstone Belt, South Africa). Precambrian Res. 2015, 266, 47–64. [Google Scholar] [CrossRef]
- Jahnert, R.J.; Collins, L.B. Characteristics, distribution and morphogenesis of subtidal microbial systems in Shark Bay, Australia. Mar. Geol. 2012, 303, 115–136. [Google Scholar] [CrossRef]
- Reid, R.P.; James, N.P.; Macintyre, I.G.; Dupraz, C.P.; Burne, R.V. Shark Bay stromatolites: Microfabrics and reinterpretation of origins. Facies 2003, 49, 299–324. [Google Scholar] [CrossRef]
- Pages, A.; Welsh, D.T.; Teasdale, P.R.; Grice, K.; Vacher, M.; Bennett, W.W.; Visscher, P.T. Diel fluctuations in solute distributions and biogeochemical cycling in a hypersaline microbial mat from Shark Bay, WA. Mar. Chem. 2014, 167, 102–112. [Google Scholar] [CrossRef]
- Arp, G.; Helms, G.; Karlinska, K.; Schumann, G.; Reimer, A.; Reitner, J.; Trichet, J. Photosynthesis versus exopolymer degradation in the formation of microbialites on the atoll of Kiritimati, Republic of Kiribati, Central Pacific. Geomicrobiol. J. 2012, 29, 29–65. [Google Scholar] [CrossRef]
- Suarez-Gonzalez, P.; Reitner, J. Ooids forming in situ within microbial mats (Kiritimati atoll, central Pacific). PalZ 2021, 95, 809–821. [Google Scholar] [CrossRef]
- Dupraz, C.; Visscher, P.T. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005, 13, 429–438. [Google Scholar] [CrossRef]
- Bontognali, T.R.; Vasconcelos, C.; Warthmann, R.J.; Bernasconi, S.M.; Dupraz, C.; Strohmenger, C.J.; McKenzie, J.A. Dolomite formation within microbial mats in the coastal sabkha of Abu Dhabi (United Arab Emirates). Sedimentology 2010, 57, 824–844. [Google Scholar] [CrossRef]
- DiLoreto, Z.A.; Bontognali, T.R.; Al Disi, Z.A.; Al-Kuwari, H.A.S.; Williford, K.H.; Strohmenger, C.J.; Sadooni, F.; Palermo, C.; Rivers, J.M.; McKenzie, J.A. Microbial community composition and dolomite formation in the hypersaline microbial mats of the Khor Al-Adaid sabkhas, Qatar. Extremophiles 2019, 23, 201–218. [Google Scholar] [CrossRef]
- Braissant, O.; Decho, A.W.; Przekop, K.M.; Gallagher, K.L.; Glunk, C.; Dupraz, C.; Visscher, P.T. Characteristics and turnover of exopolymeric substances in a hypersaline microbial mat. FEMS Microbiol. Ecol. 2009, 67, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Visscher, P.T.; Reid, R.P. Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 219, 71–86. [Google Scholar] [CrossRef]
- Baumgartner, L.K.; Reid, R.P.; Dupraz, C.; Decho, A.W.; Buckley, D.; Spear, J.; Przekop, K.M.; Visscher, P.T. Sulfate reducing bacteria in microbial mats: Changing paradigms, new discoveries. Sediment. Geol. 2006, 185, 131–145. [Google Scholar] [CrossRef]
- Birgel, D.; Meister, P.; Lundberg, R.; Horath, T.; Bontognali, T.R.; Bahniuk, A.M.; de Rezende, C.E.; Vásconcelos, C.; McKenzie, J.A. Methanogenesis produces strong 13C enrichment in stromatolites of Lagoa Salgada, Brazil: A modern analogue for Palaeo-/Neoproterozoic stromatolites? Geobiology 2015, 13, 245–266. [Google Scholar] [CrossRef]
- Ruvindy, R.; White, R.A.I.; Neilan, B.A.; Burns, B.P. Unravelling core microbial metabolisms in the hypersaline microbial mats of Shark Bay using high-throughput metagenomics. ISME J. 2016, 10, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.R.; Swart, P.K.; Eberli, G.P.; Oehlert, A.M.; Devlin, Q.; Saeid, A.; Altabet, M.A. Geochemical evidence of microbial activity within ooids. Sedimentology 2015, 62, 2090–2112. [Google Scholar] [CrossRef]
- Stolz, J.F.; Reid, R.P.; Visscher, P.T.; Decho, A.W.; Norman, R.S.; Aspden, R.J.; Bowlin, E.M.; Franks, J.; Foster, J.S.; Paterson, D.M. The microbial communities of the modern marine stromatolites at Highborne Cay, Bahamas. Atoll Res. Bull. 2009, 567, 1–29. [Google Scholar] [CrossRef]
