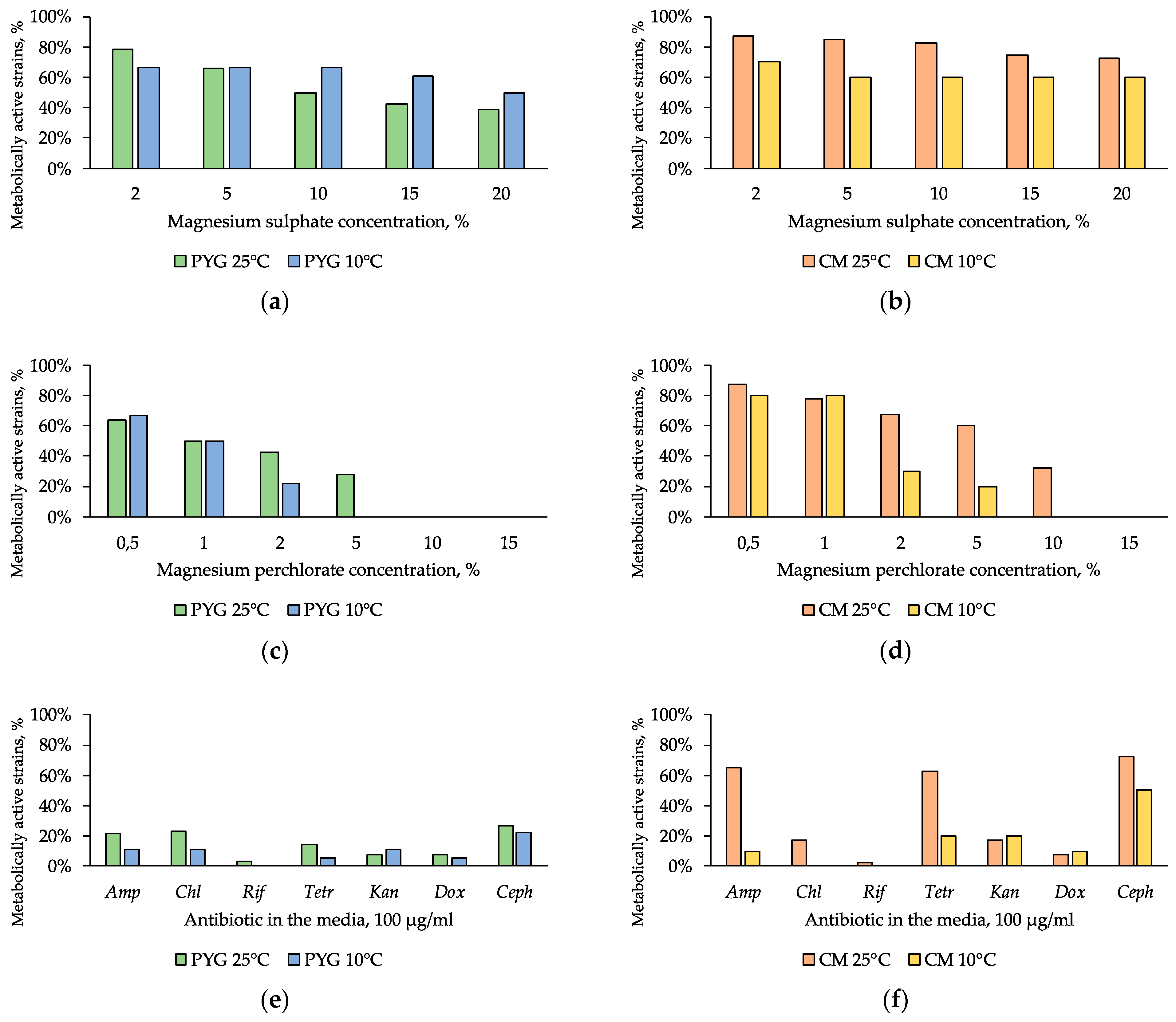Abstract
The arid Mojave Desert is one of the most significant terrestrial analogue objects for astrobiological research due to its genesis, mineralogy, and climate. However, the knowledge of culturable bacterial communities found in this extreme ecotope’s soil is yet insufficient. Therefore, our research has been aimed to fulfil this lack of knowledge and improve the understanding of functioning of edaphic bacterial communities of the Central Mojave Desert soil. We characterized aerobic heterotrophic soil bacterial communities of the central region of the Mojave Desert. A high total number of prokaryotic cells and a high proportion of culturable forms in the soil studied were observed. Prevalence of Actinobacteria, Proteobacteria, and Firmicutes was discovered. The dominance of pigmented strains in culturable communities and high proportion of thermotolerant and pH-tolerant bacteria were detected. Resistance to a number of salts, including the ones found in Martian regolith, as well as antibiotic resistance, were also estimated.
1. Introduction
One of the main tasks of modern astrobiology is studying environmental factors of different space bodies and outer space and to determine surviving and functioning limits of terrestrial life forms under extraterrestrial conditions [1,2].
It is known that potentially habitable space objects are exposed to a number of extreme abiotic factors, including high-intensity ionizing radiation, reduced pressure, contrast temperature conditions, low water availability, etc. [3,4,5]. Up to date, a large amount of data on physicochemical properties of regolith and climatic and cosmic regimes of various extraterrestrial bodies, in particular Mars, have been accumulated [6]. Based on these data, it can be stated that in today’s biosphere of the Earth there are ecosystems that possess some environmental parameters similar to those found on extraterrestrial bodies. Hot-arid and cold-arid deserts, permafrost sedimentary rocks and some arid soils are considered as terrestrial analogues of extraterrestrial regolith [7,8,9] and are used as model objects in astrobiological research.
Microorganisms of arid ecosystems are exposed to a wide spectrum of stress factors such as water and organic matter deficiency, exposure to winds and high-intensity ultraviolet (UV) radiation, and significant temperature fluctuations. However, these areas are not lifeless and are characterized by significant richness and diversity of prokaryotes [10,11]. Many astrobiological studies consider the Atacama Desert [12,13,14], the Sonora Desert [15], and the Dry Valleys of Antarctica [16] to be the main objects of research. However, it is important to investigate different ecosystems that are exposed to extreme environmental factors [17]. The Mojave Desert is one of such analogue ecosystems. It is characterized by relief and mineralogy that are close to those found on Mars [18,19,20]. There are several reasons as to why edaphic (i.e., associated with soil) bacterial communities of hot deserts should be studied, such as the lack of culturable communities’ research and the possible biotechnological potential of bacterial strains that inhabit the ecosystems and its highly valuable and significant properties for further research [21].
The Mojave Desert is located in the southwest of the United States and covers an area of approximately 150,000 km2. It is characterized by altitudes of 600–1500 m above sea level and arid-semiarid climate with an average annual precipitation of less than 300 mm (the average annual precipitation is 137 mm/year). Temperatures range from −10 to 50 °C; frequent winds are also observed in this area [22,23]. The average annual temperature is 20 ± 0.5 °C, annual heat-moisture index (AHM) is 230 ± 19 [24].
Ten thousand years ago this territory was characterized by a humid climate and well-formed soil coverage, but the aridization of weather conditions and desertification of the region have led this area to its present state [23]. The region is characterized by volcanogenic geology and topography, a high degree of weathering of surface sedimentary rocks [25] and extremely poor vegetation [19]. The total organic matter content in the soils of this desert is very low (it ranges from 145 to 260 µg of C per gram of soil). Carbon to nitrogen ratio is about 9.5 [26].
All the aforementioned properties of the Mojave Desert make it a suitable terrestrial analogue to Mars, particularly because of the climate changes that had happened here and of the ongoing geological and climatic processes [24]. The soil of this desert has a mineralogical composition close to that of the Martian regolith [27]. As well as that, it also has a similar composition of mineral salts [8,18,28]. Worth noticing is the fact that in the past the territory of modern Mojave Desert had been coated by mature soil coverage, which could contain diverse soil microbial communities. However, as time has been progressing, this region was subjected to long-lasting drought periods and microbial communities have been forced to adapt and alter their metabolism under conditions of moisture and nutrients deficiency and high-stress load. Supposedly the same happened to the Martian regolith [19,28]. The soils of this desert have already been used in model astrobiological studies and in The Mojave Mars simulant (MMS) in particular as a regolith model of Mars [29]. Bacterial isolates from the soils of this desert served as model organisms for studying lithopanspermia processes [15].
Earlier studies revealed a large number and variety of phototrophic organisms in the soil of Mojave confined to hypolith communities and able to carry out photosynthesis under aggressive environmental conditions in situ (i.e., in natural soil conditions) and in vitro (i.e., in laboratory conditions) [8,30].
Presence and metabolic activity of phototrophic and chemotrophic prokaryotes have also been confirmed, as well as presence of nitrogen-fixing bacteria and ability of those organisms to produce primary organic matter (references herein and [10,21]). Thus, it becomes obvious that along with the autotrophic community, there must be a heterotrophic community in the soil of this desert.
In general, previous studies have discovered a similar structure of edaphic bacterial communities at the phylum level in the soils of cold and hot deserts: among aerobic heterotrophic bacteria, representatives of the phyla Actinobacteria, Proteobacteria, Acidobacteria, and Bacteroidetes are the most common [10].
In the surface soil of The Mojave Desert, 48 different taxonomic units of bacteria were found by molecular cloning [31].
Biological soil crusts (BSC) confined to the surface layers of soil are common in the Mojave Desert. The next generation sequencing of 16S rRNA genes from the soil under BSC revealed the following phylogenetic structure of the bacterial community: Proteobacteria (23 ± 5%), Actinobacteria (20 ± 5%), and Chloroflexi (18 ± 3%), representatives of the genus Rubrobacter were the most dominant among actinobacteria. Acidobacteria (10 ± 2%), Bacteroidetes (6.5 ± 2.0%), and Cyanobacteria (11 ± 10%); Gemmatimonadetes, Armatimonadetes, Planctomycetes, Nitrospirae, Chlorobi, and Verrucomicrobia (<5% altogether) representatives have also been identified [32].
Proteobacteria (~45% operational taxonomic units (OTU)), Actinobacteria (~12% OTU), and Firmicutes (~10% OTU) phyla have been found as the most abundant ones by metagenomic analysis of the soils of the desert. The authors have concluded that there was a high number and high taxonomic diversity of bacteria inhabiting the arid soils of the Mojave Desert. They have also concluded that the desert soils were rich with unidentified bacteria species, supporting this with molecular-genetical data obtained [21].
Based on the high-throughput DNA sequencing and shotgun-metagenomic study of the Mojave Desert soils performed by the several research groups, a high relative abundance Proteobacteria and Cyanobacteria phyla and presence of representatives of the phyla Actinobacteria, Bacteroidetes, Verrucomicrobia, Acidobacteria, Firmicutes, and Crenarcheaota were also detected [24,33,34]. A high abundance of genes associated with spore formation processes, encoding stress proteins and amino acid metabolism, associated with osmoregulation was also discovered. Based on these results, the authors concluded that there was a significant functional diversity of desert bacterial communities [33].
Using the methods of native fluorescence and fatty acid analysis of phospholipids, the number of bacterial cells was found to be 107 bacteria per gram of soil [35,36]. Studies of culturable bacteria of this desert are less common, compared to culture-independent methods. A high number of cultured bacteria was shown up to 106 colony-forming units per gram (CFU/g) [37].
A study of the number and taxonomic composition of cultured actinomycetes in the soils of this desert revealed the dominance of Actinoplanes, Pilimelia, and Streptomyces genera representatives. The authors noted a high diversity of isolated strains, and the fatty acid methyl ester analysis of isolates indicated a significant proportion of previously undescribed actinomycete species. Extreme effects of external factors were proposed to be the driving force of speciation in desert soils, which leads to the morpho-physiological adaptations of prokaryotes inhibiting them [38].
Community level physiological profile analysis of the microbial complexes (using BIOLOG 96-well Eco-Microplates assimilation test) of soils confined to plants revealed the absence of correlation between the structure and the diversity of bacteria in communities under plant coverage and in soils with no vegetation. A positive correlation between the number of substrates consumed by the bacterial community and the presence and diversity of higher plants growing on that soil was proved. However, bacterial communities confined to areas of the desert lacking vegetation showed active assimilation of amino acids, amines, amides, and various polymers. In addition, representatives of Sinorhizobium, Bacillus, and Sphingomonas genera were isolated and identified in culture grown on selective medium for nitrogen-fixing bacteria [39].
In dry conditions, microbial communities are exposed to different extreme conditions, such as temperature and pH fluctuations, soil moisture shortage, high level of ultraviolet radiation. All of them have a significant impact on microbial communities [17].
Generally, little is known about bacterial communities of arid soils, and previous studies revealed significant taxonomic and functional differences between the bacterial communities of extreme soils and those of temperate zones [24,40]. A high diversity and a high number of cells in bacterial communities of the Mojave Desert soils in situ (i.e., in natural conditions), predominated by Proteobacteria and Actinobacteria phyla were found. The abundance and diversity of photo- and chemotrophic microorganisms were described predominantly by culture-independent methods. However, heterotrophic community studies are surprisingly rare: only individual groups of culturable bacteria, such as, for example, nitrogen-fixing bacteria and actinomycetes, have been investigated previously. Still, little is known about the bacterial communities of arid soil even though bacterial stress resistance research plays an important role when it comes to extraterrestrial analogue ecosystems in astrobiological context.
In this paper, we studied the aerobic heterotrophic bacterial community and the physiological characteristics of cultured bacteria, isolated from the surface soil sample collected in the central part of the Mojave Desert.
2. Materials and Methods
2.1. Soil Sampling
A single soil sample was collected in the central part of the Mojave Desert in the summer of 2015 in a flat area devoid of vegetation (34°55’39.0” N 115°45’33.7” W). The surface soil was taken from 0–3 cm depth with no signs of vegetation and biological soil crust and put into a sterile polypropylene container. The sample was transported and stored at room temperature in an air-dry (natural) condition. Microbiological analyses were performed within one month after the sample collection.
The central region where the sample was taken from is characterized by the most typical climate of this desert: the temperature during the year varies from −10 to 50 °C, the average annual rainfall varies from 30 to 300 mm (arid–semi-arid climate type) [22]. It has been shown that the soil of this desert can be heated up to a temperature of 67 °C [29].
The Mojave tectonic block is a part of the Basin and Range tectonic province, where many asymmetrical basins have been formed during regional extension processes. The central Mojave region was formed during localized extension which occurred between 24 and 18.5 million years ago and produced 40–60 km of extended terrain space [41]. The central Mojave Desert is characterized by a variety of geologic surfaces that includes mountain ranges of diverse rock types, playas, ancient lake deposits, lava flows, sand-dune fields, alluvial fans, and riverbeds. The region of sample studied is underlain by middle Tertiary volcanic rocks, ranging mainly from andesitic to rhyolitic compositions [42].
The sample pH was 7.2. Literature data on the soils of this region indicate the content of organic matter in the range of 0.04–0.1%, total nitrogen content varies from 0.03% to 0.09% [22]. Soil sample texture is found to be sand. Dominance of silica and iron oxide-containing minerals was visually detected and confirmed by polarized light microscopy.
2.2. Epifluorescent Microscopy
The total number of prokaryotes in the samples was determined with epifluorescence microscopy (EFM) with acridine orange dye. Cell desorption was carried out by vortexing using Heidolph Multi Reax vortex for 30 min at 2000 rpm. The preparations were produced in six replicates, fixed by heating, then stained with water solution of acridine orange (1:10,000) for 3 min, washed in still distilled water for 20 min, dried at room temperature, and viewed under the Primo Star (Zeiss, Germany) microscope with fluorescence epi-illumination system (AmScope, Irvine, CA, USA) at 700× magnification for 20 fields of vision for each replication. Cells with green fluorescence were counted. The water used for the dilution preparation was simultaneously examined as control.
The prokaryotic cells number was calculated using the equation N = (S1 × a × n)/(V × S2 × c), where N is the number of cells per gram of soil; S1 is the area of the preparation (μm2); a is the number of cells in the field of view; n is the dilution index; V is the volume of the soil suspension drop placed on the glass (mL); S2 is the field of view of the microscope (μm2); and c is the sample weight (g) [43].
2.3. Bacteria Culturing and Isolation
Peptone Yeast Glucose (PYG) medium rich in various carbon sources and nutrients (glucose—1 g/L, peptone—2 g/L, yeast extract—1 g/L, tryptone—1 g/L, agar—20 g/L) and modified Czapek (CM) medium (glucose—2 g/L, sucrose—2 g/L, starch—2 g/L, NaNO3—2 g/L, KH2PO4—1 g/L, MgSO4—0.5 g/L KCl—0.2 g/L, agar—20 g/L) [44] were used for bacteria culturing and isolation. Dilutions of the soil sample in sterile 0.01 M phosphate-saline buffer solution (pH 7.4, 0.137 M NaCl, 0.0027M KCl) in tenfold dilution step were plated on the solid nutrient media in three replicates [45].
For the more complete characterization of the cultured bacterial community, culturing was performed at temperatures of 10, 25, and 50 °C (characteristic optima of psychrophilic, mesophilic, and thermophilic bacteria [17]) until the formation of visible macrocolonies was completed (typically for 14 days at 25 and 50 °C and 30 days at 10 °C). Colonies’ morphotypes were distinguished by colour and morphology of the colonies as well as cell morphology. Isolated bacterial strains (Table A1) were deposited to the Astrobiological Collection of Microorganisms of the National Depository Bank of Live Systems “Noah’s Ark” (Available at [46]).
2.4. Amplification and Sequencing of 16S rRNA Genes
For amplification of the 16S rRNA gene, biomass of pure bacterial culture that has been cultured for 2–3 days was subjected to boiling in Tris-EDTA buffer (pH 8.0) which contained 5% Triton X-100, and homogenization by mechanical destruction with sterile glass beads (250–300 µm in diameter) using a Homogenizer Minilys (Bertin Instruments, Montigny-le-Bretonneux, France) at 5000 rpm for 30 s. The resulting homogenate was centrifuged and the resulting supernatant was used as a DNA template for PCR [47].
For amplification of the gene of interest, the primer systems 27 f + Un1492 r [48,49], 63 f + 1387 r [47], 341 f + 805 r [50], and 27 f + 537 r [48,50] were used in order of priority. Amplification process was performed with cell lysate, prepared as described above. If the procedure failed to obtain the PCR product by the first primer system, the next primer system with the same DNA matrix was used and so on until a product that can be used for further sequencing was acquired [45].
The PCR products were purified and sequenced by the Research and Production Company “Evrogen” (Moscow, Russia) using the 1100 r [50], 805 r, or 537 r [51] primers (Table A2). The editing of the nucleotide sequences was carried out using Chromas Lite 2.01 [52]. For alignment, comparison and identification of nucleotide sequences, the Clustal Omega [53] and the BLAST algorithm from the GenBank database [54] were used. The sequences were deposited in GenBank under the accession numbers MK403768—MK403884.
The phylogenetic trees were constructed using MEGA 7 software [55]. Sequences were then aligned using the Clustal Omega online alignment tool [53]. The type material sequences from GenBank were predominantly used for phylogenetic trees construction. Strain identification was based upon phylogenetic analysis and similarity of the sequences with GenBank data.
2.5. Physiological Assays
The resistance spectra of all isolated cultures (121 strains) to various factor influences were obtained by culturing isolates on the same liquid medium that was used to isolate each particular strain from a natural sample; for each of the investigated factors the experiments were conducted in three replicates. To determine resistance to temperature, cultures were incubated in thermostats at temperatures of 2, 4, 10, 25, 37, 45, and 50 °C. Phosphate (KH2PO4 + H3PO4 final concentration of 100 mM, pH 2–6) and tris-alkaline (Tris + HCl + NaOH final concentration of 100 mM, pH 7–12) buffer systems were used to determine resistance to various pH levels. Bacterial strains’ resistance to presence of salts was determined by aseptic preparation and subsequent dilution of 20% (w/v) stock solutions of NaCl, KCl, NaHCO3, MgSO4 to final concentrations of 2, 5, 10, 15, and 20%; the series of magnesium perchlorate concentrations were prepared similarly by diluting a 20% (w/v) sterile stock solution to final concentrations of 0.5, 1, 2, 5, 10, and 15%. Antibiotic resistance was determined on nutrient media containing ampicillin (Amp), cephalexin (Ceph), chloramphenicol (Chl), tetracycline (Tet), doxycycline (Dox), kanamycin (Kan), or rifampicin (Rif) (Belmed, Russia) with a final concentration of 100 µg/mL. All tests (except for resistance to temperature) were carried out at a temperature of 25 °C [44]. The growth was registered on the tenth day after inoculation for the cultures cultivated at temperatures of 10, 25, 37, 45, and 50 °C; for cultures growing at temperatures of 2 and 4 °C the incubation period was 60 and 30 days, respectively. Growth registering was performed using a Sunrise (Tecan, Menendov, Switzerland) plate photometer at a wavelength λ = 620 nm: The plates were measured immediately after inoculation and after incubation period. Detailed protocol of physiological assays is described in [44].
2.6. Multisubstrate Testing
To assess the potential metabolic activity of a microbial community, multisubstrate testing (MST) of the soil microbial community was performed [56,57]. The testing was carried out with 47 substrates including sugars, alcohols, amino acids, salts of organic acids, and polymers (all substrates are listed in [43], all tests were performed in two replicates). The soil sample was diluted (1:100) in sterile phosphate saline buffer (pH 7.4) and vortexed for 15 min at 2500 rpm using Heidolph Multi Reax vortex. The mineral particles were precipitated by centrifugation (2000 rpm, 2 min). The hydrogenase activity indicator (triphenyltetrazolium bromide) was added to the supernatant, and after mixing 200 μL aliquot was added to each well of a 96-well plate containing a set of 47 test substrates in two replicates. The plates were incubated at +28 °C for 96 h. After the incubation, the optical density of the solutions was measured photometrically at wavelength λ = 510 nm using a Sunrise (Tecan, Menendov, Switzerland) plate photometer.
2.7. Data Analysis
Statistical data processing was performed using the Microsoft Office Excel 2011 software package (Redmond, Washington, D.C., USA). For data on cell numbers, confidence intervals of the mean were calculated using Student’s test at p < 0.05. When determining the physiological spectra, the results obtained in three replicates was considered a positive reaction. Indices of similarity and biodiversity of microbial communities were calculated according to Chernov and Lysak [58] at genera level of taxonomic affiliation.
3. Results
3.1. Cell Numbers
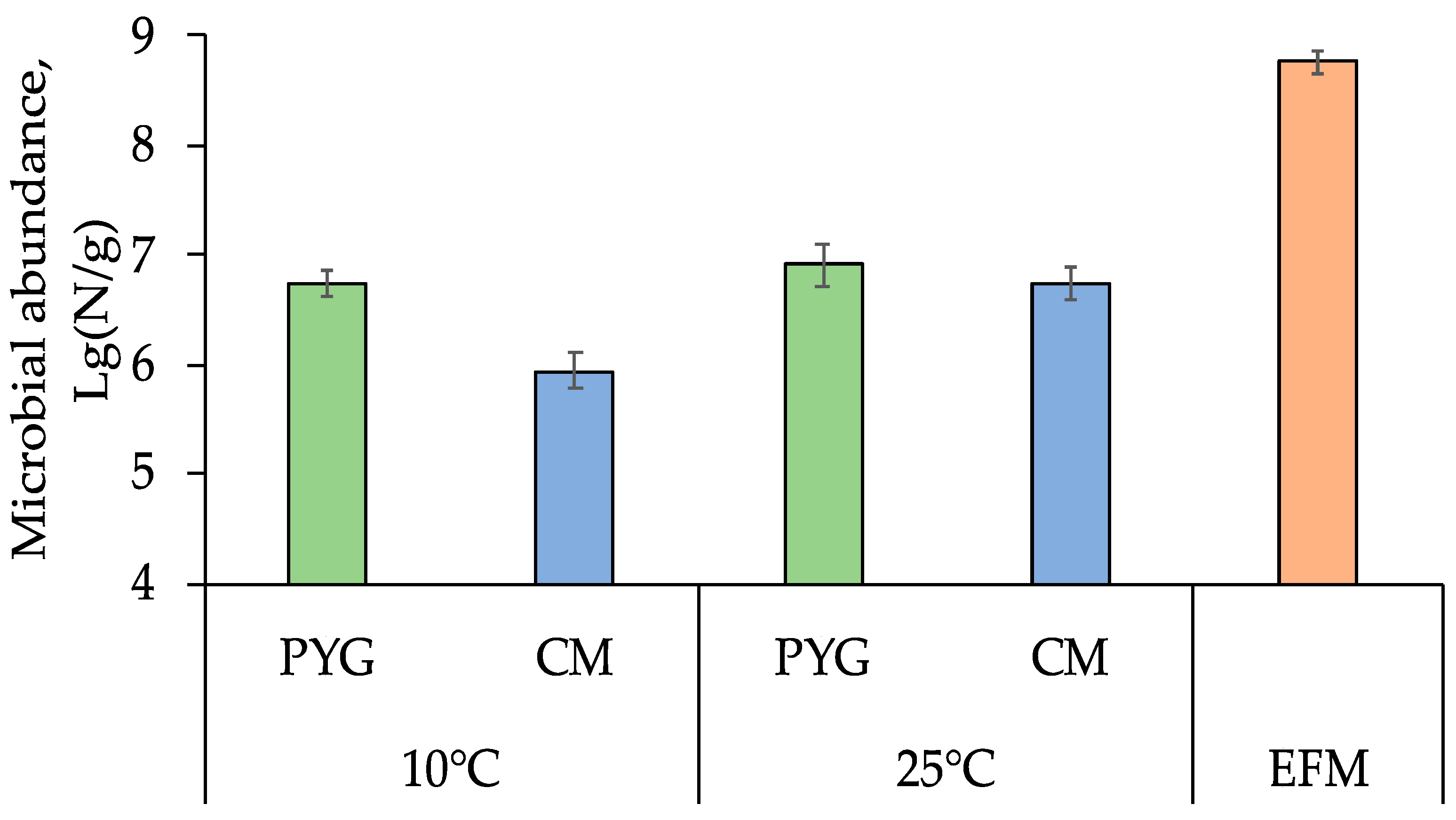
The total number of prokaryotic cells in the soil sample, obtained by epifluorescence microscopy method, was (5.8 ± 1.7) × 108 cells per gram of soil. Cultured bacteria on PYG medium were detected in the amount of (8.0 ± 0.5) × 106 colony-forming units (CFU) per gram and (5.5 ± 0.2) × 106 CFU/g at 25 °C and 10 °C, respectively. On the CM medium (8.8 ± 0.4) × 105 and (5.5 ± 0.2) × 105 CFU/g were found at 25 °C and 10 °C, respectively (Figure 1). Colony-forming units were not detected after incubation at 50 °C.
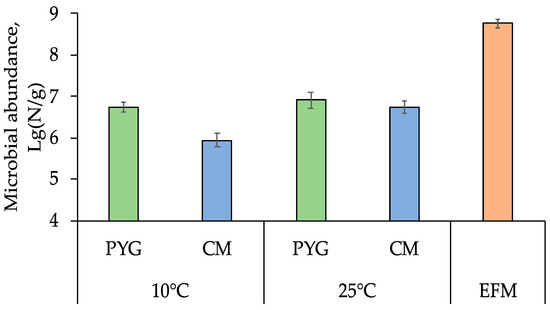
Figure 1.
Total prokaryotic cells count obtained by epifluorescent microscopy (EFM) and culturable bacteria count on peptone-yeast-glucose (PYG) and Czapek modified (CM) media in the examined soil sample. Error bars are within the standard deviation, p < 0.05.
3.2. Culturable Communities’ Structure
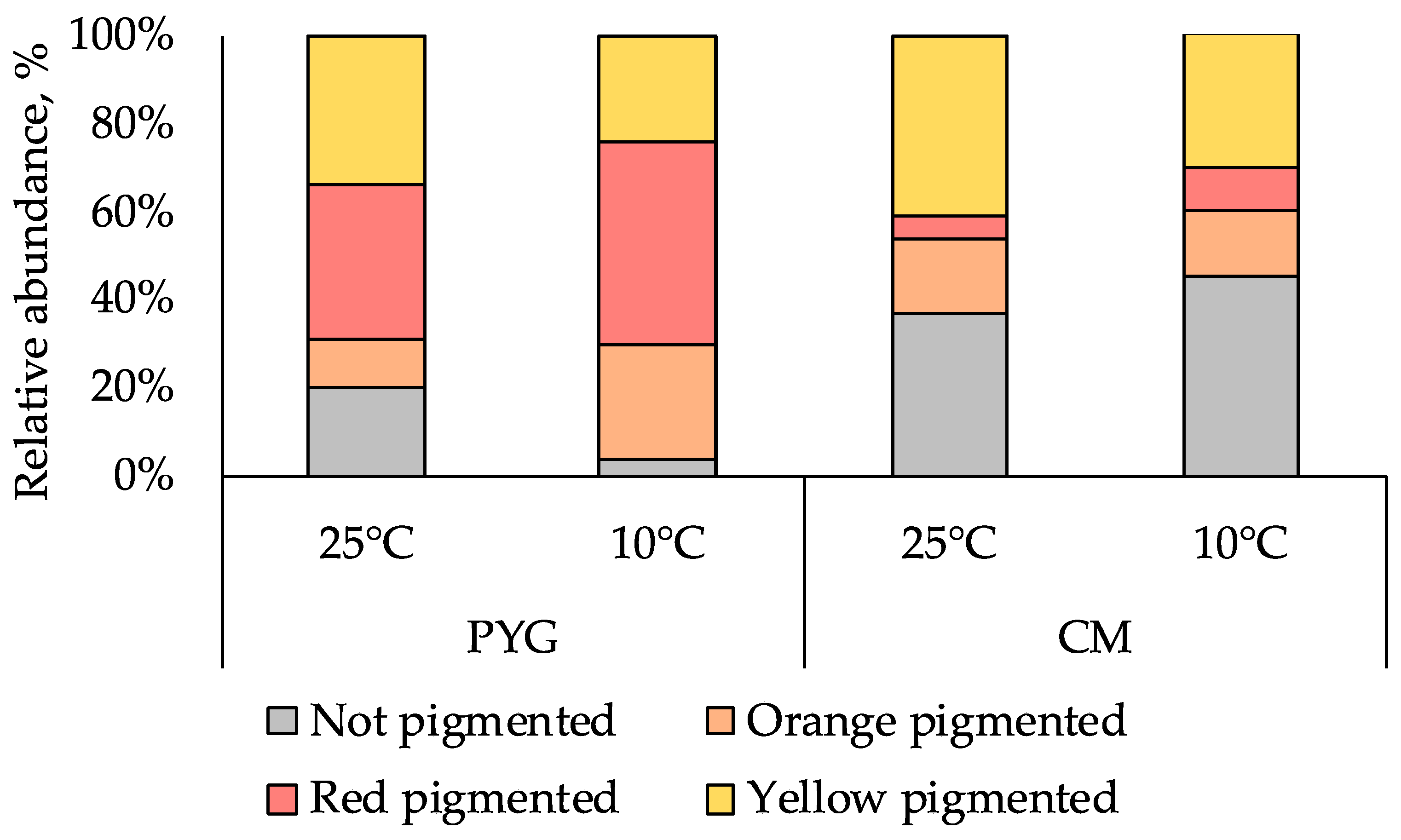
Pigmented forms of microorganisms dominated in all the cultured communities isolated from the sample. In the PYG medium plated communities, which were characterized by a higher cell counts, red-pigmented colonies were predominant and yellow-pigmented colonies were dominant on CM medium (Figure 2). Maximum diversity of colony morphotypes was observed on PYG medium: 61 morphotypes were described in the course of culturing at mesophilic optimum; on CM medium 28 morphotypes were isolated under similar temperature conditions. At low temperature (10 °C), diversity decreased: 22 and 10 colony morphotypes were isolated on PYG and CM media, respectively (Figure S1). In total 121 strains of aerobic heterotrophic bacteria were isolated from the investigated soil sample and subjected to further analysis.
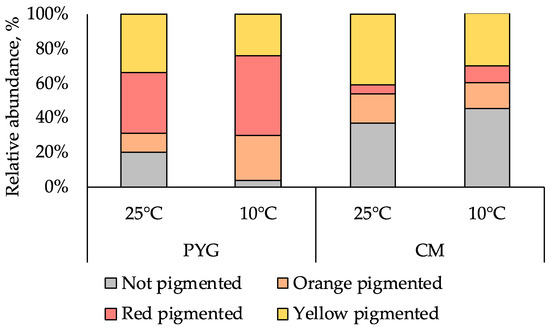
Figure 2.
Proportion of pigmented colonies in the communities studied.
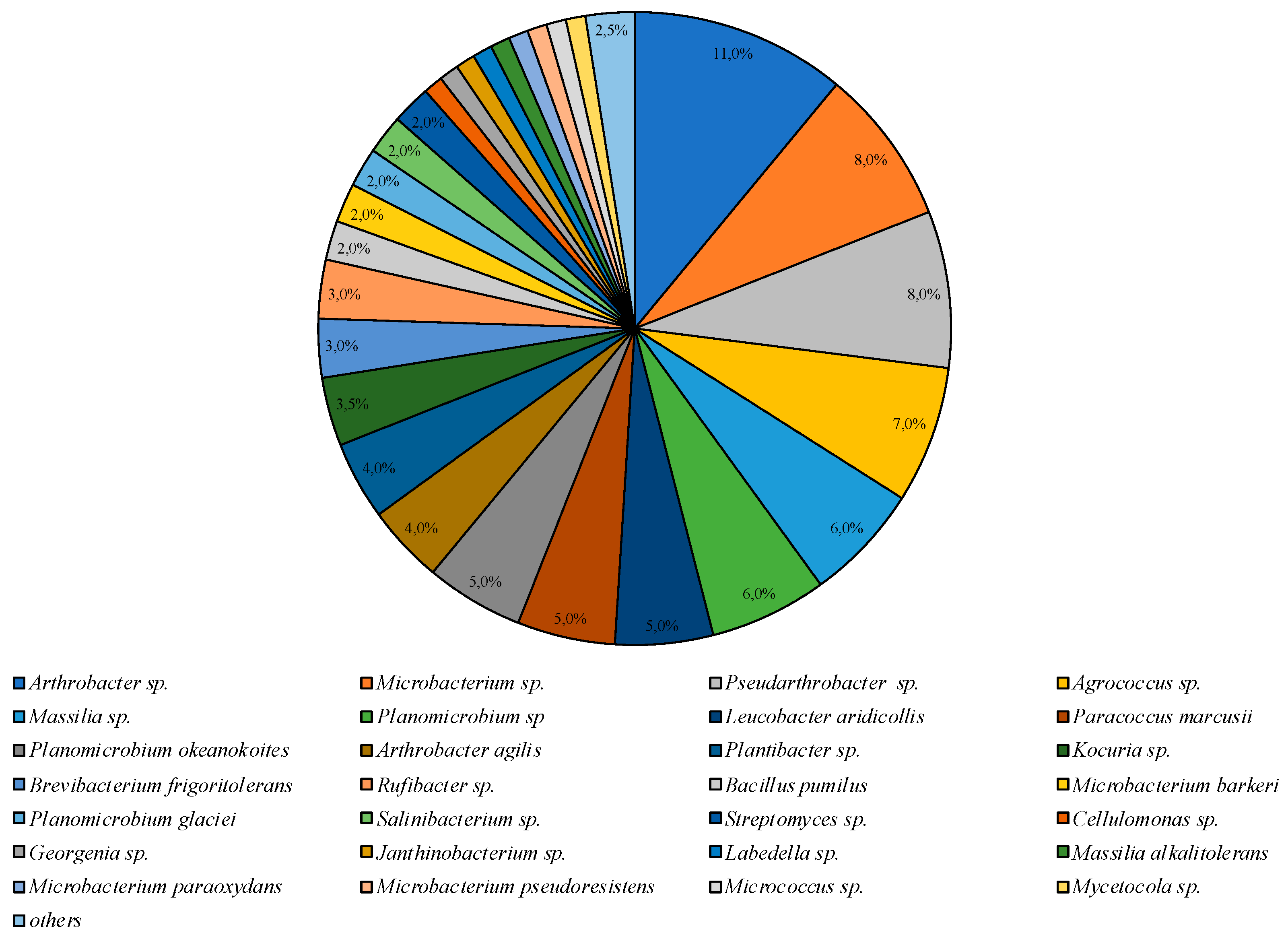
The greatest taxonomic diversity of cultured bacteria was recorded in the microbial community isolated on PYG medium under mesophilic conditions (25 °C). Representatives of 24 genera of bacteria were identified: Agrococcus, Arthrobacter, Bacillus, Brachybacterium, Brevibacterium, Cellulomonas, Georgenia, Janthinobacterium, Kocuria, Labedella, Leucobacter, Massilia, Microbacterium, Micrococcus, Mycetocola, Paracoccus, Planomicrobium, Pseudarthrobacter, Rufibacter, Salinibacterium, and Streptomyces. Bacteria of the species Arthrobacter sp. were dominant and the subdominant positions were occupied by Microbacterium sp., Pseudoarthrobacter sp., and Agrococcus sp. representatives (Figure 3).