- Wong, H.L.; MacLeod, F.I.; White, R.A.; Visscher, P.T.; Burns, B.P. Microbial dark matter filling the niche in hypersaline microbial mats. Microbiome 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Wong, H.L.; Smith, D.-L.; Visscher, P.T.; Burns, B.P. Niche differentiation of bacterial communities at a millimeter scale in Shark Bay microbial mats. Sci. Rep. 2015, 5, 15607. [Google Scholar] [CrossRef]
- Foster, J.S.; Green, S.J. Microbial diversity in modern stromatolites. In Stromatolites: Interaction of Microbes with Sediments; Springer: Dordrecht, The Netherlands, 2011; pp. 383–405. [Google Scholar]
- Ley, R.E.; Harris, J.K.; Wilcox, J.; Spear, J.R.; Miller, S.R.; Bebout, B.M.; Maresca, J.A.; Bryant, D.A.; Sogin, M.L.; Pace, N.R. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 2006, 72, 3685–3695. [Google Scholar] [CrossRef]
- Puckett, M.K.; McNeal, K.S.; Kirkland, B.L.; Corley, M.E.; Ezell, J.E. Biogeochemical stratification and carbonate dissolution-precipitation in hypersaline microbial mats (Salt Pond, San Salvador, The Bahamas). Aquat. Geochem. 2011, 17, 397–418. [Google Scholar] [CrossRef]
- Visscher, P.T.; Stolz, J.F. Microbial mats as bioreactors: Populations, processes, and products. In Geobiology: Objectives, Concepts, Perspectives; Elsevier: Amsterdam, The Netherlands, 2005; pp. 87–100. [Google Scholar]
- Babilonia, J.; Conesa, A.; Casaburi, G.; Pereira, C.; Louyakis, A.S.; Reid, R.P.; Foster, J.S. Comparative metagenomics provides insight into the ecosystem functioning of the Shark Bay Stromatolites, Western Australia. Front. Microbiol. 2018, 9, 1359. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.L.; White, R.A.; Visscher, P.T.; Charlesworth, J.C.; Vázquez-Campos, X.; Burns, B.P. Disentangling the drivers of functional complexity at the metagenomic level in Shark Bay microbial mat microbiomes. ISME J. 2018, 12, 2619–2639. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Grice, K.; Visscher, P.T.; Morris, T.; Wong, H.L.; White, R.A.I.; Burns, B.P.; Coolen, M.J. Functional gene expression in Shark Bay hypersaline microbial mats: Adaptive responses. Front. Microbiol. 2020, 2741. [Google Scholar] [CrossRef]
- Skoog, E.J.; Moore, K.R.; Gong, J.; Ciccarese, D.; Momper, L.; Cutts, E.; Bosak, T. Metagenomic, (bio)chemical and microscopic analyses reveal the potential for the cycling of sulfated EPS in Shark Bay pustular mats. ISME J. In Press.
- Van Lith, Y.; Warthmann, R.; Vasconcelos, C.; McKenzie, J.A. Sulphate-reducing bacteria induce low-temperature Ca-dolomite and high Mg-calcite formation. Geobiology 2003, 1, 71–79. [Google Scholar] [CrossRef]
- Morse, J.W.; Zullig, J.J.; Bernstein, L.D.; Millero, F.J.; Milne, P.; Mucci, A.; Choppin, G.R. Chemistry of calcium carbonate-rich shallow water sediments in the Bahamas. Am. J. Sci. 1985, 285, 147–185. [Google Scholar] [CrossRef]
- Meister, P. Two opposing effects of sulfate reduction on carbonate precipitation in normal marine, hypersaline, and alkaline environments. Geology 2013, 41, 499–502. [Google Scholar] [CrossRef]
- Ku, T.; Walter, L.; Coleman, M.; Blake, R.; Martini, A.M. Coupling between sulfur recycling and syndepositional carbonate dissolution: Evidence from oxygen and sulfur isotope composition of pore water sulfate, South Florida Platform, USA. Geochim. Et Cosmochim. Acta 1999, 63, 2529–2546. [Google Scholar] [CrossRef]
- Visscher, P.T.; Reid, R.P.; Bebout, B.M.; Hoeft, S.E.; Macintyre, I.G.; Thompson, J.A. Formation of lithified micritic laminae in modern marine stromatolites (Bahamas); the role of sulfur cycling. Am. Mineral. 1998, 83, 1482–1493. [Google Scholar] [CrossRef]
- Vile, M.A.; Wieder, R.K. Alkalinity generation by Fe(III) reduction versus sulfate reduction in wetlands constructed for acid mine drainage treatment. Water Air Soil Pollut. 1993, 69, 425–441. [Google Scholar] [CrossRef]
- Achal, V.; Pan, X. Characterization of urease and carbonic anhydrase producing bacteria and their role in calcite precipitation. Curr. Microbiol. 2011, 62, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Kenward, P.; Goldstein, R.; Gonzalez, L.; Roberts, J. Precipitation of low-temperature dolomite from an anaerobic microbial consortium: The role of methanogenic Archaea. Geobiology 2009, 7, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Bennett, P.C.; Gonzalez, L.A.; Macpherson, G.L.; Milliken, K.L. Microbial precipitation of dolomite in methanogenic groundwater. Geology 2004, 32, 277–280. [Google Scholar] [CrossRef]
- Roberts, J.A.; Kenward, P.A.; Fowle, D.A.; Goldstein, R.H.; González, L.A.; Moore, D.S. Surface chemistry allows for abiotic precipitation of dolomite at low temperature. Proc. Natl. Acad. Sci. USA 2013, 110, 14540–14545. [Google Scholar] [CrossRef]
- Decho, A.W. Overview of biopolymer-induced mineralization: What goes on in biofilms? Ecol. Eng. 2010, 36, 137–144. [Google Scholar] [CrossRef]
- Bosak, T.; Newman, D.K. Microbial nucleation of calcium carbonate in the Precambrian. Geology 2003, 31, 577–580. [Google Scholar] [CrossRef]
- Bosak, T.; Newman, D.K. Microbial kinetic controls on calcite morphology in supersaturated solutions. J. Sediment. Res. 2005, 75, 190–199. [Google Scholar] [CrossRef]
- Daye, M.; Higgins, J.; Bosak, T. Formation of ordered dolomite in anaerobic photosynthetic biofilms. Geology 2019, 47, 509–512. [Google Scholar] [CrossRef]
- Bontognali, T.R.; McKenzie, J.A.; Warthmann, R.J.; Vasconcelos, C. Microbially influenced formation of Mg-calcite and Ca-dolomite in the presence of exopolymeric substances produced by sulphate-reducing bacteria. Terra Nova 2014, 26, 72–77. [Google Scholar] [CrossRef]
- Braissant, O.; Cailleau, G.; Dupraz, C.; Verrecchia, E.P. Bacterially induced mineralization of calcium carbonate in terrestrial environments: The role of exopolysaccharides and amino acids. J. Sediment. Res. 2003, 73, 485–490. [Google Scholar] [CrossRef]
- Krause, S.; Liebetrau, V.; Gorb, S.; Sánchez-Román, M.; McKenzie, J.A.; Treude, T. Microbial nucleation of Mg-rich dolomite in exopolymeric substances under anoxic modern seawater salinity: New insight into an old enigma. Geology 2012, 40, 587–590. [Google Scholar] [CrossRef]
- Sánchez-Román, M.; Vasconcelos, C.; Schmid, T.; Dittrich, M.; McKenzie, J.A.; Zenobi, R.; Rivadeneyra, M.A. Aerobic microbial dolomite at the nanometer scale: Implications for the geologic record. Geology 2008, 36, 879–882. [Google Scholar] [CrossRef]
- Dupraz, C.; Visscher, P.T.; Baumgartner, L.; Reid, R. Microbe–mineral interactions: Early carbonate precipitation in a hypersaline lake (Eleuthera Island, Bahamas). Sedimentology 2004, 51, 745–765. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Decho, A.W. A laboratory investigation of cyanobacterial extracellular polymeric secretions (EPS) in influencing CaCO3 polymorphism. J. Cryst. Growth 2002, 240, 230–235. [Google Scholar] [CrossRef]
- Perri, E.; Tucker, M.E.; Słowakiewicz, M.; Whitaker, F.; Bowen, L.; Perrotta, I.D. Carbonate and silicate biomineralization in a hypersaline microbial mat (Mesaieed sabkha, Qatar): Roles of bacteria, extracellular polymeric substances and viruses. Sedimentology 2018, 65, 1213–1245. [Google Scholar] [CrossRef]
- Perri, E.; Tucker, M.E.; Spadafora, A. Carbonate organo-mineral micro- and ultrastructures in sub-fossil stromatolites: Marion lake, South Australia. Geobiology 2012, 10, 105–117. [Google Scholar] [CrossRef]
- Spadafora, A.; Perri, E.; McKenzie, J.A.; Vasconcelos, C. Microbial biomineralization processes forming modern Ca:Mg carbonate stromatolites. Sedimentology 2010, 57, 27–40. [Google Scholar] [CrossRef]