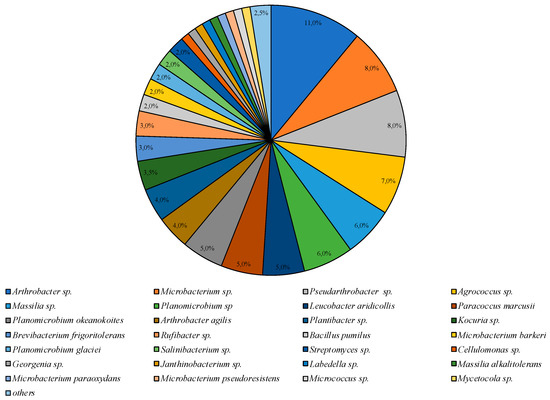
Figure 3.
Structure of the culturable bacterial community isolated on PYG medium at 25 °C. Hidden percentage corresponds to 1%; the “others” group includes Bacillus sp., Brachybacterium sp., Massilia varians, Microbacterium aurantiacum, and Pseudarthrobacter oxydans represented by 0.5% each.
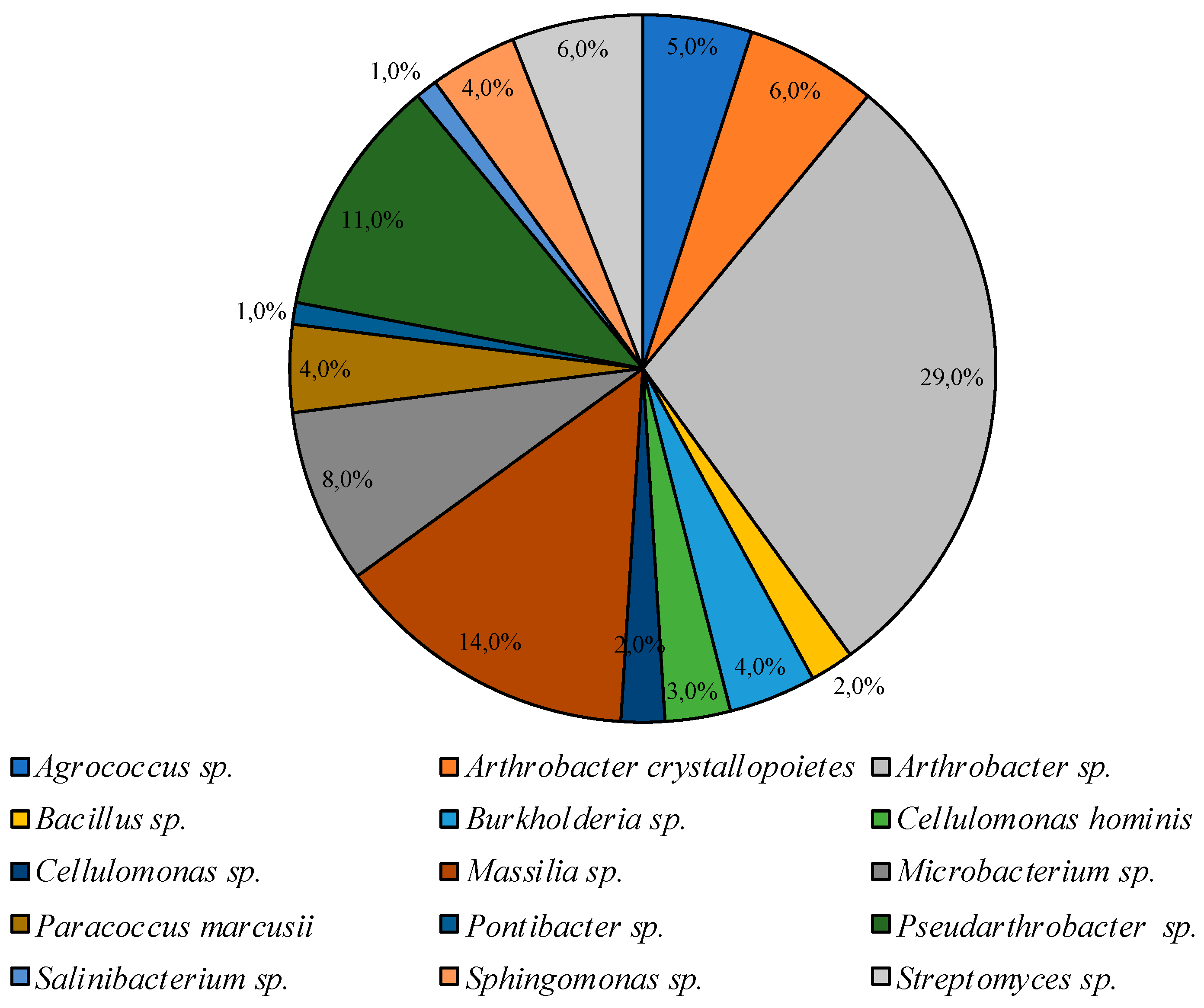
Under similar conditions on CM medium, representatives of the genera Agrococcus Arthrobacter, Bacillus, Burkholderia, Cellulomonas, Massilia, Microbacterium, Paracoccus, Pontibacter, Pseudoarthrobacter, Salinibacterium, Sphingomonas, and Streptomyces were identified (listed in descending order of representation degree in the community, Figure 4).
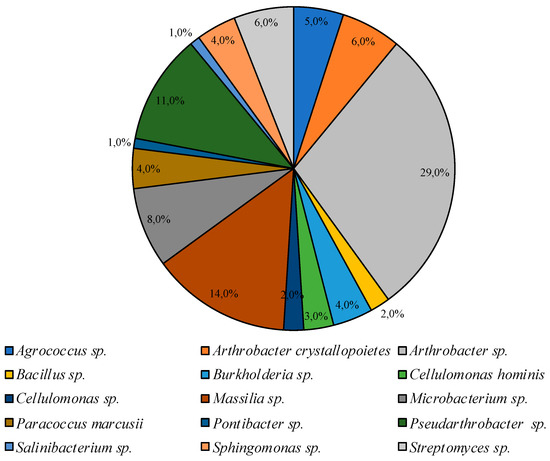
Figure 4.
Structure of the culturable bacterial community isolated on CM medium at 25 °C.
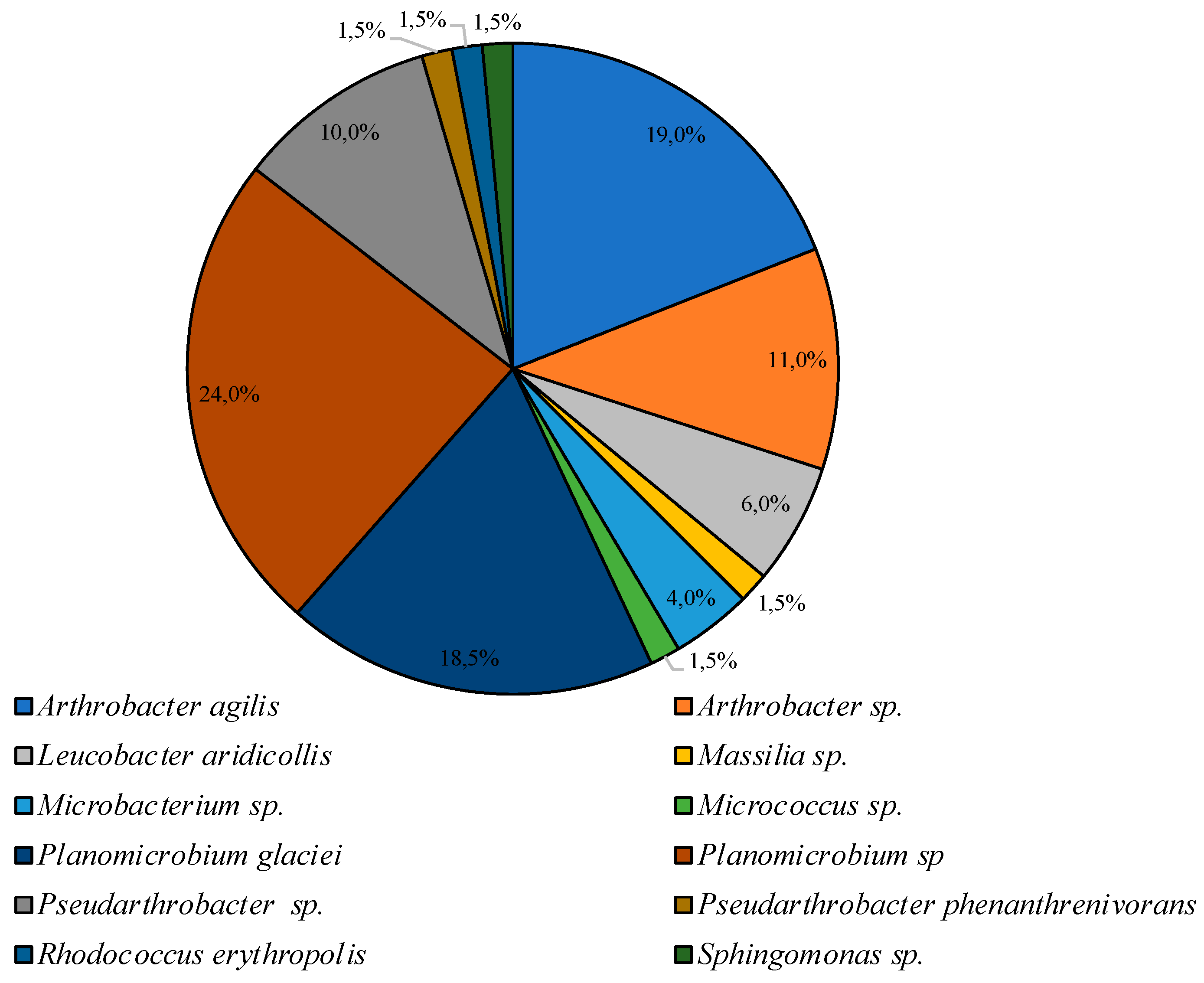
Smaller diversity was observed in the community incubated under low temperature conditions. Bacteria of the genus Planomicrobium were dominant on PYG medium, subdominant positions were represented by species Arthrobacter agilis, and Planomicrobium glaciei, and representatives of Arthrobacter, Pseudarthrobacter, Leucobacter, Microbacterium, Massilia, Micrococcus, Pseudarthrobacter, Rhodococcus, and Sphingomonas genera were minor components of the culturable bacterial community (Figure 5).
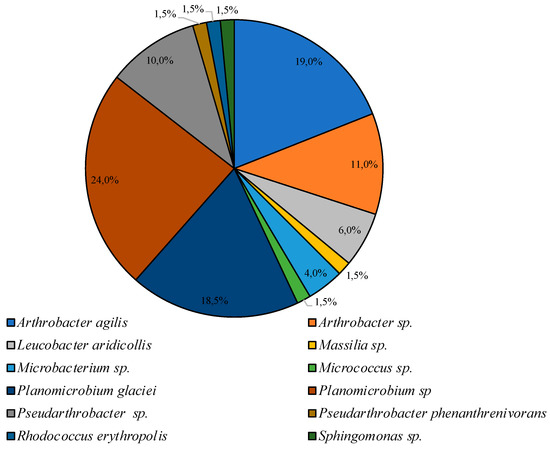
Figure 5.
Structure of the culturable bacterial community isolated on PYG medium at 10 °C.
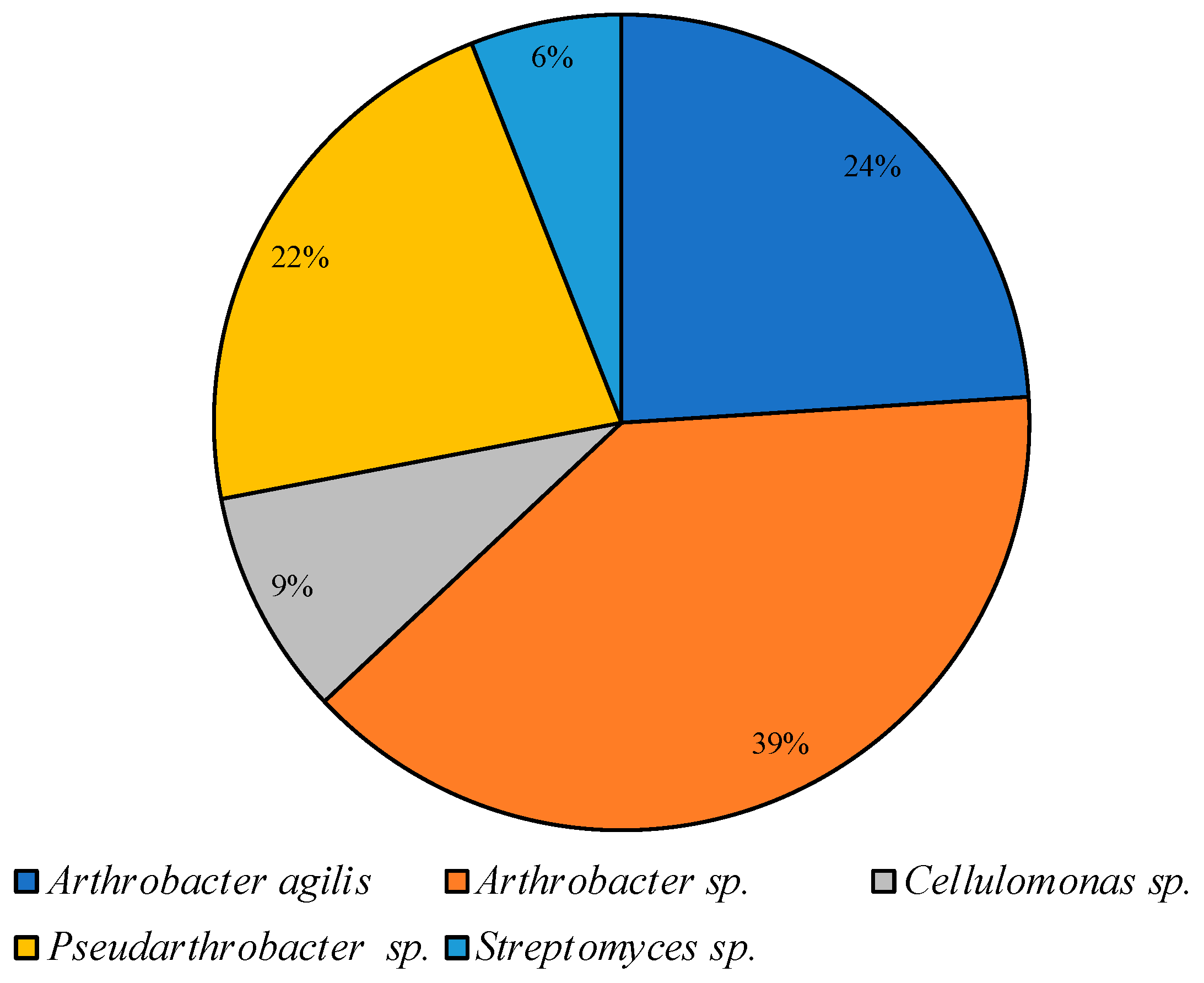
On CM medium at low temperature, representatives of the Arthrobacter and Pseudarthrobacter genera accounted for 85% of all colonies cultured under these conditions; representatives of Cellulomonas and Streptomyces were identified as minor components (Figure 6).
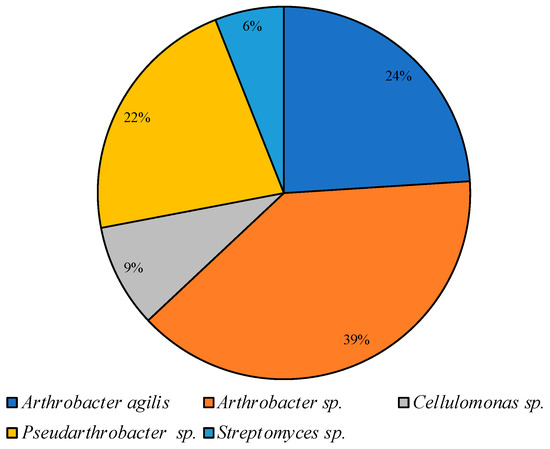
Figure 6.
Structure of the culturable bacterial community isolated on CM medium at 10 °C.
The coefficients of biological diversity, evenness, and similarity of bacterial communities, isolated from the examined site, are presented in Table 1 and Table 2. The summarized communities’ diversity data obtained are presented in Table A3. The phylogenetic trees of isolated strains are presented in Supplementary materials Figures S2–S4.

Table 1.
Ecological indices characterised of cultured communities.

Table 2.
Ecological indices of cultured communities’ similarity.
3.3. Multisubstrate Testing
Multisubstrate testing revealed a wide range of substrates suitable for development of the investigated microbial community. The most active substrate assimilation was observed on simple sugars, easily accessible polysaccharides (pullulan, starch, sucrose), lactate, Tween 80, and some amino acids (alanine, glutamine, and proline) (Table 3).

Table 3.
Substrate assimilation obtained by multisubstrate testing.