- Pace, A.; Bourillot, R.; Bouton, A.; Vennin, E.; Braissant, O.; Dupraz, C.; Duteil, T.; Bundeleva, I.; Patrier, P.; Galaup, S. Formation of stromatolite lamina at the interface of oxygenic–anoxygenic photosynthesis. Geobiology 2018, 16, 378–398. [Google Scholar] [CrossRef]
- Stanley, W.; Southam, G. The effect of Gram-positive (Desulfosporosinus orientis) and Gram-negative (Desulfovibrio desulfuricans) sulfate-reducing bacteria on iron sulfide mineral precipitation. Can. J. Microbiol. 2018, 64, 629–637. [Google Scholar] [CrossRef]
- Al Disi, Z.A.; Zouari, N.; Dittrich, M.; Jaoua, S.; Al-Kuwari, H.A.S.; Bontognali, T.R. Characterization of the extracellular polymeric substances (EPS) of Virgibacillus strains capable of mediating the formation of high Mg-calcite and protodolomite. Mar. Chem. 2019, 216, 103693. [Google Scholar] [CrossRef]
- Braissant, O.; Decho, A.W.; Dupraz, C.; Glunk, C.; Przekop, K.M.; Visscher, P.T. Exopolymeric substances of sulfate-reducing bacteria: Interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology 2007, 5, 401–411. [Google Scholar] [CrossRef]
- Moore, K.R.; Gong, J.; Pajusalu, M.; Skoog, E.J.; Xu, M.; Feliz Soto, T.; Sojo, V.; Matreux, T.; Baldes, M.J.; Braun, D. A new model for silicification of cyanobacteria in Proterozoic tidal flats. Geobiology 2021, 19, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.R.; Pajusalu, M.; Gong, J.; Sojo, V.; Matreux, T.; Braun, D.; Bosak, T. Biologically mediated silicification of marine cyanobacteria and implications for the Proterozoic fossil record. Geology 2020, 48, 862–866. [Google Scholar] [CrossRef]
- Moore, K.R.; Daye, M.; Gong, J.; Williford, K.; Konhauser, K.O.; Bosak, T. The record of biological-environmental interactions hosted in Proterozoic carbonate-hosted chert. Geobiology, submitted.
- Arp, G.; Hofmann, J.; Reitner, J. Microbial fabric formation in spring mounds (“microbialites”) of alkaline salt lakes in the Badain Jaran sand sea, PR China. Palaios 1998, 13, 581–592. [Google Scholar] [CrossRef]
- Litchfield, C.; Gillevet, P. Microbial diversity and complexity in hypersaline environments: A preliminary assessment. J. Ind. Microbiol. Biotechnol. 2002, 28, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Burns, B.P.; Goh, F.; Allen, M.; Neilan, B.A. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 2004, 6, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Papineau, D.; Walker, J.J.; Mojzsis, S.J.; Pace, N.R. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 2005, 71, 4822–4832. [Google Scholar] [CrossRef]
- Yannarell, A.C.; Steppe, T.F.; Paerl, H.W. Genetic variance in the composition of two functional groups (diazotrophs and cyanobacteria) from a hypersaline microbial mat. Appl. Environ. Microbiol. 2006, 72, 1207–1217. [Google Scholar] [CrossRef]
- Mobberley, J.M.; Khodadad, C.L.; Foster, J.S. Metabolic potential of lithifying cyanobacteria-dominated thrombolitic mats. Photosynth. Res. 2013, 118, 125–140. [Google Scholar] [CrossRef]
- Foster, J.S.; Green, S.J.; Ahrendt, S.R.; Golubic, S.; Reid, R.P.; Hetherington, K.L.; Bebout, L. Molecular and morphological characterization of cyanobacterial diversity in the stromatolites of Highborne Cay, Bahamas. ISME J. 2009, 3, 573–587. [Google Scholar] [CrossRef]
- Yannarell, A.C.; Steppe, T.F.; Paerl, H.W. Disturbance and recovery of microbial community structure and function following Hurricane Frances. Environ. Microbiol. 2007, 9, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, J.M.; Edgcomb, V.P.; Visscher, P.T.; McIntyre-Wressnig, A.; Summons, R.E.; Bouxsein, M.L.; Louis, L.; Jeglinski, M. Insights into foraminiferal influences on microfabrics of microbialites at Highborne Cay, Bahamas. Proc. Natl. Acad. Sci. USA 2013, 110, 9830–9834. [Google Scholar] [CrossRef] [PubMed]