3.4. Physiological Assays
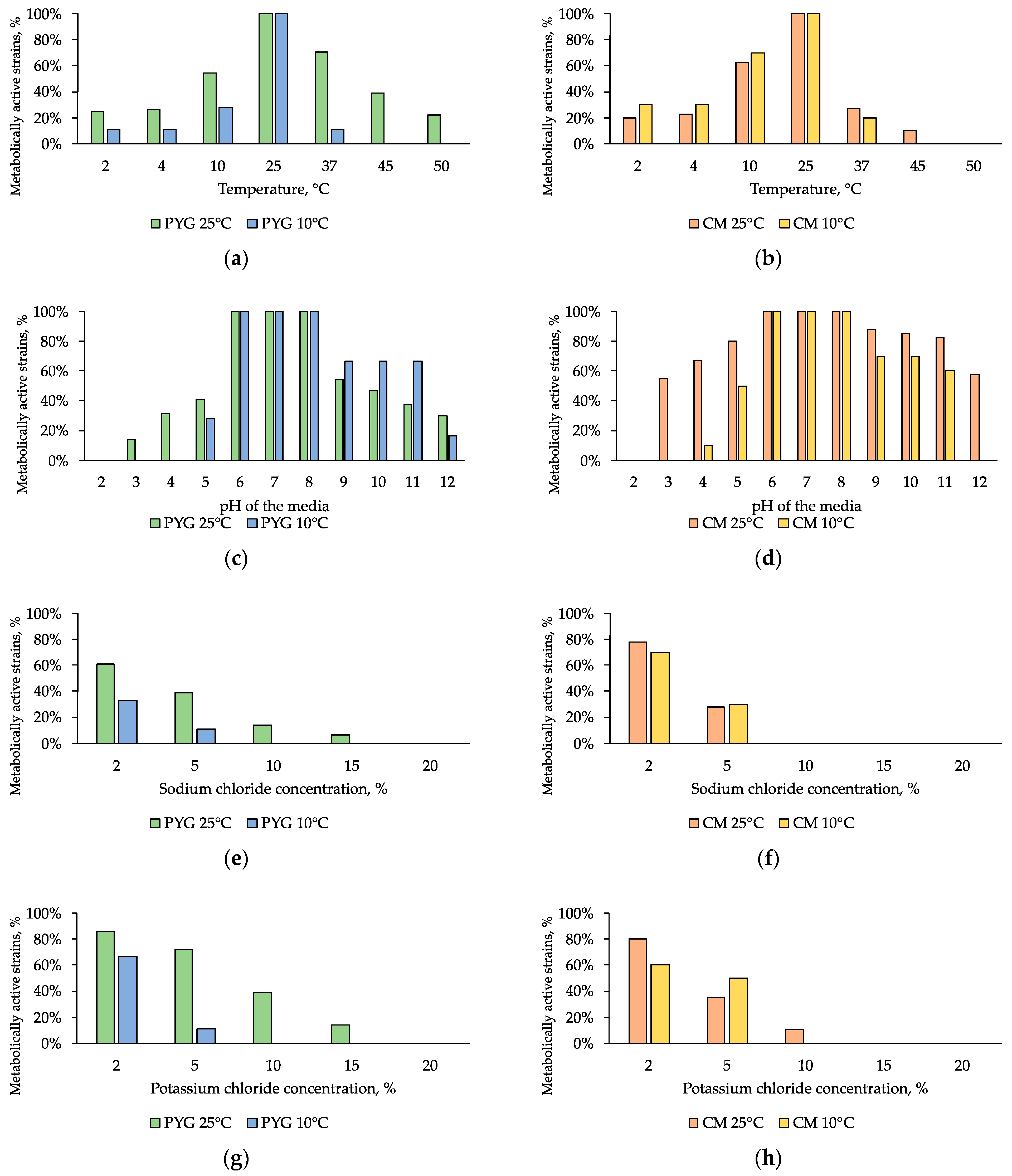
Physiological assays were performed for all isolated 121 strains. Strains that were able to metabolize at low temperatures (up to 2 °C) were found to be more common than those that were able to grow at high temperatures. Thermotolerant strains that were able to metabolize at a temperature of 45 °C were found in communities isolated on both media at 25 °C. These strains made up 39% and 10% of the total number of strains in communities cultured on PYG and CM media, respectively. The strains capable to grow at 50 °C were only found in the community isolated on a rich medium at 25°C, and made up 22% of the total number of isolates under these conditions (Figure 7a,b). Twenty percent of the strains isolated on PYG at 25 °C were able to metabolize at 2 °C and the same percentage was able to grow at 50 °C. At the same time, less than 20% of the community isolated on CM medium under similar conditions were metabolicaly active at 2 °C and only 10% of the community were able to grow at 45 °C. Higher levels of resistance to low temperatures were detected for strains isolated on CM medium at 10 °C. Amongst all investigated bacteria 10 strains were able to metabolize in extremely wide temperature range (2–4 – 45–50 °C). Detailed characteristics of strain resistance are presented in Table S1.
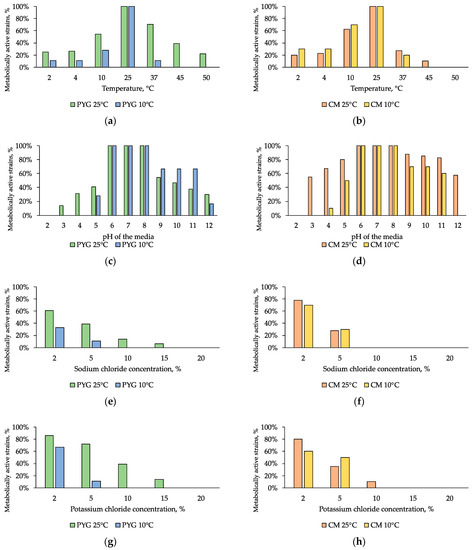
Figure 7.
Isolated strains resistance to: (a,b)—cultivation temperature; (c,d)—pH of the media; (e,f)—sodium chloride, (g,h)—potassium chloride. Left column: strains isolated on PYG medium; right column—strains isolated on CM medium.
All of the isolated strains were able to metabolize in the pH range of 6–8. The greatest resistance to pH of the medium was observed in the community isolated on CM medium at 25 °C: more than a half of the isolates were able to metabolize in a medium with pH of 3 and the same percentage was observed for pH of 12. Among communities isolated at 10 °C, the community isolated on CM medium (Figure 7c,d) was also characterized by a higher (by 20% on average) proportion of pH-tolerant strains. The pH range of strains’ metabolic activity was shifted to the alkaline region and ranged from 3–12 pH units. Among all investigated bacteria 44 strains were able to metabolize in extremely wide pH range (3–4 – 12 units). Detailed characteristics of strains’ resistance are presented in Table S1.
Tests on media with addition of sodium chloride or potassium chloride revealed a moderate halotolerance of all the studied communities. Less than 50% of the strains retained metabolic activity in the presence of 5% NaCl. At the same time, 14% of the community isolated on PYG medium at 25 °C were capable to grow in the presence of 10% sodium chloride and 6% of the same community were able to metabolize on media containing 15% sodium chloride. In addition, almost the same level of resistance was revealed for the communities isolated on CM medium at 10 °C and 25 °C (Figure 7e,f).
Potassium chloride had a smaller inhibitory effect (on average two times smaller) on the examined communities: 14% of the strains isolated on PYG at 25 °C metabolized in a medium containing 15% of this salt; similarly to sodium chloride resistance, 80% of the strains isolated on CM at 25 °C were resistant to the presence of 2% KCl, and 10% were resistant in 10% solution (Figure 7g,h).
The presence of magnesium sulphate in the medium had a low inhibitory effect on all of the studied communities: 39% and 50% of the strains isolated on PYG medium at 25 °C and 10 °C respectively retained metabolic activity in presence of 20% magnesium sulphate. Seventy-three percent and 60% of the strains isolated on CM medium at 25 °C and 10 °C retained metabolic activity in the same conditions (Figure 8a,b).
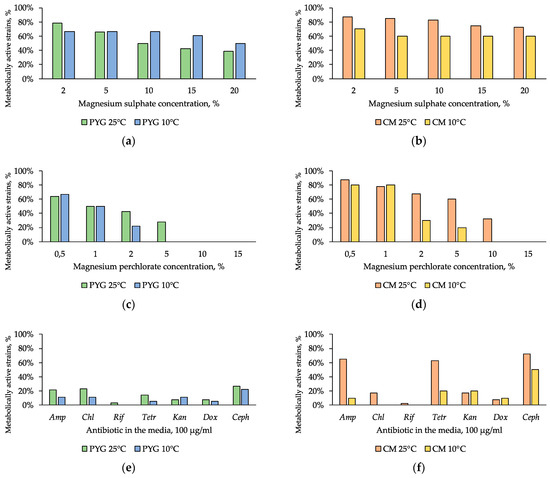
Figure 8.
Isolated strains resistance to: (a,b)—magnesium sulphate; (c,d)—magnesium perchlorate, (e,f)—antibiotics. Left column: strains isolated on PYG medium; right column—strains isolated on CM medium. Amp—ampicillin; Chl—chloramphenicol; Rif—rifampicin; Tetr—tetracycline; Kan—kanamycin; Dox—doxycycline; Ceph—cephalexin.
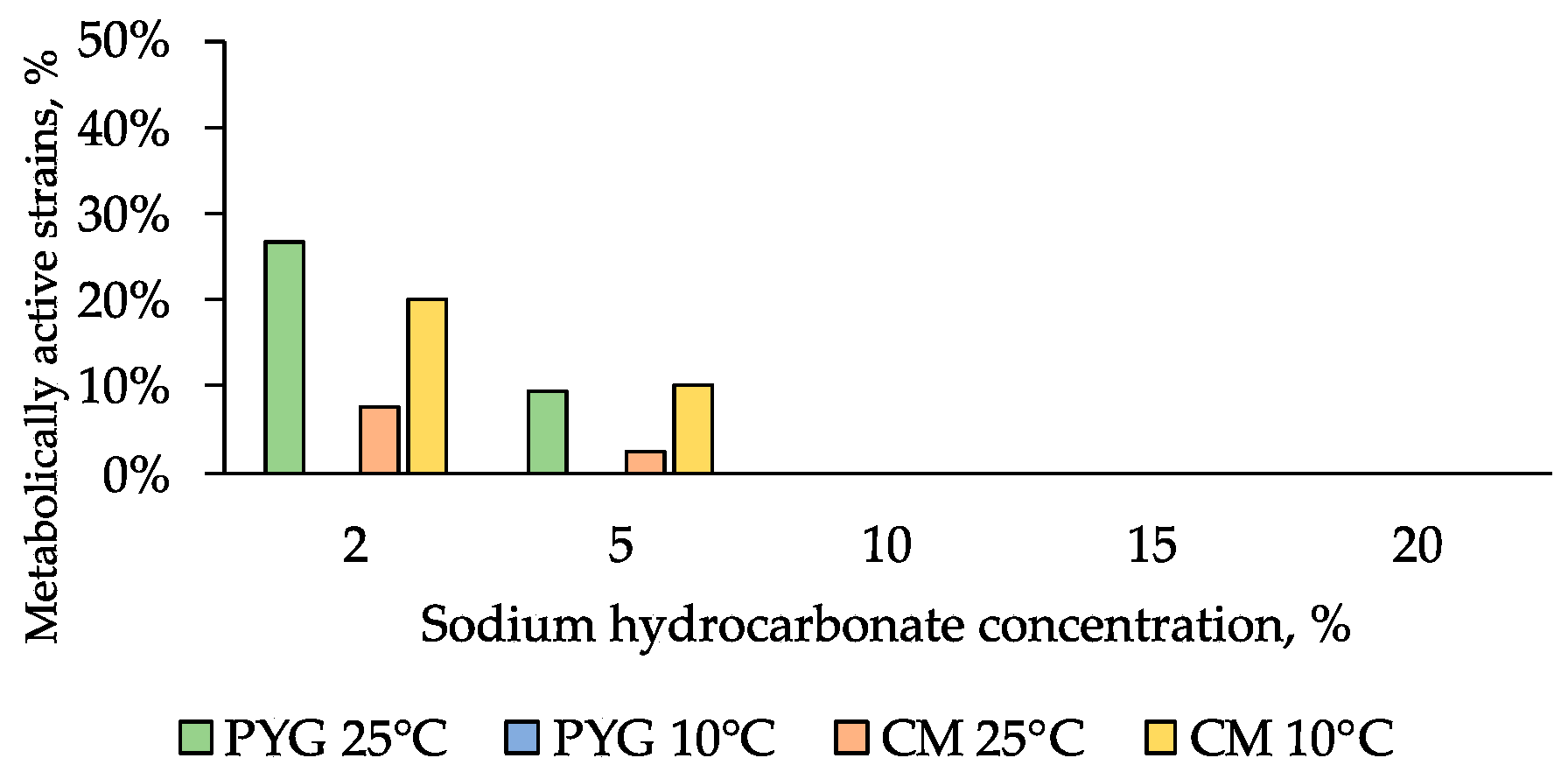
Sodium bicarbonate had the highest inhibitory effect among the studied water-soluble salts: few strains resistant to 5% of this salt in the culture medium were detected among the communities isolated on PYG and CM media at 25 °C (9% and 3% of strains, respectively) and among the community isolated on CM at 10 °C (10% of strains) (Figure 9).
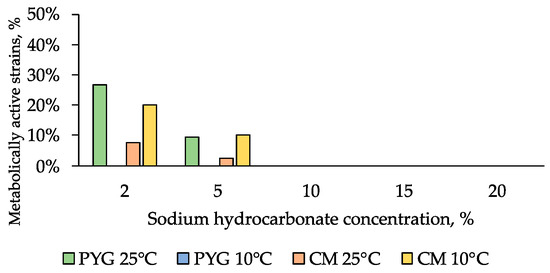
Figure 9.
NaHCO3 resistance diagram of strains isolated on PYG medium and CM medium.
Strains of the community isolated on CM medium at 25 °C (33% of strains) were resistant to the presence of 10% magnesium perchlorate. The community isolated on PYG at 25 °C was resistant to concentrations of up to 5% of magnesium perchlorate (28% of strains) (Figure 8c,d). Furthermore, higher (on average by 20%) proportion of the strains isolated on CM medium showed high resistance to the presence of this salt in the medium compared to the strains isolated on PYG medium.
The individual ranges of resistance of the isolated strains to physicochemical stress factors are presented in supplementary materials (Table S1).
In general, the investigated communities showed low resistance to the presence of antibiotics—about 20% of isolates in each of the four communities were capable to grow in the presence of a single clinical antibiotic.
High resistance of strains isolated on CM medium at 25 °C to ampicillin, tetracycline, and cephalexin is noteworthy (Figure 8e,f). In all the communities the strains characterized with multiple antibiotic resistance were found. These strains were related to the genera Bacillus, Agrococcus, Arthrobacter, Leucobacter, Pseudarthrobacter, Planomicrobium, Massilia, Microbacterium, and Pontibacter (Table S2).
4. Discussion
The total number of prokaryotes and numbers of culturable bacteria revealed in soil studied are in accordance with the previously obtained data for the soils of other regions of the Mojave Desert [32,36,37]. Different cell numbers observed in different regions of the desert could be caused by the high heterogeneity of soil composition and the high heterogeneity of soil resources in this desert, as well as past and current climatic conditions or plant associations which influence bacterial communities [59,60]. At the same time, considering the climatic regime of the studied region, the absence of growth during the isolation of culturable bacteria at high (50 °C) temperature is worth noticing, whereas some strains isolated at lower temperatures (10 °C, 25 °C) were capable of growing at this temperature in axenic culture. This is probably due to the conditions of culturing and intra- and interpopulation interactions between microorganisms, which can complicate the transition from the state of metabolic dormancy to grow under stressful conditions [61,62,63]. It is notable that the numbers of bacteria cultured at 10 °C and 25 °C were virtually equal. Based on these data, it is possible to assume that there is a high proportion of strains with psychrotolerant properties in the investigated soil, as was previously found for bacteria isolated from the Sahara and the Gibson Desert soils [44].
Lower numbers of CFUs for CM nutrient medium could be explained by lack of vegetation coverage at the sampling site and, as a result, the rareness of plant origin carbon sources (such as starch and sucrose in CM medium) in the soil in situ [39]. In addition, these compounds require preliminary hydrolysis for further assimilation by the cells. However, not all bacteria can produce hydrolytic enzymes for assimilation of these substrates [61].
The predominance of pigmented bacteria in the studied surface soil of the Mojave Desert is not surprising due to the role of pigments in protecting cells from damaging ultraviolet radiation [63,64]. The maximum diversity of the community detected on the rich PYG medium under the conditions of mesophilic temperature optimum indicates a predominantly mesophilic strategy of development for the majority of the microbial community studied. Diversity decreases along with decreasing culturing temperatures and limiting of easily available carbon sources and growth factors (such as amino acids and vitamins) in the composition of the nutrient medium. Mesophilic development strategy common for the acquired isolates is probably due to the climatic conditions experienced by the soil sample. The development of psychrotolerant properties could be caused by a high contrast of both diurnal and seasonal temperatures and can be considered to be an adaptive feature under these conditions. The higher biodiversity of cultured bacteria on PYG medium is due to the diversity of carbon sources and the presence of growth factors (such as vitamins and amino acids), which can be used by a larger number of bacteria. The presence of growth factors may also contribute to the reactivation of cells that were in a state of metabolic rest [62].
Representatives of the Actinobacteria phylum were predominant in all of the cultured communities, representatives of the Proteobacteria phylum were the second most common ones, Firmicutes and Bacteriodetes were identified as minor components. The dominance of representatives of Proteobacteria phylum in the edaphic bacterial communities of the Mojave Desert has been shown based on the molecular-genetic tests [21,24]. Simultaneously, the results we obtained reveal the similar structure of bacterial communities in the studied soil of this desert, consistent with culture-independent data published earlier [10,32,33]. The predominance of Actinobacteria in the community is quite natural, considering the fact that many species among them are resistant to desiccation, UV radiation, and other stress effects [22]. Low abundance of representatives of the Firmicutes phylum was previously detected in the Mojave Desert [24]. Among all isolated strains low relative abundance of spore-forming bacteria was observed (seven isolates only), while a high abundance of genes associated with spore formation processes was found in situ by culture-independent methods [33].
The bacteria of genera Arthrobacter, Bacillus, Kocuria, Leucobacter, Massilia, Mycetocola, Microbacterium, Paracoccus, Planomicrobium, Pseudarthrobacter, Rufibacter, Sphingomonas, and Streptomyces were previously isolated from other hot deserts [44,63,64,65]. Bacteria of the genus Agrococcus were previously cultured from cold deserts [66]. Among the representatives of the genus Brachybacterium several species connected to the root systems of halophilic plants were described and characterized by having plant growth-promoting potential [67]. Members of the genus Plantibacter, which have not been previously isolated from deserts, are also considered to be associated with herbaceous plants [68]. Representatives of the genus Cellulomonas are often isolated from soil [69] with some of them known to be halotolerant species [70]. Bacteria of the genus Pontibacter have been previously isolated from arid soils [71,72].
In this research, bacteria of the genera Janthinobacterium, which were found dominant in the meltwaters of glaciers [73], Salinibacterium, previously found in marine ecosystems and glaciers [74,75], and Labedella were isolated from desert soil for the first time.
Noteworthy is the discovery of the bacteria Planomicrobium glaciei in the desert soil. This species was first isolated from glacier, and after that from desert, ecosystems [76]. This allows us to assume that the intracellular mechanisms that determine the resistance and survivability of the species turn out to be adaptive in both cold and hot deserts.
According to community structure diagrams and ecological indices of communities’ similarity, all the obtained communities differ from each other significantly. It indicates that culturing conditions (such as medium composition and cultivation temperature) are the main factors influencing the structure of the communities isolated from studied soil. Nevertheless, the genera represented in all cultured communities are quite similar.
It should be considered that transport of microorganisms by winds is possible [15], which is especially important in case of surface soil sampling. According to this fact, the presence of introduced bacteria could not being excluded. Both culture-dependent and culture-independent methods could not separate indigenous and introduced microorganisms. However, the sampling site is located in the central part of the Mojave Desert, thus minimizing the chance to long distant cell transport from non-desert regions. Thus, even if some of the isolates were not indigenous, they probably were introduced from the other desert sites. Moreover, virtually all representatives of genera isolated were previously detected in different desert soils. The resistance data obtained also testifies to the high adaptation to the desert conditions and confirms predominantly desert-associated strains were isolated.
Substrates on which the most active development of the examined community was observed, could be produced as metabolites of cyanobacteria [8]. This suggests a functional relationship between the previously shown metabolically active cyanobacterial communities in the studied soil of the Mojave Desert and the heterotrophic community studied by us that exists due to the presence of the primary organic substance of cyanobacterial origin. Based on the aforementioned facts and data on the taxonomic and morphological diversity of isolates, it can be presumed that the culturable bacterial community of the studied site in central Mojave Desert, which is considered to be a Mars analogue site, is functionally differentiated, and is highly adapted to harsh physicochemical conditions. These results are consistent with the data obtained previously in the other regions of this desert [32,33].
The analysis of the resistance of isolates to temperature reveals the thermotolerant properties of communities cultured at 25 °C. The expansion of the range of temperatures suitable for growth occurs mainly in the region of low temperatures, which is probably caused by daily and seasonal temperature fluctuations [19]. The highest resistance was recorded for the community isolated on PYG media at 25 °C, which is probably due to the high number of cells and diversity of this community.
The wide pH ranges in which the metabolic activity is maintained can be explained by contrast temperature conditions and precipitation regimes leading to a change in the solubility of the substances in the soil solution [22]. Higher levels of pH resistance of strains isolated on CM could be caused by their hydrolytic activity, often associated with changes in the pH of the solution [61].
The studied region is characterized by a low content of water-soluble salts [19,20,24], which could mean that the abundance of salts present in the studied sample is low. It explains the moderate general resistance of isolated communities to the presence of sodium chloride or potassium chloride. However, strains with high resistance (up to 15%) to sodium chloride were found. It was previously shown that the same physiological mechanism provides resistance for both desiccation and salinization, which could explains the obtained result [77,78].
High resistance to magnesium sulphate was previously shown for bacterial communities isolated from desert soils [44]. It is probably associated with the assimilation of the components of this salt as elements of mineral nutrition. Low resistance to sodium bicarbonate is associated with the hydrolytic alkaline nature of this salt and its local distribution in the desert in the form of soda lakes. The presence of resistant forms is probably caused by a slight aeolian transfer of sodium bicarbonate and bacteria adapted to its presence, as well as by the formation of this salt in situ during weathering processes [42,44].
High resistance to magnesium perchlorate is particularly important in the astrobiology-oriented review of the obtained results. Maintenance of metabolic activity in vitro in the presence of perchlorates, at concentrations significantly exceeding those found in the regolith of Mars [79], testifies their non-limiting role in the regolith. Resistance to oxidizing agents is also likely due to the physiological adaptation of bacteria to arid conditions [78]. The higher level of resistance of the community isolated on a rich medium (PYG) at 10 °C, compared with the one isolated at 25 °C on the same medium, can be explained by the physiological connection of processes that ensure adaptation to low temperatures and the presence of oxidizers [80]. It has previously been shown that bacterial communities of other desert soils are resistant to high perchlorate concentrations [44].
The highest levels of resistance to physicochemical stress factors were found in representatives of the genera Arthrobacter, Bacillus, Kocuria, Leucobacter, Microbacterium, Planomicrobium, and Pontibacter (Table S1). Representatives of these genera are widely distributed in desert soils and have previously demonstrated high resistance to various types of stress effects [15,17,22,25,44,59,66].
For the representatives of majority genera isolated from soil studied resistance to ionizing and UV radiation was shown ([22,43,81,82,83,84,85,86,87,88,89] and references therein). It is in accordance with pigmented colonies’ high proportion data and it allows to suggest the presence of high UV- and radioresistance of the strains studied. Data on the resistance to physicochemical factors combined with the literature data on UV and ionizing radiation resistance could be considered as additional evidence of potential survivability of this bacterial community in Martian-like conditions. UV- and radioresistance of the isolates should be confirmed in further research.
The low resistance of the studied strains to antibiotics suggests that there is no active antibiotics-dependant competition in the communities. It is possible that the communities are realizing a co-metabolism strategy [90], as indirectly confirmed by the results of culturing on PYG medium and MST analysis: they revealed the most intensive growth of a cultured community on peptone containing various oligopeptides, amino acids, and vitamins. A similar result was obtained earlier in the study of the number of antibiotic resistance genes copies in the soils of the Mojave Desert [33]. At the same time, the discovery of a high number of antibiotic-resistant bacteria in the community isolated on CM medium at 25 °C is probably due to the signal role of antibiotics as quorum sensing agents [91,92] for activation of hydrolytic processes in the community. We have not found any previously published data on antibiotic resistance of representatives of Planomicrobium, Salinibacterium, and Labedella genera.
5. Conclusions
Stress effects, such as low water availability, nutrients deficiencies, high levels of solar radiation, and wide temperature fluctuations in the surface soil of the Mojave Desert lead to the formation of physiological survival strategies for prokaryotes existing under these conditions [2,3]. Our study revealed a high diversity (both taxonomic and morphophysiological) of culturable bacteria in the investigated soil. Species typical for various soils, including arid soils, and species that have not been previously isolated from hot arid soils were found in isolated bacterial communities. This suggests that the mechanisms of adaptation to cryo-arid and xero-arid conditions include similar physiological processes and adaptations. The obtained spectra of metabolic activity in the gradients of physicochemical factors indicate predominantly mesophilic and neutrophilic optima of all the studied strains, mostly capable of maintaining metabolic activity in a wide range of conditions, up to extreme ones for individual strains. Extreme resistance levels were identified in bacteria of Arthrobacter, Microbacterium, Bacillus, Planomicrobium, Kocuria, Leucobacter, and Pontibacter genera.
The obtained results, considered together with the data of previous works [25,41,64], testify in favour of viability, functionality, and taxonomical diversity of soil bacterial communities in the central part of the Mojave Desert studied, which is considered as a terrestrial analogue of Martian regolith. The isolated strains require further characterization and description for potential biotechnological applications. Due to the low similarity of the nucleotide sequences of some individual strains to those found in the database, these isolates could be the representatives of undescribed species of culturable bacteria. The most resistant strains that have been found could be used as model objects in further astrobiological studies.
As mentioned earlier, the Mojave Desert has undergone processes similar to those that took place on Mars and formed its current state. The properties of culturable bacterial communities revealed in this study indicate that they have adapted to survive in aggressive changing environmental conditions. Consequently, such communities could be considered as a model for further research of the Martian regolith habitability and the ability of terrestrial bacteria to adapt to extraterrestrial conditions.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3263/9/4/166/s1; Figure S1. Number of colonies’ morphotypes in the communities studied; Figure S2a. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Mojave Desert soil, which were sequenced using 1100r primer; Figure S2b. Phylogenetic analysis subtree of 16S rRNA genes of the strains isolated from the Mojave Desert soil, which were sequenced using 1100 r primer; Figure S2c. Phylogenetic analysis subtree of 16S rRNA genes of the strains isolated from the Mojave Desert soil, which were sequenced using 1100 r primer; Figure S3. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Mojave Desert soil, which were sequenced using 805r primer; Figure S4. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Mojave Desert soil, which were sequenced using 537r primer; Table S1. Physiological characteristics of isolated bacteria, Table S2. Antibiotic resistance spectra of isolated strains.
Author Contributions
A.A.B. and V.S.C. designed the study, all authors performed the study, A.A.B. and V.S.C. prepared draft manuscript, all authors edited and commented on the manuscript.
Funding
This research was funded by Russian Foundation for Basic Research according to the research project No 18-34-00331, and partially by PRAS no. 17 (Evolution of organic world. Role and influence of planetary processes), subprogram no. 1 (in the part of study of bacterial resistance to perchlorates).
Acknowledgments
The authors express their respect to T. Yu. Ulyanova, who provided the soil sample from the Mojave Desert. The authors thank anonymous reviewers for the help in improving the article. The authors thank George A. Abashin for the help in language improving the article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing the manuscript, and in the decision to publish the results.
Appendix A

Table A1.
Taxonomic affiliation of isolated strains.
Table A1.
Taxonomic affiliation of isolated strains.
| Strain–Isolation Conditions–GenBank Accession Number | BLAST Search Results | Taxonomic Affiliation |
|---|---|---|
| KBP.AS.17–PYG 25 °C–MK403789 | Pseudarthrobacter siccitolerans [MF682005]—99.0% Pseudarthrobacter oxydans [MF681862]—99.0% Pseudarthrobacter oxydans [MF681851]—99.0% | Pseudarthrobacter oxydans |
| KBP.AS.62–PYG 25 °C–MK403790 | Arthrobacter sp. [KX928444]—99.5% Arthrobacter sp. [KT944733]—99.5% Arthrobacter sp. [KM507593]—99.5% | Arthrobacter sp. |
| KBP.AS.75–PYG 25 °C–MK403794 | Brachybacterium sp. [MH518254]—99.3% Brachybacterium paraconglomeratum [KY622837]—99.3% Brachybacterium conglomeratum [LN995481]—99.3% | Brachybacterium sp. |
| KBP.AS.105–PYG 25 °C–MK403787 | Janthinobacterium sp. [KT715033]—98.7% Janthinobacterium sp. [FJ605429]—98.7% Janthinobacterium sp. [FJ605404]—98.7% | Janthinobacterium sp. |
| KBP.AS.160–PYG 25 °C–MK403788 | Arthrobacter sp. [HM216918]—99.2% Uncultured Arthrobacter sp. [JN129287]—99.0% Arthrobacter agilis [JN009621]—99.0% | Arthrobacter sp. |
| KBP.AS.747–PYG 25 °C–MK403791 | Microbacterium aurantiacum [MH734538]—99.2% Microbacterium aurantiacum [MH715198]—99.2% Microbacterium sp. [MH702463]—99.2% | Microbacterium aurantiacum |
| KBP.AS.748–PYG 25 °C–MK403792 | Pseudarthrobacter siccitolerans [MF682005]—99.22% Pseudarthrobacter oxydans [MF681862]—99.2% Pseudarthrobacter oxydans [MF681851]—99.2% | Pseudarthrobacter sp. |
| KBP.AS.749–PYG 25 °C–MK403793 | Brevibacterium frigoritolerans [MK318239]—99.9% Brevibacterium frigoritolerans [MK318233]—99.9% Brevibacterium frigoritolerans [MK318218]—99.9% | Brevibacterium frigoritolerans |
| KBP.AS.750–PYG 25 °C–MK403795 | Planomicrobium okeanokoites [KF749386]—99.3% Planomicrobium okeanokoites [KF749394]—99.3% Planomicrobium okeanokoites [KF724965]—99.3% | Planomicrobium okeanokoites |
| KBP.AS.751–PYG 25 °C–MK403796 | Agrococcus lahaulensis [KF318382]—100% Agrococcus sp. [JX949729]—100% Agrococcus jenensis [EF672044]—100% | Agrococcus sp. |
| KBP.AS.752–PYG 25 °C–MK403797 | Agrococcus sp. [KC702716]—99.5% Agrococcus sp. [JX949729]—99.5% Agrococcus jenensis [EF672044]—99.4% | Agrococcus sp. |
| KBP.AS.753–PYG 25 °C–MK403798 | Agrococcus sp. [MG232346]—99.5% Agrococcus jenensis [AJ717350]—99.5% Agrococcus sp. [HF954438]—99.5% | Agrococcus sp. |
| KBP.AS.754–PYG 25 °C–MK403799 | Kocuria sp. [DQ192212]—99.3% Kocuria sp. [EF612289]—99.2% Kocuria sp. [DQ180950]—99.0% | Kocuria sp. |
| KBP.AS.755–PYG 25 °C–MK403800 | Planomicrobium glaciei [KF387709]—100% Planomicrobium sp. [JX949538]—100% Planomicrobium glaciei HQ232419]—100% | Planomicrobium glaciei |
| KBP.AS.756–PYG 25 °C–MK403801 | Leucobacter sp. [KY623368]—99.6% Leucobacter aridicollis [KR827428]—99.6% Leucobacter sp. [KP152582]—99.6% | Leucobacter aridicollis |
| KBP.AS.757–PYG 25 °C–MK403802 | Planomicrobium okeanokoites [KF724965]—99.5% Planomicrobium okeanokoites [KF749394]—99.5% Planomicrobium okeanokoites [KF749386]—99.5% | Planomicrobium okeanokoites |
| KBP.AS.758–PYG 25 °C–MK403803 | Microbacterium esteraromaticum [GU111572]—98.9% Microbacterium sp. [KU221074]—98.6% Microbacterium sp. [KU221073]—98.6% | Microbacterium sp. |
| KBP.AS.759–PYG 25 °C–MK403804 | Bacillus pumilus [MH910167]—99.9% Bacillus pumilus [MH910156]—99.9% Bacillus pumilus [MH908782]—99.9% | Bacillus pumilus |
| KBP.AS.760–PYG 25 °C–MK403805 | Microbacterium esteraromaticum [GU111572]—98.9% Microbacterium sp. MH231521]—98.2% Microbacterium sp. [MH231520]—98.2% | Microbacterium sp. |
| KBP.AS.761–PYG 25 °C–MK403806 | Rufibacter sp. [MH050957]—97.6% Rufibacter immobilis [MH050949]—97.6% Uncultured bacterium clone [JQ376150]—98.9% | Rufibacter sp. |
| KBP.AS.762–PYG 25 °C–MK403807 | Microbacterium esteraromaticum [GU111572]—99.2% Microbacterium sp. [MH231521]—98.5% Microbacterium sp. [MH231520]—98.5% | Microbacterium sp. |
| KBP.AS.763–PYG 25 °C–MK403808 | Massilia niabensis [NR_044571]—98.9% Oxalobacter sp. [DQ196473]—99.2% Massilia suwonensis [LN774622]—98.8% | Massilia sp. |
| KBP.AS.764–PYG 25 °C–MK403809 | Bacillus sp. [MF990764]—98.2% Bacillus pumilus [JX988407]—97.8% Bacillus pumilus [EU795022]—97.8% | Bacillus sp. |
| KBP.AS.765–PYG 25 °C–MK403810 | Massilia alkalitolerans [KY010279]—99.7% Uncultured bacterium clone [KU515079]—99.7% Massilia alkalitolerans [KP282807]—99.7% | Massilia alkalitolerans |