- Edgcomb, V.P.; Bernhard, J.M.; Summons, R.E.; Orsi, W.; Beaudoin, D.; Visscher, P.T. Active eukaryotes in microbialites from Highborne Cay, Bahamas, and Hamelin Pool (Shark Bay), Australia. ISME J. 2014, 8, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Ahrendt, S.R.; Mobberley, J.M.; Visscher, P.T.; Koss, L.L.; Foster, J.S. Effects of elevated carbon dioxide and salinity on the microbial diversity in lithifying microbial mats. Minerals 2014, 4, 145–169. [Google Scholar] [CrossRef]
- Allen, M.; Goh, F.; Burns, B.; Neilan, B. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology 2009, 7, 82–96. [Google Scholar] [CrossRef]
- Goh, F.; Allen, M.A.; Leuko, S.; Kawaguchi, T.; Decho, A.W.; Burns, B.P.; Neilan, B.A. Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 2009, 3, 383–396. [Google Scholar] [CrossRef]
- Myshrall, K.; Mobberley, J.; Green, S.; Visscher, P.; Havemann, S.; Reid, R.; Foster, J. Biogeochemical cycling and microbial diversity in the thrombolitic microbialites of Highborne Cay, Bahamas. Geobiology 2010, 8, 337–354. [Google Scholar] [CrossRef]
- Havemann, S.A.; Foster, J.S. Comparative characterization of the microbial diversities of an artificial microbialite model and a natural stromatolite. Appl. Environ. Microbiol. 2008, 74, 7410–7421. [Google Scholar] [CrossRef]
- Baumgartner, L.K.; Dupraz, C.; Buckley, D.H.; Spear, J.R.; Pace, N.R.; Visscher, P.T. Microbial species richness and metabolic activities in hypersaline microbial mats: Insight into biosignature formation through lithification. Astrobiology 2009, 9, 861–874. [Google Scholar] [CrossRef]
- Baumgartner, L.K.; Spear, J.R.; Buckley, D.H.; Pace, N.R.; Reid, R.P.; Dupraz, C.; Visscher, P.T. Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ. Microbiol. 2009, 11, 2710–2719. [Google Scholar] [CrossRef]
- Casaburi, G.; Duscher, A.A.; Reid, R.P.; Foster, J.S. Characterization of the stromatolite microbiome from Little Darby Island, The Bahamas using predictive and whole shotgun metagenomic analysis. Environ. Microbiol. 2016, 18, 1452–1469. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.G.; Wronkiewicz, D.J.; Mormile, M.R.; Foster, J.S. Mineralogy and microbial diversity of the microbialites in the hypersaline Storr’s Lake, The Bahamas. Astrobiology 2016, 16, 282–300. [Google Scholar] [CrossRef] [PubMed]
- Long, R.A.; Eveillard, D.; Franco, S.L.M.; Reeves, E.; Pinckney, J.L. Antagonistic interactions between heterotrophic bacteria as a potential regulator of community structure of hypersaline microbial mats. FEMS Microbiol. Ecol. 2013, 83, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Kirk Harris, J.; Gregory Caporaso, J.; Walker, J.J.; Spear, J.R.; Gold, N.J.; Robertson, C.E.; Hugenholtz, P.; Goodrich, J.; McDonald, D.; Knights, D. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J. 2013, 7, 50–60. [Google Scholar] [CrossRef]
- Schneider, D.; Arp, G.; Reimer, A.; Reitner, J.; Daniel, R. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, Central Pacific. PLoS ONE 2013, 8, e66662. [Google Scholar] [CrossRef]
- Khodadad, C.L.; Foster, J.S. Metagenomic and metabolic profiling of nonlithifying and lithifying stromatolitic mats of Highborne Cay, The Bahamas. PLoS ONE 2012, 7, e38229. [Google Scholar] [CrossRef]
- Louyakis, A.S.; Gourlé, H.; Casaburi, G.; Bonjawo, R.M.; Duscher, A.A.; Foster, J.S. A year in the life of a thrombolite: Comparative metatranscriptomics reveals dynamic metabolic changes over diel and seasonal cycles. Environ. Microbiol. 2018, 20, 842–861. [Google Scholar] [CrossRef]
- Louyakis, A.S.; Mobberley, J.M.; Vitek, B.E.; Visscher, P.T.; Hagan, P.D.; Reid, R.P.; Kozdon, R.; Orland, I.J.; Valley, J.W.; Planavsky, N.J. A study of the microbial spatial heterogeneity of Bahamian thrombolites using molecular, biochemical, and stable isotope analyses. Astrobiology 2017, 17, 413–430. [Google Scholar] [CrossRef]