| KBP.AS.766–PYG 25 °C–MK403811 | Pseudarthrobacter siccitolerans [MF682005]—98.1% Pseudarthrobacter oxydans [MF681862]—98.1% Pseudarthrobacter oxydans [MF681851]—98.1% | Pseudarthrobacter sp. |
| KBP.AS.767–PYG 25 °C–MK403812 | Massilia varians [MF077216]—98.4% Massilia varians [MF077215]—98.4% Massilia varians [MF077214]—98.5% | Massilia varians |
| KBP.AS.768–PYG 25 °C–MK403813 | Planococcus donghaensis [LN774491]—100% Planomicrobium okeanokoites [KF749394]—100% Planomicrobium okeanokoites [KF749386]—100% | Planomicrobium sp. |
| KBP.AS.769–PYG 25 °C–MK403814 | Microbacterium pseudoresistens [LN890047]—98.9% Microbacterium pseudoresistens [KF687021]—98.9% Microbacterium sp. [MH671545]—98,2% | Microbacterium pseudoresistens |
| KBP.AS.770–PYG 25 °C–MK403815 | Arthrobacter agilis [KF924209]—99.3% Arthrobacter sp. [JX949695]—99.3% Arthrobacter sp. [JX949646]—99.3% | Arthrobacter sp. |
| KBP.AS.771–PYG 25 °C–MK403816 | Arthrobacter sp. [MH714643]—99.5% Arthrobacter sp. [MG757950]—99.5% Arthrobacter sp. [JX949321]—99.5% | Arthrobacter sp. |
| KBP.AS.772–PYG 25 °C–MK403817 | Planomicrobium glaciei [MG025800]—100% Planomicrobium glaciei [KF387709]—100% Planomicrobium sp. [JX949538]—100% | Planomicrobium glaciei |
| KBP.AS.773–PYG 25 °C–MK403818 | Uncultured bacterium [LC026838]—98.9% Cellulomonas sp. [KP126821]—98.4% Cellulomonas cellasea [KR922256]—98.4% | Cellulomonas sp. |
| KBP.AS.774–PYG 25 °C–MK403819 | Planomicrobium okeanokoites [KF749394]—99.8% Planomicrobium okeanokoites [KF749386]—99.8% Planomicrobium okeanokoites [KF724965]—99.8% | Planomicrobium okeanokoites |
| KBP.AS.775–PYG 25 °C–MK403820 | Planomicrobium okeanokoites [KF724965]—99.9% Planomicrobium okeanokoites [KF749394]—99.9% Planomicrobium okeanokoites [KF749386]—99.9% | Planomicrobium okeanokoites |
| KBP.AS.776–PYG 25 °C–MK403821 | Georgenia sp. [EF512742]—99.6% Georgenia sp. [AY880044]—99.5% Georgenia satyanarayanai [NR_117051]—99.5% | Georgenia sp. |
| KBP.AS.777–PYG 25 °C–MK403822 | Microbacterium barkeri [MG706019]—98.4% Microbacterium sp. [MG203882]—98.4% Microbacterium barkeri [KY928100]—98.4% | Microbacterium barkeri |
| KBP.AS.778–PYG 25 °C–MK403823 | Salinibacterium sp. [MH714650]—99.0% Salinibacterium sp. [MH790149]—99.0% Salinibacterium sp. [MH299814]—99.0% | Salinibacterium sp. |
| KBP.AS.779–PYG 25 °C–MK403824 | Brevibacterium frigoritolerans [MK318239]—100% Brevibacterium frigoritolerans [MK318233]—100% Brevibacterium frigoritolerans [MK318218]—100% | Brevibacterium frigoritolerans |
| KBP.AS.780–PYG 25 °C–MK403825 | Micrococcus sp. [JQ229694]—99.3% Micrococcus sp. [JQ229689]—99.3% Micrococcus sp. [KM613156]—99.2% | Micrococcus sp. |
| KBP.AS.781–PYG 25 °C–MK403826 | Kocuria sp. [KT900591]—98.9% Kocuria sediminis [MH178354]—98.85% Kocuria turfanensis [MF769333]—98.8% | Kocuria sp. |
| KBP.AS.782–PYG 25 °C–MK403827 | Labedella sp. [KM253033]—99.2% Labedella gwakjiensis [LK021191]—98.2% Labedella sp. [JX273669]—98.2% | Labedella sp. |
| KBP.AS.783–PYG 25 °C–MK403828 | Plantibacter flavus [CP019402]—100% Plantibacter sp. [HE662660]—100% Plantibacter sp. [HE662693]—100% | Plantibacter sp. |
| KBP.AS.784–PYG 25 °C–MK403829 | Arthrobacter agilis [KT804924]—99.9% Arthrobacter agilis [MF767263]—99.8% Arthrobacter agilis [KR811202]—99.8% | Arthrobacter agilis |
| KBP.AS.785–PYG 25 °C–MK403830 | Microbacterium esteraromaticum [GU111572]—99.1% Microbacterium aureliae [NR152652]—98.6% Microbacterium aureliae [KF793922]—98.6% | Microbacterium sp. |
| KBP.AS.786–PYG 25 °C–MK403831 | Pseudarthrobacter siccitolerans [MF682005]—99.0% Pseudarthrobacter oxydans [MF681862]—99.0% Pseudarthrobacter oxydans [MF681851]—99.0% | Pseudarthrobacter sp. |
| KBP.AS.787–CM 25 °C–MK403832 | Uncultured bacterium [KX507906]—98.2% Massilia niabensis [JF496256]—98.0% Massilia alkalitolerans [KY010279]—97.8% | Massilia sp. |
| KBP.AS.788–CM 25 °C–None | Was identified by morphology | Bacillus sp. |
| KBP.AS.789–CM 25 °C–MK403833 | Burkholderia sp. [KU060146] —95.7% Burkholderia sp. [KU060136]—95.7% Burkholderia sp. [KU060135]—95.7% | Burkholderia sp. |
| KBP.AS.790–CM 25 °C–MK403834 | Cellulomonas hominis [KF817811]—100% Cellulomonas hominis [MF928361]—100% Cellulomonas sp. [LC133615]—100% | Cellulomonas hominis |
| KBP.AS.791–CM 25 °C–MK403835 | Pseudarthrobacter siccitolerans [MF682005]—99.5% Pseudarthrobacter oxydans [MF681862]—99.5% Pseudarthrobacter oxydans [MF681851]—99.5% | Pseudarthrobacter sp. |
| KBP.AS.792–CM 25 °C–MK403836 | Massilia sp. [JQ511858]—100% Massilia sp. [KY635898]—99.6% Massilia sp. [KY635895]—99.6% | Massilia sp. |
| KBP.AS.793–CM 25 °C–MK403837 | Uncultured actinobacterium clone [KC994764]—98.8% Salinibacterium sp. [MH299814]—98.7% Salinibacterium sp. [CP026951]—98.7% | Salinibacterium sp. |
| KBP.AS.794–CM 25 °C–MK403838 | Arthrobacter sp. [LN871743]—99.7% Arthrobacter sp. [KU951449]—99.7% Arthrobacter sp. [KJ191079]—99.7% | Arthrobacter sp. |
| KBP.AS.795–CM 25 °C–MK403839 | Arthrobacter sp. [LN871743]—99.2% Arthrobacter sp. [KU951449]—99.2% Arthrobacter sp. [KJ191079]—99.2% | Arthrobacter sp. |
| KBP.AS.816–PYG 25 °C–MK403840 | Arthrobacter sp. [KJ191079]—99.8% Arthrobacter sp. [KJ191059]—99.8% Arthrobacter sp. [KJ191035]—99.8% | Arthrobacter sp. |
| KBP.AS.842–PYG 25 °C–MK403841 | Uncultured bacterium clone [KX507404]—99.1% Massilia suwonensis [NR116872]—99.1% Massilia sp. [KM187344]—98.8% | Massilia sp. |
| KBP.AS.843–PYG 25 °C–MK403842 | Mycetocola sp. [MH299815]—98.4% Mycetocola sp. [MH298813]—98.4% Mycetocola sp. [CP026949]—98.4% | Mycetocola sp. |
| KBP.AS.844–PYG 25 °C–MK403843 | Arthrobacter agilis [KT804924]—98.9% Arthrobacter agilis [MF767263]—98.8% Arthrobacter agilis [KR811202]—98.8% | Arthrobacter agilis |
| KBP.AS.845–PYG 25 °C–None | Was identified by morphology | Streptomyces sp. |
| KBP.AS.846–PYG 25 °C–MK403844 | Arthrobacter pascens [KY880876]—100% Pseudarthrobacter defluvii [KY880845]—100% Arthrobacter sp. [LM994042]—100% | Arthrobacter sp. |
| KBP.AS.847–CM 25 °C–MK403845 | Cellulomonas sp. [MH046844]—99.2% Cellulomonas sp. [HM587947]—99.2% Cellulomonas flavigena [KY951360]—99.0% | Cellulomonas sp. |
| KBP.AS.848–PYG 25 °C–MK403846 | Massilia alkalitolerans [KY010279]—99.6% Uncultured bacterium clone [KU515079]—99.6% Massilia alkalitolerans [KP282807]—99.6% | Massilia sp. |
| KBP.AS.849–PYG 25 °C–MK403847 | Microbacterium esteraromaticum [GU111572]—98.8% Microbacterium aureliae [NR152652]—98.2% Microbacterium aureliae [KF793922]—98.2% | Microbacterium sp. |
| KBP.AS.850–PYG 25 °C–MK403848 | Massilia sp. [MH734560]—99.6% Massilia sp. [KY635898]—99.6% Massilia sp. [KY635895]—99.6% | Massilia sp. |
| KBP.AS.854–CM 25 °C–MK403849 | Paracoccus marcusii [MH744726]—99.9% Paracoccus marcusii [MH725412]—99.9% Paracoccus marcusii [MH725411]—99.9% | Paracoccus marcusii |
| KBP.AS.855–PYG 25 °C–None | Were identified by morphology | Rufibacter sp. |
| KBP.AS.858–CM 25 °C–MK403850 | Arthrobacter crystallopoietes [CP018864]—98.7% Arthrobacter crystallopoietes [CP018863]—98.7% Arthrobacter sp. [KX645729]—98.7% | Arthrobacter crystallopoietes |
| KBP.AS.862–PYG 25 °C–MK403851 | Arthrobacter sulfonivorans [KR233773]—99.9% Uncultured soil bacterium clone [JX490026]—99.8% Uncultured bacterium clone [KC554813]—99.7% | Arthrobacter sp. |
| KBP.AS.866–CM 25 °C–MK403852 | Uncultured bacterium clone [EU647527]—98.8% Sphingomonas xinjiangensis [NR_108386]—98.6% Sphingomonas mucosissima [KM502884]—98.3% | Sphingomonas sp. |
| KBP.AS.867–PYG 25 °C–MK403853 | Microbacterium paraoxydans [MH819720]—99.9% Microbacterium paraoxydans [MH281749]—99.9% Microbacterium sp. [MH558380]—99.9% | Microbacterium paraoxydans |
| KBP.AS.868–PYG 25 °C–MK403854 | Pseudarthrobacter siccitolerans [MF682005]—99.2% Pseudarthrobacter oxydans [MF681862]—99.2% Pseudarthrobacter oxydans [MF681851]—99.2% | Pseudarthrobacter sp. |
| KBP.AS.869–PYG 25 °C–MK403855 | Kocuria rosea [KY194315]—98.8% Kocuria sp. [KY194287]—98.8% Kocuria turfanensis [KY194236]—98.8% | Kocuria sp. |
| KBP.AS.876–PYG 25 °C–MK403856 | Arthrobacter agilis [CP024915]—99.1% Arthrobacter agilis [MG279727]—99.1% Arthrobacter sp. [KJ191029]—99.1% | Arthrobacter agilis |
| KBP.AS.877–PYG 25 °C–MK403857 | Salinibacterium sp. [MH790149]—99.0% Salinibacterium sp. [MH299814]—99.0% Salinibacterium sp. [CP026951]—99.0% | Salinibacterium sp. |
| KBP.AS.899–CM 25 °C–MK403858 | Streptomyces sp. [MG930074]—100% Streptomyces sp. [MG930073]—100% Streptomyces atrovirens [MF662228]—100% | Streptomyces sp. |
| KBP.AS.916–PYG 10 °C–MK403859 | Planomicrobium glaciei [MG025800]—99.1% Planomicrobium glaciei [KY307898]—99.1% Uncultured bacterium clone [KU514918]—99.1% | Planomicrobium glaciei |
| KBP.AS.926–PYG 10 °C–MK403860 | Sphingomonas xinjiangensis [NR108386]—98.9% Uncultured bacterium clone [EU647527]—98.6% Sphingomonas mucosissima [KM502884]—98.6% | Sphingomonas sp. |
| KBP.AS.927–PYG 10 °C–MK403861 | Arthrobacter agilis [MG279728]—99.4% Arthrobacter agilis [MF767263]—99.4% Arthrobacter agilis [KU884338]—99.4% | Arthrobacter agilis |
| KBP.AS.928–PYG 10 °C–MK403862 | Uncultured bacterium [KX509715]—99.5% Uncultured bacterium [KX508022]—99.5% Massilia niabensis [NR044571]—99.5% | Massilia sp. |
| KBP.AS.929–CM 10 °C–MK403863 | Pseudarthrobacter siccitolerans [MF682005]—98.5% Pseudarthrobacter oxydans [MF681862]—98.5% Pseudarthrobacter oxydans [MF681851]—98.5% | Pseudarthrobacter sp. |
| KBP.AS.930–CM 10 °C–MK403864 | Pseudarthrobacter siccitolerans [MF682005]—98.7% Pseudarthrobacter oxydans [MF681862]—98.7% Pseudarthrobacter oxydans [MF681851]—98.7% | Pseudarthrobacter sp. |
| KBP.AS.931–CM 10 °C–MK403865 | Arthrobacter boritolerans [MH828349]—99.7% Paenarthrobacter sp. [MH734752]—99.7% Arthrobacter sp. [LC416398]—99.7% | Arthrobacter sp. |
| KBP.AS.932–CM 10 °C–MK403866 | Pseudarthrobacter sp. [MG860452]—99.0% Pseudarthrobacter sp. [MG860451]—99.0% Arthrobacter sp. [MG860340]—99.0% | Pseudarthrobacter sp. |
| KBP.AS.936–PYG 10 °C–MK403867 | Pseudarthrobacter phenanthrenivorans [MF077156]—99.8% Pseudarthrobacter phenanthrenivorans [MF077155]—99.8% Arthrobacter phenanthrenivorans [KC789777]—99.8% | Pseudarthrobacter phenanthrenivorans |
| KBP.AS.937–PYG 10 °C–MK403868 | Arthrobacter agilis [KF876857]—99.9% Arthrobacter agilis [KT763365]—99.8% Uncultured Arthrobacter sp. [JN129287]—99.7% | Arthrobacter agilis |
| KBP.AS.938–CM 25 °C–MK403869 | Streptomyces sp. [KU182931]—99.8% Streptomyces sp. [MG820052]—99.6% Streptomyces sp. [MG930074]—99.6% | Streptomyces sp. |
| KBP.AS.939–CM 25 °C–MK403870 | Uncultured bacterium clone [KU515164]—99.0% Uncultured bacterium clone [KX509026]—98.9% Massilia sp. [MG733567]—98.6% | Massilia sp. |
| KBP.AS.940–CM 25 °C–MK403871 | Microbacterium esteraromaticum [GU111572]—99.2% Microbacterium sp. [MH231521]—98.8% Microbacterium sp. [KU221074]—98.8% | Microbacterium sp. |
| KBP.AS.941–CM 25 °C–MK403872 | Pseudarthrobacter siccitolerans [MF682005]—99.4% Pseudarthrobacter oxydans [MF681862]—99.4% Pseudarthrobacter oxydans [MF681851]—99.4% | Pseudarthrobacter sp. |
| KBP.AS.943–CM 10 °C–MK403873 | Arthrobacter agilis [JQ684255]—99.8% Arthrobacter agilis [CP024915]—99.7% Arthrobacter agilis [MG279727]—99.7% | Arthrobacter agilis |
| KBP.AS.947–PYG 10 °C–MK403874 | Microbacterium esteraromaticumstrain [GU111572]—99.3% Microbacterium sp. [MH231521]—98.5% Microbacterium sp. [MH231520]—98.5% | Microbacterium sp. |
| KBP.AS.954–PYG 10 °C–MK403875 | Uncultured Microbacterium sp. Clone [MH894291]—99.9% Microbacterium sp. [MH813404]—99.9% Microbacterium sp. [MH714660]—99.9% | Microbacterium sp. |
| KBP.AS.955–PYG 10 °C–MK403876 | Pseudarthrobacter siccitolerans [MF682005]—99.5% Pseudarthrobacter oxydans [MF681862]—99.5% Pseudarthrobacter oxydans [MF681851]—99.5% | Pseudarthrobacter sp. |
| KBP.AS.956–PYG 10°C–MK403877 | Planomicrobium glaciei [KF387709]—99.9% Planomicrobium sp. [JX949538]—99.9% Planomicrobium sp. [JQ618344]—99.9% | Planomicrobium sp. |
| KBP.AS.957–CM 10 °C–MK403878 | Arthrobacter sp. [KX645729]—99.5% Arthrobacter sp. [KF889389]—99.5% Arthrobacter crystallopoietes [KM257018]—99.5% | Arthrobacter sp. |
| KBP.AS.958–CM 10 °C–MK403879 | Arthrobacter sp. [KY173031]—99.6% Arthrobacter sp. [LN614607]—99.6% Arthrobacter oryzae [KC934820]—99.6% | Arthrobacter sp. |
| KBP.AS.960–CM 25 °C–MK403880 | Microbacterium esteraromaticum [GU111572]—99.3% Microbacterium sp. [MH231521]—98.6% Microbacterium sp. [MH231520]—98.6% | Microbacterium sp. |
| KBP.AS.961–CM 10 °C–MK403881 | Cellulomonas hominis [KT992126]—99.1% Cellulomonas sp. [Y09658]—99.1% Cellulomonas sp. [MF361869]—98.9% | Cellulomonas sp. |
| KBP.AS.962–CM 10 °C–None | Was identified by morphology | Streptomyces sp. |
| KBP.AS.963–CM 10 °C–MK403882 | Arthrobacter agilis [CP024915]—99.7% Arthrobacter agilis [MG279727]—99.7% Arthrobacter sp. [KY476520]—99.7% | Arthrobacter agilis |
| KBP.AS.972–PYG 10 °C–MK403883 | Micrococcus sp. [MH202946]—99.5% Micrococcus sp. [AB188213]—99.5% Micrococcus sp. [EF540464] —99.5% | Micrococcus sp. |
| KBP.AS.973–PYG 10 °C–MK403884 | Rhodococcus sp. [MH285872]—100% Rhodococcus sp. [CP034152]—100% Rhodococcus erythropolis [MH251258]—100% | Rhodococcus erythropolis |
| KBP.AS.1003–PYG 10 °C–MK403768 | Pseudarthrobacter phenanthrenivorans [NR_074770]—100% Arthrobacter sp. [LN871743]—100% Arthrobacter sp. [KU951449]—100% | Arthrobacter sp. |
| KBP.AS.1005–PYG 10 °C–MK403769 | Leucobacter sp. [KY623368]—99.9% Leucobacter aridicollis [KR827428]—99.9% Leucobacter sp. [KP152582]—99.9% | Leucobacter aridicollis |
| KBP.AS.1007–PYG 10 °C–MK403770 | Arthrobacter sp. [KT944733]—100% Arthrobacter sp. [KM507593]—100% Arthrobacter pityocampae [MH472818]—100% | Arthrobacter sp. |
| KBP.AS.1008–PYG 10 °C–MK403771 | Uncultured bacterium clone [KU514918]—99.3% Planomicrobium sp. [JX949538]—99.0% Planomicrobium glaciei [JF411320]—99.0% | Planomicrobium glaciei |
| KBP.AS.1009–PYG 10 °C–MK403772 | Pseudarthrobacter siccitolerans [MF682005]—99.8% Pseudarthrobacter oxydans [MF681862]—99.8% Pseudarthrobacter oxydans [MF681851]—99.8% | Pseudarthrobacter sp. |
| KBP.AS.1010–PYG 10 °C–MK403773 | Planomicrobium glaciei [KF387709]—99.7% Planomicrobium sp. [JX949538]—99.7% Planomicrobium glaciei [HQ232419]—99.7% | Planomicrobium glaciei |
| KBP.AS.1011–PYG 10 °C–MK403774 | Arthrobacter phenanthrenivorans [JX840975]—99.3% Uncultured soil bacterium clone [DQ248289]—99.3% Arthrobacter sp. [AY238502]—99.3% | Arthrobacter sp. |
| KBP.AS.1012–PYG 10 °C–MK403775 | Planococcus donghaensis [LN774491]—100% Planomicrobium okeanokoites [KF749394]—100% Planomicrobium okeanokoites [KF724965]—100% | Planomicrobium sp. |
| KBP.AS.1021–PYG 10 °C–MK403776 | Arthrobacter sp. [KT424968]—100% Arthrobacter agilis [KM036066]—100% Arthrobacter agilis [JN377650]—99.9% | Arthrobacter agilis |
| KBP.AS.1024–PYG 10 °C–MK403777 | Pseudarthrobacter phenanthrenivorans [NR_074770]—100% Arthrobacter sp. [LN871743]—100% Arthrobacter phenanthrenivorans [KR085846]—100% | Arthrobacter sp. |
| KBP.AS.1026–CM 25 °C–MK403778 | Uncultured bacterium [AB696407]—98.5% Massilia sp. [JQ511857]—98.0% Massilia sp. [JQ511858]—97.8% | Massilia sp. |
| KBP.AS.1028–CM 25 °C–MK403779 | Agrococcus lahaulensis [KF318382]—99.8% Agrococcus jenensis [EF672044]—99.8% Agrococcus lahaulensis [MF351823]—99.6% | Agrococcus sp. |
| KBP.AS.1029–CM 25 °C–MK403780 | Uncultured bacterium clone [DQ125573]—99.5% Pseudarthrobacter oxydans [MH304399]—99.3% Arthrobacter sp. [MH714681]—99.3% | Arthrobacter sp. |
| KBP.AS.1030–CM 25 °C–MK403781 | Pseudarthrobacter siccitolerans [MF682005]—99.9% Pseudarthrobacter oxydans [MF681862]—99.9% Pseudarthrobacter oxydans [MF681851]—99.9% | Pseudarthrobacter sp. |
| KBP.AS.1031–CM 25 °C–MK403782 | Massilia sp. [MH707216]—99.9% Massilia alkalitolerans [KY010279]—99.9% Massilia varians [KX784920]—99.9% | Massilia sp. |
| KBP.AS.1032–CM 25 °C–MK403783 | Arthrobacter agilis [KF924209]—99.8% Arthrobacter sp. [JX949695]—99.8% Arthrobacter sp. [JX949646]—99.8% | Arthrobacter sp. |
| KBP.AS.1033–CM 25 °C–MK403784 | Microbacterium sp. [LT601250]—99.5% Uncultured bacterium clone [MG805031]—99.5% Microbacterium sp. [MF526604]—99.5% | Microbacterium sp. |
| KBP.AS.1037–CM 25 °C–MK403785 | Massilia varians [KC429599]—99.7% Massilia alkalitolerans [KY010279]—99.6% Massilia alkalitolerans [KP282807]—99.6% | Massilia sp. |
| KBP.AS.1044–CM 25 °C–MK403786 | Uncultured bacterium clone [KC442649]—87.5% Pontibacter sp. [HM579810]—87.5% Pontibacter diazotrophicus [NR_126288]—87.2% | Pontibacter sp. |
Colour highlight indicates strains characterized by 100% sequence similarity.