- Chen, R.; Wong, H.L.; Kindler, G.S.; MacLeod, F.I.; Benaud, N.; Ferrari, B.C.; Burns, B.P. Discovery of an abundance of biosynthetic gene clusters in Shark Bay microbial mats. Front. Microbiol. 2020, 11, 1950. [Google Scholar] [CrossRef]
- Diaz, M.; Van Norstrand, J.; Eberli, G.; Piggot, A.; Zhou, J.; Klaus, J. Functional gene diversity of oolitic sands from Great Bahama Bank. Geobiology 2014, 12, 231–249. [Google Scholar] [CrossRef]
- Diaz, M.R.; Eberli, G.P.; Blackwelder, P.; Phillips, B.; Swart, P.K. Microbially mediated organomineralization in the formation of ooids. Geology 2017, 45, 771–774. [Google Scholar] [CrossRef]
- Mobberley, J.; Khodadad, C.; Visscher, P.; Reid, R.; Hagan, P.; Foster, J. Inner workings of thrombolites: Spatial gradients of metabolic activity as revealed by metatranscriptome profiling. Sci. Rep. 2015, 5, 12601. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.A.; Coolen, M.J.; Visscher, P.T.; Morris, T.; Grice, K. Structure and function of Shark Bay microbial communities following tropical cyclone Olwyn: A metatranscriptomic and organic geochemical perspective. Geobiology 2021, 19, 642–664. [Google Scholar] [CrossRef] [PubMed]
- Pruss, S.B.; Bosak, T.; Macdonald, F.A.; McLane, M.; Hoffman, P. Microbial facies in an early Cryogenian (Sturtian) cap carbonate, the Rasthof Formation, Otavi Group, northern Namibia. Precambrian Res. 2010, 181, 187–198. [Google Scholar] [CrossRef]
- Siahi, M.; Hofmann, A.; Hegner, E.; Master, S. Sedimentology and facies analysis of Mesoarchaean stromatolitic carbonate rocks of the Pongola Supergroup, South Africa. Precambrian Res. 2016, 278, 244–264. [Google Scholar] [CrossRef]
- Siahi, M.; Hofmann, A.; Master, S.; Mueller, C.; Gerdes, A. Carbonate ooids of the Mesoarchaean Pongola Supergroup, South Africa. Geobiology 2017, 15, 750–766. [Google Scholar] [CrossRef]
- Sibley, D.F. Secular changes in the amount and texture of dolomite. Geology 1991, 19, 151–154. [Google Scholar] [CrossRef]
- Burns, S.J.; Mckenzie, J.A.; Vasconcelos, C. Dolomite formation and biogeochemical cycles in the Phanerozoic. Sedimentology 2000, 47, 49–61. [Google Scholar] [CrossRef]
- Arvidson, R.S.; Mackenzie, F.T. The dolomite problem; control of precipitation kinetics by temperature and saturation state. Am. J. Sci. 1999, 299, 257–288. [Google Scholar] [CrossRef]
- Land, L.S. The origin of massive dolomite. J. Geol. Educ. 1985, 33, 112–125. [Google Scholar] [CrossRef]
- Lippmann, F. Sedimentary Carbonate Minerals; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 6. [Google Scholar]
- Bontognali, T.R.; Vasconcelos, C.; Warthmann, R.J.; Lundberg, R.; McKenzie, J.A. Dolomite-mediating bacterium isolated from the sabkha of Abu Dhabi (UAE). Terra Nova 2012, 24, 248–254. [Google Scholar] [CrossRef]
- Warthmann, R.; Vasconcelos, C.; Sass, H.; McKenzie, J.A. Desulfovibrio brasiliensis sp. nov., a moderate halophilic sulfate-reducing bacterium from Lagoa Vermelha (Brazil) mediating dolomite formation. Extremophiles 2005, 9, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Al Disi, Z.A.; Jaoua, S.; Bontognali, T.R.; Attia, E.S.; Al-Kuwari, H.A.; Zouari, N. Evidence of a role for aerobic bacteria in high magnesium carbonate formation in the evaporitic environment of Dohat Faishakh Sabkha in Qatar. Front. Environ. Sci. 2017, 5, 1. [Google Scholar] [CrossRef]
- Diloreto, Z.A.; Garg, S.; Bontognali, T.R.; Dittrich, M. Modern dolomite formation caused by seasonal cycling of oxygenic phototrophs and anoxygenic phototrophs in a hypersaline sabkha. Sci. Rep. 2021, 11, 4170. [Google Scholar] [CrossRef] [PubMed]