Appendix B

Table A2.
The primers used for 16S rRNA gene amplification and sequencing.
Table A2.
The primers used for 16S rRNA gene amplification and sequencing.
| GenBank Accession Number | Strain | Taxonomic Affiliation | Primers Used for Amplification | Primers Used for Sequencing |
|---|---|---|---|---|
| MK403789 | KBP.AS.17 | Pseudarthrobacter oxydans | 27 f + Un1492 r | 1100 r |
| MK403790 | KBP.AS.62 | Arthrobacter sp. | 341 f + 805 r | 805 r |
| MK403794 | KBP.AS.75 | Brachybacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403787 | KBP.AS.105 | Janthinobacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403788 | KBP.AS.160 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403791 | KBP.AS.747 | Microbacterium aurantiacum | 27 f + 537 r | 537 r |
| MK403792 | KBP.AS.748 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403793 | KBP.AS.749 | Brevibacterium frigoritolerans | 27 f + Un1492 r | 1100 r |
| MK403795 | KBP.AS.750 | Planomicrobium okeanokoites | 341 f + 805 r | 805 r |
| MK403796 | KBP.AS.751 | Agrococcus sp. | 27 f + Un1492 r | 1100 r |
| MK403797 | KBP.AS.752 | Agrococcus sp. | 27 f + Un1492 r | 1100 r |
| MK403798 | KBP.AS.753 | Agrococcus sp. | 27 f + 537 r | 537 r |
| MK403799 | KBP.AS.754 | Kocuria sp. | 27 f + Un1492 r | 1100 r |
| MK403800 | KBP.AS.755 | Planomicrobium glaciei | 27 f + Un1492 r | 1100 r |
| MK403801 | KBP.AS.756 | Leucobacter aridicollis | 27 f + Un1492 r | 1100 r |
| MK403802 | KBP.AS.757 | Planomicrobium okeanokoites | 341 f + 805 r | 805 r |
| MK403803 | KBP.AS.758 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403804 | KBP.AS.759 | Bacillus pumilus | 27 f + Un1492 r | 1100 r |
| MK403805 | KBP.AS.760 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403806 | KBP.AS.761 | Rufibacter sp. | 27 f + 537 r | 537 r |
| MK403807 | KBP.AS.762 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403808 | KBP.AS.763 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403809 | KBP.AS.764 | Bacillus sp. | 341 f + 805 r | 805 r |
| MK403810 | KBP.AS.765 | Massilia alkalitolerans | 27 f + Un1492 r | 1100 r |
| MK403811 | KBP.AS.766 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403812 | KBP.AS.767 | Massilia varians | 27 f + Un1492r | 1100r |
| MK403813 | KBP.AS.768 | Planomicrobium sp. | 27 f + Un1492 r | 1100r |
| MK403814 | KBP.AS.769 | Microbacterium pseudoresistens | 27 f + 537 r | 537 r |
| MK403815 | KBP.AS.770 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403816 | KBP.AS.771 | Arthrobacter sp. | 27 f+Un1492 r | 1100 r |
| MK403817 | KBP.AS.772 | Planomicrobium glaciei | 27 f + Un1492 r | 1100 r |
| MK403818 | KBP.AS.773 | Cellulomonas sp. | 27 f + Un1492 r | 1100 r |
| MK403819 | KBP.AS.774 | Planomicrobium okeanokoites | 27 f + Un1492 r | 1100 r |
| MK403820 | KBP.AS.775 | Planomicrobium okeanokoites | 27 f + Un1492 r | 1100 r |
| MK403821 | KBP.AS.776 | Georgenia sp. | 27 f + Un1492 r | 1100 r |
| MK403822 | KBP.AS.777 | Microbacterium barkeri | 27 f + Un1492 r | 1100 r |
| MK403823 | KBP.AS.778 | Salinibacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403824 | KBP.AS.779 | Brevibacterium frigoritolerans | 27 f + Un1492 r | 1100 r |
| MK403825 | KBP.AS.780 | Micrococcus sp. | 27 f + Un1492 r | 1100 r |
| MK403826 | KBP.AS.781 | Kocuria sp. | 27 f + Un1492 r | 1100 r |
| MK403827 | KBP.AS.782 | Labedella sp. | 27 f + Un1492 r | 1100 r |
| MK403828 | KBP.AS.783 | Plantibacter sp. | 27 f+ 537 r | 537 r |
| MK403829 | KBP.AS.784 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403830 | KBP.AS.785 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403831 | KBP.AS.786 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403832 | KBP.AS.787 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| None | KBP.AS.788 | Bacillus sp. | Was identified by morphology | |
| MK403833 | KBP.AS.789 | Burkholderia sp. | 341 f + 805 r | 805 r |
| MK403834 | KBP.AS.790 | Cellulomonas hominis | 27 f + Un1492 r | 1100 r |
| MK403835 | KBP.AS.791 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403836 | KBP.AS.792 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403837 | KBP.AS.793 | Salinibacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403838 | KBP.AS.794 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403839 | KBP.AS.795 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403840 | KBP.AS.816 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403841 | KBP.AS.842 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403842 | KBP.AS.843 | Mycetocola sp. | 27 f + Un1492 r | 1100 r |
| MK403843 | KBP.AS.844 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| None | KBP.AS.845 | Streptomyces sp. | Was identified by morphology | |
| MK403844 | KBP.AS.846 | Arthrobacter sp. | 27 f+ 537 r | 537 r |
| MK403845 | KBP.AS.847 | Cellulomonas sp. | 27 f + Un1492 r | 1100 r |
| MK403846 | KBP.AS.848 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403847 | KBP.AS.849 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403848 | KBP.AS.850 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403849 | KBP.AS.854 | Paracoccus marcusii | 27 f + Un1492 r | 1100r |
| None | KBP.AS.855 | Rufibacter sp. | Was identified by morphology | |
| MK403850 | KBP.AS.858 | Arthrobacter crystallopoietes | 27 f + Un1492 r | 1100 r |
| MK403851 | KBP.AS.862 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403852 | KBP.AS.866 | Sphingomonas sp. | 27 f + Un1492 r | 1100 r |
| MK403853 | KBP.AS.867 | Microbacterium paraoxydans | 27 f + Un1492 r | 1100 r |
| MK403854 | KBP.AS.868 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403855 | KBP.AS.869 | Kocuria sp. | 341 f + 805 r | 805 r |
| MK403856 | KBP.AS.876 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403857 | KBP.AS.877 | Salinibacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403858 | KBP.AS.899 | Streptomyces sp. | 27 f + Un1492 r | 1100 r |
| MK403859 | KBP.AS.916 | Planomicrobium glaciei | 27 f + Un1492 r | 1100 r |
| MK403860 | KBP.AS.926 | Sphingomonas sp. | 27 f + Un1492 r | 1100 r |
| MK403861 | KBP.AS.927 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403862 | KBP.AS.928 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403863 | KBP.AS.929 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403864 | KBP.AS.930 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403865 | KBP.AS.931 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403866 | KBP.AS.932 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403867 | KBP.AS.936 | Pseudarthrobacter phenanthrenivorans | 27 f + Un1492 r | 1100 r |
| MK403868 | KBP.AS.937 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403869 | KBP.AS.938 | Streptomyces sp. | 27 f + Un1492 r | 1100 r |
| MK403870 | KBP.AS.939 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403871 | KBP.AS.940 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403872 | KBP.AS.941 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403873 | KBP.AS.943 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403874 | KBP.AS.947 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403875 | KBP.AS.954 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403876 | KBP.AS.955 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403877 | KBP.AS.956 | Planomicrobium sp | 27 f + Un1492 r | 1100 r |
| MK403878 | KBP.AS.957 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403879 | KBP.AS.958 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403880 | KBP.AS.960 | Microbacterium sp. | 27 f + Un1492 r | 1100 r |
| MK403881 | KBP.AS.961 | Cellulomonas sp. | 27 f + Un1492 r | 1100 r |
| None | KBP.AS.962 | Streptomyces sp. | Was identified by morphology | |
| MK403882 | KBP.AS.963 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403883 | KBP.AS.972 | Micrococcus sp. | 27 f + Un1492 r | 1100 r |
| MK403884 | KBP.AS.973 | Rhodococcus erythropolis | 27 f + Un1492 r | 1100 r |
| MK403768 | KBP.AS.1003 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403769 | KBP.AS.1005 | Leucobacter aridicollis | 27 f + Un1492 r | 1100 r |
| MK403770 | KBP.AS.1007 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403771 | KBP.AS.1008 | Planomicrobium glaciei | 27 f + Un1492 r | 1100 r |
| MK403772 | KBP.AS.1009 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403773 | KBP.AS.1010 | Planomicrobium glaciei | 27 f + Un1492 r | 1100 r |
| MK403774 | KBP.AS.1011 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403775 | KBP.AS.1012 | Planomicrobium sp | 341 f + 805 r | 805 r |
| MK403776 | KBP.AS.1021 | Arthrobacter agilis | 27 f + Un1492 r | 1100 r |
| MK403777 | KBP.AS.1024 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403778 | KBP.AS.1026 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403779 | KBP.AS.1028 | Agrococcus sp. | 27 f + Un1492 r | 1100 r |
| MK403780 | KBP.AS.1029 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403781 | KBP.AS.1030 | Pseudarthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403782 | KBP.AS.1031 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403783 | KBP.AS.1032 | Arthrobacter sp. | 27 f + Un1492 r | 1100 r |
| MK403784 | KBP.AS.1033 | Microbacterium sp. | 341 f + 805 r | 805 r |
| MK403785 | KBP.AS.1037 | Massilia sp. | 27 f + Un1492 r | 1100 r |
| MK403786 | KBP.AS.1044 | Pontibacter sp. | 341 f + 805 r | 805 r |
Colour highlight indicates strains characterized by 100% sequence similarity.
Appendix C

Table A3.
The bacterial genera cultured from the Mojave Desert soil sample on different media. The table summarizes data of community structure diagrams and allows to compare bacterial diversity in a simpler view.
Table A3.
The bacterial genera cultured from the Mojave Desert soil sample on different media. The table summarizes data of community structure diagrams and allows to compare bacterial diversity in a simpler view.
| PYG 25 °C | PYG 10 °C | CM 25 °C | CM 10 °C |
|---|---|---|---|
| Agrococcus | Agrococcus | ||
| Arthrobacter | Arthrobacter | Arthrobacter | Arthrobacter |
| Bacillus | Bacillus | ||
| Brachybacterium | |||
| Burkholderia | |||
| Cellulomonas | Cellulomonas | Cellulomonas | |
| Georgenia | |||
| Janthinobacterium | |||
| Kocuria | |||
| Labedella | |||
| Leucobacter | Leucobacter | ||
| Massilia | Massilia | Massilia | |
| Microbacterium | Microbacterium | Microbacterium | |
| Micrococcus | |||
| Mycetocola | |||
| Paracoccus | Paracoccus | ||
| Planomicrobium | Planomicrobium | ||
| Plantibacter | |||
| Pontibacter | |||
| Pseudarthrobacter | Pseudarthrobacter | Pseudarthrobacter | |
| Rhodococcus | |||
| Rufibacter | |||
| Salinibacterium | Salinibacterium | ||
| Sphingomonas | Sphingomonas | ||
| Streptomyces | Streptomyces | Streptomyces |
References
- Chyba, C.F.; Hand, K.P. Astrobiology: The study of the living universe. Annu. Rev. Astron. Astrophys. 2005, 43, 31–74. [Google Scholar] [CrossRef]
- Cockell, C.S.; Schwendner, P.; Perras, A.; Rettberg, P.; Beblo-Vranesevic, K.; Bohmeier, M.; Rabbow, E.; Moissl-Eichinger, C.; Wink, L.; Marteinsson, V.; et al. Anaerobic microorganisms in astrobiological analogue environments: From field site to culture collection. Int. J. Astrobiol. 2017, 1–15. [Google Scholar] [CrossRef]
- Cloutis, E.A.; Craig, M.A.; Kruzelecky, R.V.; Jamroz, W.R.; Scott, A.; Hawthorne, F.C.; Mertzman, S.A. Spectral reflectance properties of minerals exposed to simulated Mars surface conditions. Icarus 2008, 195, 140–168. [Google Scholar] [CrossRef]
- Horneck, G.; Klaus, D.M.; Mancinelli, R.L. Space microbiology. Microbiol. Mol. Biol. Rev. 2010, 74, 121–156. [Google Scholar] [CrossRef]
- Rummel, J.D.; Beaty, D.W.; Jones, M.A.; Bakermans, C.; Barlow, N.G.; Boston, P.J.; Chevrier, V.F.; Clark, B.C.; de Vera, J.-P.P.; Gough, R.V.; et al. A new analysis of Mars “special regions”: Findings of the second MEPAG Special Regions Science Analysis Group (SR-SAG2). Astrobiology 2014, 14, 887–968. [Google Scholar] [CrossRef]
- Carr, M.H. The Surface of Mars; Cambridge University Press: Cambridge, UK, 2007; Volume 6. [Google Scholar]
- Gilichinsky, D.; Rivkina, E.; Shcherbakova, V.; Laurinavichuis, K.; Tiedje, J. Supercooled water brines within permafrost—an unknown ecological niche for microorganisms: A model for astrobiology. Astrobiology 2003, 3, 331–341. [Google Scholar] [CrossRef]
- Smith, H.D.; Baqué, M.; Duncan, A.G.; Lloyd, C.R.; McKay, C.P.; Billi, D. Comparative analysis of cyanobacteria inhabiting rocks with different light transmittance in the Mojave Desert: A Mars terrestrial analogue. Int. J. Astrobiol. 2014, 13, 271–277. [Google Scholar] [CrossRef]
- Dartnell, L.R.; Desorgher, L.; Ward, J.M.; Coates, A.J. Modelling the surface and subsurface martian radiation environment: Implications for astrobiology. Geophys. Res. Lett. 2007, 34, L02207. [Google Scholar] [CrossRef]
- Bay, S.; Ferrari, B.; Greening, C. Life without water: How do bacteria generate biomass in desert ecosystems? Microbiol. Aust. 2018, 39, 28–32. [Google Scholar] [CrossRef]
- Paenibacilius, B. Microbial diversity of extreme regions: An unseen heritage and wealth. Indian. J. Plant Genet. Resour. 2016, 29, 246–248. [Google Scholar] [CrossRef]
- Wierzchos, J.; Ascaso, C.; McKay, C.P. Endolithic cyanobacteria in halite rocks from the hyperarid core of the Atacama Desert. Astrobiology 2006, 6, 415–422. [Google Scholar] [CrossRef]
- McKay, C.P.; Friedmann, E.I.; Gómez-Silva, B.; Cáceres-Villanueva, L.; Andersen, D.T.; Landheim, R. Temperature and moisture conditions for life in the extreme arid region of the Atacama Desert: Four years of observations including the El Nino of 1997–1998. Astrobiology 2003, 3, 393–406. [Google Scholar] [CrossRef]
- Vítek, P.; Jehlička, J.; Edwards, H.G.; Hutchinson, I.; Ascaso, C.; Wierzchos, J. The miniaturized Raman system and detection of traces of life in halite from the Atacama Desert: Some considerations for the search for life signatures on Mars. Astrobiology 2012, 12, 1095–1099. [Google Scholar] [CrossRef]
- Benardini, J.N.; Sawyer, J.; Venkateswaran, K.; Nicholson, W.L. Spore UV and acceleration resistance of endolithic Bacillus pumilus and Bacillus subtilis isolates obtained from Sonoran desert basalt: Implications for lithopanspermia. Astrobiology 2003, 3, 709–717. [Google Scholar] [CrossRef]
- Doran, P.T.; Lyons, W.B.; McKnight, D.M. (Eds.) Life in Antarctic Deserts and Other Cold Dry Environments: Astrobiological Analogs; Cambridge University Press: Cambridge, UK, 2010; Volume 5, ISBN 978-0-521-88919-3. [Google Scholar]
- Rampelotto, P.H. Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustainability 2010, 2, 1602–1623. [Google Scholar] [CrossRef]
- Vítek, P.; Jehlička, J.; Edwards, H.G.; Hutchinson, I.; Ascaso, C.; Wierzchos, J. Miniaturized Raman instrumentation detects carotenoids in Mars-analogue rocks from the Mojave and Atacama deserts. Phil. Trans. R. Soc. A 2014, 372, 20140196. [Google Scholar] [CrossRef]
- Salas, E.; Abbey, W.; Bhartia, R.; Beegle, L.W. The Mojave Desert: A Martian Analog Site for Future Astrobiology Themed Missions; Jet Propulsion Laboratory, NASA: Pasadena, CA, USA, 2011. Available online: http://hdl.handle.net/2014/43479 (accessed on 9 April 2019).