- Daye, M.; Klepac-Ceraj, V.; Pajusalu, M.; Rowland, S.; Farrell-Sherman, A.; Beukes, N.; Tamura, N.; Fournier, G.; Bosak, T. Light-driven anaerobic microbial oxidation of manganese. Nature 2019, 576, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Balci, N.; Gunes, Y.; Kaiser, J.; On, S.A.; Eris, K.; Garczynski, B.; Horgan, B.H.N. Biotic and abiotic imprints on Mg-rich stromatolites: Lessons from lake Salda, SW Turkey. Geomicrobiol. J. 2020, 37, 401–425. [Google Scholar] [CrossRef]
- Power, I.M.; Wilson, S.A.; Thom, J.M.; Dipple, G.M.; Southam, G. Biologically induced mineralization of dypingite by cyanobacteria from an alkaline wetland near Atlin, British Columbia, Canada. Geochem. Trans. 2007, 8, 13. [Google Scholar] [CrossRef]
- White III, R.A.; Visscher, P.T.; Burns, B.P. Between a rock and a soft place: The role of viruses in lithification of modern microbial mats. Trends Microbiol. 2021, 29, 204–213. [Google Scholar] [CrossRef]
- Carreira, C.; Staal, M.; Middelboe, M.; Brussaard, C.P.D.; Wommack, K.E. Counting viruses and bacteria in photosynthetic microbial mats. Appl. Environ. Microbiol. 2015, 81, 2149–2155. [Google Scholar] [CrossRef][Green Version]
- Carreira, C.; Piel, T.; Staal, M.; Stuut, J.-B.W.; Middelboe, M.; Brussaard, C.P.D. Microscale spatial distributions of microbes and viruses in intertidal photosynthetic microbial mats. SpringerPlus 2015, 4, 239. [Google Scholar] [CrossRef]
- De Wit, R.; Gautret, P.; Bettarel, Y.; Roques, C.; Marlière, C.; Ramonda, M.; Nguyen Thanh, T.; Tran Quang, H.; Bouvier, T. Viruses occur incorporated in biogenic high-Mg calcite from hypersaline microbial mats. PLoS ONE 2015, 10, e0130552. [Google Scholar] [CrossRef] [PubMed]
- Słowakiewicz, M.; Borkowski, A.; Syczewski, M.D.; Perrota, I.D.; Owczarek, F.; Sikora, A.; Detman, A.; Perri, E.; Tucker, M.E. Newly-discovered interactions between bacteriophages and the process of calcium carbonate precipitation. Geochim. Et Cosmochim. Acta 2021, 292, 482–498. [Google Scholar] [CrossRef]
- Pacton, M.; Wacey, D.; Corinaldesi, C.; Tangherlini, M.; Kilburn, M.R.; Gorin, G.E.; Danovaro, R.; Vasconcelos, C. Viruses as new agents of organomineralization in the geological record. Nat. Commun. 2014, 5, 4298. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M.; Yamamoto, T.; Nagakubo, T.; Morinaga, K.; Obana, N.; Nomura, N.; Toyofuku, M. Phage genes induce quorum sensing signal release through membrane vesicle formation. Microbes Environ. 2022, 37, ME21067. [Google Scholar] [CrossRef]
- Devos, S.; Van Putte, W.; Vitse, J.; Van Driessche, G.; Stremersch, S.; Van Den Broek, W.; Raemdonck, K.; Braeckmans, K.; Stahlberg, H.; Kudryashev, M. Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ. Microbiol. 2017, 19, 3930–3937. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- Barbeyron, T.; Brillet-Guéguen, L.; Carré, W.; Carrière, C.; Caron, C.; Czjzek, M.; Hoebeke, M.; Michel, G. Matching the diversity of sulfated biomolecules: Creation of a classification database for sulfatases reflecting their substrate specificity. PLoS ONE 2016, 11, e0164846. [Google Scholar] [CrossRef]
- Rani, A.; Babu, S. Environmental proteomic studies: Closer step to understand bacterial biofilms. World J. Microbiol. Biotechnol. 2018, 34, 1–14. [Google Scholar] [CrossRef]
- Řezanka, T.; Kolouchová, I.; Gharwalová, L.; Palyzová, A.; Sigler, K. Lipidomic analysis: From archaea to mammals. Lipids 2018, 53, 5–25. [Google Scholar] [CrossRef]
- Amicucci, M.J.; Nandita, E.; Galermo, A.G.; Castillo, J.J.; Chen, S.; Park, D.; Smilowitz, J.T.; German, J.B.; Mills, D.A.; Lebrilla, C.B. A nonenzymatic method for cleaving polysaccharides to yield oligosaccharides for structural analysis. Nat. Commun. 2020, 11, 3963. [Google Scholar] [CrossRef] [PubMed]
- Caleb Bagley, M.; Garrard, K.P.; Muddiman, D.C. The development and application of matrix assisted laser desorption electrospray ionization: The teenage years. Mass Spectrom. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.R.; DeLaney, K.; Johnson, J.; Li, L. Mass spectrometry imaging: A review of emerging advancements and future insights. Anal. Chem. 2018, 90, 240. [Google Scholar] [CrossRef] [PubMed]
- Kostopoulos, I.; Aalvink, S.; Kovatcheva-Datchary, P.; Nijsse, B.; Bäckhed, F.; Knol, J.; de Vos, W.M.; Belzer, C. A continuous battle for host-derived glycans between a mucus specialist and a glycan generalist in vitro and in vivo. Front. Microbiol. 2021, 12, 1518. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Biewend, M.; Rana, S.; Binder, W.H. The CuAAC: Principles, homogeneous and heterogeneous catalysts, and novel developments and applications. Macromol. Rapid Commun. 2020, 41, 1900359. [Google Scholar] [CrossRef]
- Anderson, C.T.; Wallace, I.S.; Somerville, C.R. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc. Natl. Acad. Sci. USA 2012, 109, 1329–1334. [Google Scholar] [CrossRef]
- Dar, D.; Dar, N.; Cai, L.; Newman, D.K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 2021, 373, eabi4882. [Google Scholar] [CrossRef]
- Xia, C.; Fan, J.; Emanuel, G.; Hao, J.; Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA 2019, 116, 19490–19499. [Google Scholar] [CrossRef]
- Flintrop, C.M.; Rogge, A.; Miksch, S.; Thiele, S.; Waite, A.M.; Iversen, M.H. Embedding and slicing of intact in situ collected marine snow. Limnol. Oceanogr. Methods 2018, 16, 339–355. [Google Scholar] [CrossRef]
- Tanabe, Y.; Okazaki, Y.; Yoshida, M.; Matsuura, H.; Kai, A.; Shiratori, T.; Ishida, K.-I.; Nakano, S.-I.; Watanabe, M.M. A novel alphaproteobacterial ectosymbiont promotes the growth of the hydrocarbon-rich green alga Botryococcus braunii. Sci. Rep. 2015, 5, 10467. [Google Scholar] [CrossRef] [PubMed]
- Reintjes, G.; Arnosti, C.; Fuchs, B.M.; Amann, R. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 2017, 11, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Bennke, C.M.; Neu, T.R.; Fuchs, B.M.; Amann, R. Mapping glycoconjugate-mediated interactions of marine Bacteroidetes with diatoms. Syst. Appl. Microbiol. 2013, 36, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Bosak, T.; Liang, B.; Wu, T.D.; Templer, S.; Evans, A.; Vali, H.; Guerquin-Kern, J.L.; Klepac-Ceraj, V.; Sim, M.; Mui, J. Cyanobacterial diversity and activity in modern conical microbialites. Geobiology 2012, 10, 384–401. [Google Scholar] [CrossRef] [PubMed]
- Petroff, A.P.; Wu, T.-D.; Liang, B.; Mui, J.; Guerquin-Kern, J.-L.; Vali, H.; Rothman, D.H.; Bosak, T. Reaction–diffusion model of nutrient uptake in a biofilm: Theory and experiment. J. Theor. Biol. 2011, 289, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Rumpel, C.; Baumann, K.; Remusat, L.; Dignac, M.-F.; Barré, P.; Deldicque, D.; Glasser, G.; Lieberwirth, I.; Chabbi, A. Nanoscale evidence of contrasted processes for root-derived organic matter stabilization by mineral interactions depending on soil depth. Soil Biol. Biochem. 2015, 85, 82–88. [Google Scholar] [CrossRef]
- Liang, B.; Wu, T.-D.; Sun, H.-J.; Vali, H.; Guerquin-Kern, J.-L.; Wang, C.-H.; Bosak, T. Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS ONE 2014, 9, e88142. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Alvarez, D.; Hudak, J.E.; Reading, N.C.; Erturk-Hasdemir, D.; Dasgupta, S.; von Andrian, U.H.; Kasper, D.L. In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 2015, 21, 1091–1100. [Google Scholar] [CrossRef]
- Tateno, H.; Nakamura-Tsuruta, S.; Hirabayashi, J. Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology 2009, 19, 527–536. [Google Scholar] [CrossRef]
- Gowthaman, S.; Nawarathna, T.H.K.; Nayanthara, P.G.N.; Nakashima, K.; Kawasaki, S. The amendments in typical microbial induced soil stabilization by low-grade chemicals, biopolymers and other additives: A review. Build. Mater. Sustain. Ecol. Environ. 2021, 49–72. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).