- Bishop, J.L.; Schelble, R.T.; McKay, C.P.; Brown, A.J.; Perry, K.A. Carbonate rocks in the Mojave Desert as an analogue for Martian carbonates. Int. J. Astrobiol. 2011, 10, 349–358. [Google Scholar] [CrossRef]
- Purohit, H.J.; Kaila, V.C.; Vaidya, V.C.; Khardenavis, A.A. Optimization and Applicability of Bioprocesses; Springer Nature: Singapore, 2017; ISBN 978-981-10-6863-8. [Google Scholar]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Abbey, W.; Salas, E.; Bhartia, R.; Beegle, L.W. The Mojave vadose zone: A subsurface biosphere analogue for Mars. Astrobiology 2013, 13, 637–646. [Google Scholar] [CrossRef]
- McHugh, T.A.; Compson, Z.; van Gestel, N.; Hayer, M.; Ballard, L.; Haverty, M.; Hines, J.; Irvine, N.; Krassner, D.; Lyons, T.; et al. Climate controls prokaryotic community composition in desert soils of the southwestern United States. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Preston, L.J.; Dartnell, L.R. Planetary habitability: Lessons learned from terrestrial analogues. Int. J. Astrobiol. 2014, 13, 81–98. [Google Scholar] [CrossRef]
- Navarro-González, R.; Navarro, K.F.; de la Rosa, J.; Iñiguez, E.; Molina, P.; Miranda, L.D.; Morales, P.; Cienfuegos, E.; Coll, P.; Raulin, F.; et al. The limitations on organic detection in Mars-like soils by thermal volatilization–gas chromatography–MS and their implications for the Viking results. Proc. Natl. Acad. Sci. USA 2006, 103, 16089–16094. [Google Scholar] [CrossRef]
- Greeley, R.; Bridges, N.T.; Kuzmin, R.O.; Laity, J.E. Terrestrial analogs to wind-related features at the Viking and Pathfinder landing sites on Mars. J. Geophys. Res. Planets 2002, 107, 5-1–5-22. [Google Scholar] [CrossRef]
- Davila, A.F.; Schulze-Makuch, D. The last possible outposts for life on Mars. Astrobiology 2016, 16, 159–168. [Google Scholar] [CrossRef]
- Peters, G.H.; Abbey, W.; Bearman, G.H.; Mungas, G.S.; Smith, J.A.; Anderson, R.C.; Douglas, S.; Beegle, L.W. Mojave Mars simulant—Characterization of a new geologic Mars analog. Icarus 2008, 197, 470–479. [Google Scholar] [CrossRef]
- Schlesinger, W.H.; Pippen, J.S.; Wallenstein, M.D.; Hofmockel, K.S.; Klepeis, D.M.; Mahall, B.E. Community composition and photosynthesis by photoautotrophs under quartz pebbles, southern Mojave Desert. Ecology 2003, 84, 3222–3231. [Google Scholar] [CrossRef]
- Navarro-González, R.; Rainey, F.A.; Molina, P.; Bagaley, D.R.; Hollen, B.J.; de la Rosa, J.; Small, A.M.; Quinn, R.C.; Grunthaner, F.J.; Cáceres, L.; et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 2003, 302, 1018–1021. [Google Scholar] [CrossRef]
- Mogul, R.; Vaishampayan, P.; Bashir, M.; McKay, C.P.; Schubert, K.; Bornaccorsi, R.; Gomez, E.; Payton, G.; Capra, J.; Bacon, L.; et al. Microbial community and biochemical dynamics of biological soil crusts across a gradient of surface coverage in the central Mojave Desert. Front. Microbiol. 2017, 8, 1974. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Schelble, R.T.; McDonald, G.D.; Hall, J.A.; Nealson, K.H. Community structure comparison using FAME analysis of desert varnish and soil, Mojave Desert, California. Geomicrobiol. J. 2005, 22, 353–360. [Google Scholar] [CrossRef]
- Ewing, S.A.; Southard, R.J.; Macalady, J.L.; Hartshorn, A.S.; Johnson, M.J. Soil microbial fingerprints, carbon, and nitrogen in a Mojave Desert creosote-bush ecosystem. Soil Sci. Soc. Am. J. 2007, 71, 469–475. [Google Scholar] [CrossRef]
- Smith, H.D.; Duncan, A.G.; Neary, P.L.; Lloyd, C.R.; Anderson, A.J.; Sims, R.C.; McKay, C.P. In situ microbial detection in Mojave Desert soil using native fluorescence. Astrobiology 2012, 12, 247–257. [Google Scholar] [CrossRef]
- Bryant, E.; Rech, S. The effect of moisture on soil microbial communities in the Mojave Desert. Astrobiology 2008, 8, 427. [Google Scholar] [CrossRef]
- Garrity, G.M.; Heimbuch, B.K.; Gagliardi, M. Isolation of zoosporogenous actinomycetes from desert soils. J. Ind. Microbiol. 1996, 17, 260–267. [Google Scholar] [CrossRef]
- Garcia, D.E.; Lopez, B.R.; de-Bashan, L.E.; Hirsch, A.M.; Maymon, M.; Bashan, Y. Functional metabolic diversity of the bacterial community in undisturbed resource island soils in the southern Sonoran Desert. Land. Degrad. Dev. 2018, 29, 1467–1477. [Google Scholar] [CrossRef]
- Pointing, S.B.; Belnap, J. Microbial colonization and controls in dryland systems. Nat. Rev. Microbiol. 2012, 10, 551. [Google Scholar] [CrossRef]
- Loughney, K.M.; Badgley, C. Facies, Environments, and Fossil Preservation in the Barstow Formation, Mojave Desert, California. Palaios 2017, 32, 396–412. [Google Scholar] [CrossRef]
- Reynolds, R.L.; Reheis, M.; Yount, J.; Lamothe, P. Composition of aeolian dust in natural traps on isolated surfaces of the central Mojave Desert—Insights to mixing, sources, and nutrient inputs. J. Arid Environ. 2006, 66, 42–61. [Google Scholar] [CrossRef]
- Cheptsov, V.S.; Vorobyova, E.A.; Osipov, G.A.; Manucharova, N.A.; Polyanskaya, L.M.; Gorlenko, M.V.; Pavlov, A.K.; Rosanova, M.S.; Lomasov, V.N. Microbial activity in Martian analog soils after ionizing radiation: Implications for the preservation of subsurface life on Mars. AIMS Microbiol. 2018, 4, 541–562. [Google Scholar] [CrossRef]
- Belov, A.A.; Cheptsov, V.S.; Vorobyova, E.A. Soil bacterial communities of Sahara and Gibson deserts: Physiological and taxonomical characteristics. AIMS Microbiol. 2018, 4, 685–710. [Google Scholar] [CrossRef]
- Schinner, F.; Öhlinger, R.; Kandeler, E.; Margesin, R. (Eds.) Methods in Soil Biology; Springer Science & Business Media: Berlin, Germany, 2012; ISBN 978-3-642-60966-4. [Google Scholar]
- Depository of Live Systems. Available online: https://depo.msu.ru (accessed on 8 April 2019).
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing, In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Chromas and ChromasPro DNA Sequencing Software—Technelysium Pty Ltd. Available online: http://www.technelysium.com.au (accessed on 8 April 2019).
- Clustal Omega < Multiple Sequence Alignment < EMBL-EBI. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 8 April 2019).
- BLAST: Basic Local Alignment Search Tool. Available online: http: //blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 8 April 2019).
- Mega Home. Available online: https://www.megasoftware.net/ (accessed on 8 April 2019).
- Gorlenko, M.V.; Majorova, T.N.; Kozhevin, P.A. Disturbances and their influence on substrate utilization patterns in soil microbial communities. In Microbial Communities; Springer: Berlin, Germany, 1997; pp. 84–93. [Google Scholar]
- Gorlenko, M.V.; Kozhevin, P.A. Multisubstrate Testing of Natural Microbial Communities; MAKS Press: Moscow, Russia, 2005. (In Russian) [Google Scholar]
- Chernov, I.Y.; Lysak, L.V. Methodical Materials for Seminars on the Course-General Ecology; MAKS Press: Moscow, Russia, 2005. (In Russian) [Google Scholar]
- Titus, J.H.; Nowak, R.S.; Smith, S.D. Soil resource heterogeneity in the Mojave Desert. J. Arid Environ. 2002, 52, 269–292. [Google Scholar] [CrossRef]
- Hereford, R.; Webb, R.H.; Longpre, C.I. Precipitation history and ecosystem response to multidecadal precipitation variability in the Mojave Desert region, 1893–2001. J. Arid Environ. 2006, 67, 13–34. [Google Scholar] [CrossRef]
- Manucharova, N.A.; Vlasenko, A.N.; Men’ko, E.V.; Zvyagintsev, D.G. Specificity of the chitinolytic microbial complex of soils incubated at different temperatures. Microbiology 2011, 80, 205–215. [Google Scholar] [CrossRef]
- Barer, M.R. Viable but non-culturable and dormant bacteria: Time to resolve an oxymoron and a misnomer? J. Med. Microbiol. 1997, 46, 629–631. [Google Scholar] [CrossRef]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Zagorodnov, V.; Sandman, K.; Reeve, J.N. Recovery and identification of viable bacteria immured in glacial ice. Icarus 2000, 144, 479–485. [Google Scholar] [CrossRef]
- Rainey, F.A.; Ray, K.; Ferreira, M.; Gatz, B.Z.; Nobre, M.F.; Bagaley, D.; Small, A.M. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 2005, 71, 5225–5235. [Google Scholar] [CrossRef]
- Luo, X.; Wang, J.; Zeng, X.C.; Wang, Y.; Zhou, L.; Nie, Y.; Dai, J.; Fang, C. Mycetocola manganoxydans sp. nov., an actinobacterium isolated from the Taklamakan desert. Int. J. Syst. Evol. Microbiol. 2012, 62, 2967–2970. [Google Scholar] [CrossRef]
- Mayilraj, S.; Suresh, K.; Schumann, P.; Kroppenstedt, R.M.; Saini, H.S. Agrococcus lahaulensis sp. nov., isolated from a cold desert of the Indian Himalayas. Int. J. Syst. Evol. Microbiol. 2006, 56, 1807–1810. [Google Scholar] [CrossRef]
- Gontia, I.; Kavita, K.; Schmid, M.; Hartmann, A.; Jha, B. Brachybacterium saurashtrense sp. nov., a halotolerant root-associated bacterium with plant growth-promoting potential. Int. J. Syst. Evol. Microbiol. 2011, 61, 27992804. [Google Scholar] [CrossRef]
- Lin, Y.C.; Yokota, A. Plantibacter auratus sp. nov., in the family Microbacteriaceae. Int. J. Syst. Evol. Microbiol. 2006, 56, 2337–2339. [Google Scholar] [CrossRef]
- Hatayama, K.; Esaki, K.; Ide, T. Cellulomonas soli sp. nov. and Cellulomonas oligotrophica sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 60–65. [Google Scholar] [CrossRef]
- Ahmed, I.; Kudo, T.; Abbas, S.; Ehsan, M.; Iino, T.; Fujiwara, T.; Ohkuma, M. Cellulomonas pakistanensis sp. nov., a moderately halotolerant Actinobacteria. Int. J. Syst. Evol. Microbiol. 2014, 64, 2305–2311. [Google Scholar] [CrossRef]
- Cheptsov, V.; Vorobyova, E.; Belov, A.; Pavlov, A.; Tsurkov, D.; Lomasov, V.; Bulat, S. Survivability of soil and permafrost microbial communities after irradiation with accelerated electrons under simulated Martian and open space conditions. Geosciences 2018, 8, 298. [Google Scholar] [CrossRef]
- Mahato, N.K.; Tripathi, C.; Nayyar, N.; Singh, A.K.; Lal, R. Pontibacter ummariensis sp. nov., isolated from a hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1080–1087. [Google Scholar] [CrossRef]
- Rassner, S.M.E.; Anesio, A.M.; Girdwood, S.E.; Hell, K.; Gokul, J.K.; Whitworth, D.E.; Edwards, A. Can the bacterial community of a high Arctic glacier surface escape viral control? Front. Microbiol. 2016, 7, 956. [Google Scholar] [CrossRef]
- Zhang, D.C.; Liu, H.C.; Xin, Y.H.; Yu, Y.; Zhou, P.J.; Zhou, Y.G. Salinibacterium xinjiangense sp. nov., a psychrophilic bacterium isolated from the China No. 1 glacier. Int. J. Syst. Evol. Microbiol. 2008, 58, 2739–2742. [Google Scholar] [CrossRef]
- Han, S.K.; Nedashkovskaya, O.I.; Mikhailov, V.V.; Kim, S.B.; Bae, K.S. Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int. J. Syst. Evol. Microbiol. 2003, 53, 2061–2066. [Google Scholar] [CrossRef]
- Zhang, D.C.; Liu, H.C.; Xin, Y.H.; Yu, Y.; Zhou, P.J.; Zhou, Y.G. Planomicrobium glaciei sp. nov., a psychrotolerant bacterium isolated from a glacier. Int. J. Syst. Evol. Microbiol. 2009, 59, 1387–1390. [Google Scholar] [CrossRef]
- Fagliarone, C.; Mosca, C.; Ubaldi, I.; Verseux, C.; Baqué, M.; Wilmotte, A.; Billi, D. Avoidance of protein oxidation correlates with the desiccation and radiation resistance of hot and cold desert strains of the cyanobacterium Chroococcidiopsis. Extremophiles 2017, 21, 981–991. [Google Scholar] [CrossRef]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: Correlation between stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef]
- Clark, B.C.; van Hart, D.C. The salts of Mars. Icarus 1981, 45, 370–378. [Google Scholar] [CrossRef]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 175–190. [Google Scholar] [CrossRef]
- Cheptsov, V.S.; Vorobyova, E.A.; Manucharova, N.A.; Gorlenko, M.V.; Pavlov, A.K.; Vdovina, M.A.; Lomasov, V.N.; Bulat, S.A. 100 kGy gamma-affected microbial communities within the ancient Arctic permafrost under simulated Martian conditions. Extremophiles 2017, 21, 1057–1067. [Google Scholar] [CrossRef]
- Musilova, M.; Wright, G.; Ward, J.M.; Dartnell, L.R. Isolation of radiation-resistant bacteria from Mars analog Antarctic Dry Valleys by preselection, and the correlation between radiation and desiccation resistance. Astrobiology 2015, 15, 1076–1090. [Google Scholar] [CrossRef]
- Gholami, M.; Etemadifar, Z.; Bouzari, M. Isolation a new strain of Kocuria rosea capable of tolerating extreme conditions. J. Environ. Radioact. 2015, 144, 113–119. [Google Scholar] [CrossRef]
- Yu, L.Z.H.; Luo, X.S.; Liu, M.; Huang, Q. Diversity of ionizing radiation-resistant bacteria obtained from the Taklimakan Desert. J. Basic Microbiol. 2015, 55, 135–140. [Google Scholar] [CrossRef]
- Mao, J.; Tang, Q.; Zhang, Z.; Wang, W.; Wei, D.; Huang, Y.; Liu, Z.; Shi1, Y.; Goodfellow, M. Streptomyces radiopugnans sp. nov., a radiation-resistant actinomycete isolated from radiation-polluted soil in China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2578–2582. [Google Scholar] [CrossRef]
- Marizcurrena, J.J.; Morel, M.A.; Braña, V.; Morales, D.; Martinez-López, W.; Castro-Sowinski, S. Searching for novel photolyases in UVC-resistant Antarctic bacteria. Extremophiles 2017, 21, 409–418. [Google Scholar] [CrossRef]
- Farías, M.E.; Fernández-Zenoff, V.; Flores, R.; Ordóñez, O.; Estévez, C. Impact of solar radiation on bacterioplankton in Laguna Vilama, a hypersaline Andean lake (4650 m). J. Geophys. Res. Biogeosci. 2009, 114. [Google Scholar] [CrossRef]
- Mojib, N.; Farhoomand, A.; Andersen, D.T.; Bej, A.K. UV and cold tolerance of a pigment-producing Antarctic Janthinobacterium sp. Ant5-2. Extremophiles 2013, 17, 367–378. [Google Scholar] [CrossRef]
- Vallalar, B. Investigation of the Growth and Survival of Bacteria from Mars Analog Environments When Exposed to Mars-like Conditions. Master of Science (MS), Bharathi Vallalar, Louisiana State University and Agricultural and Mechanical College. Available online: https://digitalcommons.lsu.edu/gradschool_theses/3601 (accessed on 9 April 2019).
- Horvath, R.S. Microbial co-metabolism and the degradation of organic compounds in nature. Bacteriol. Rev. 1972, 36, 146–155. [Google Scholar]
- Romero, D.; Traxler, M.F.; López, D.; Kolter, R. Antibiotics as signal molecules. Chem. Rev. 2011, 111, 5492–5505. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).