Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach
Abstract
:1. Introduction
2. Study Design
2.1. Pre-Clinical Studies
2.2. Clinical Study
2.2.1. Participants
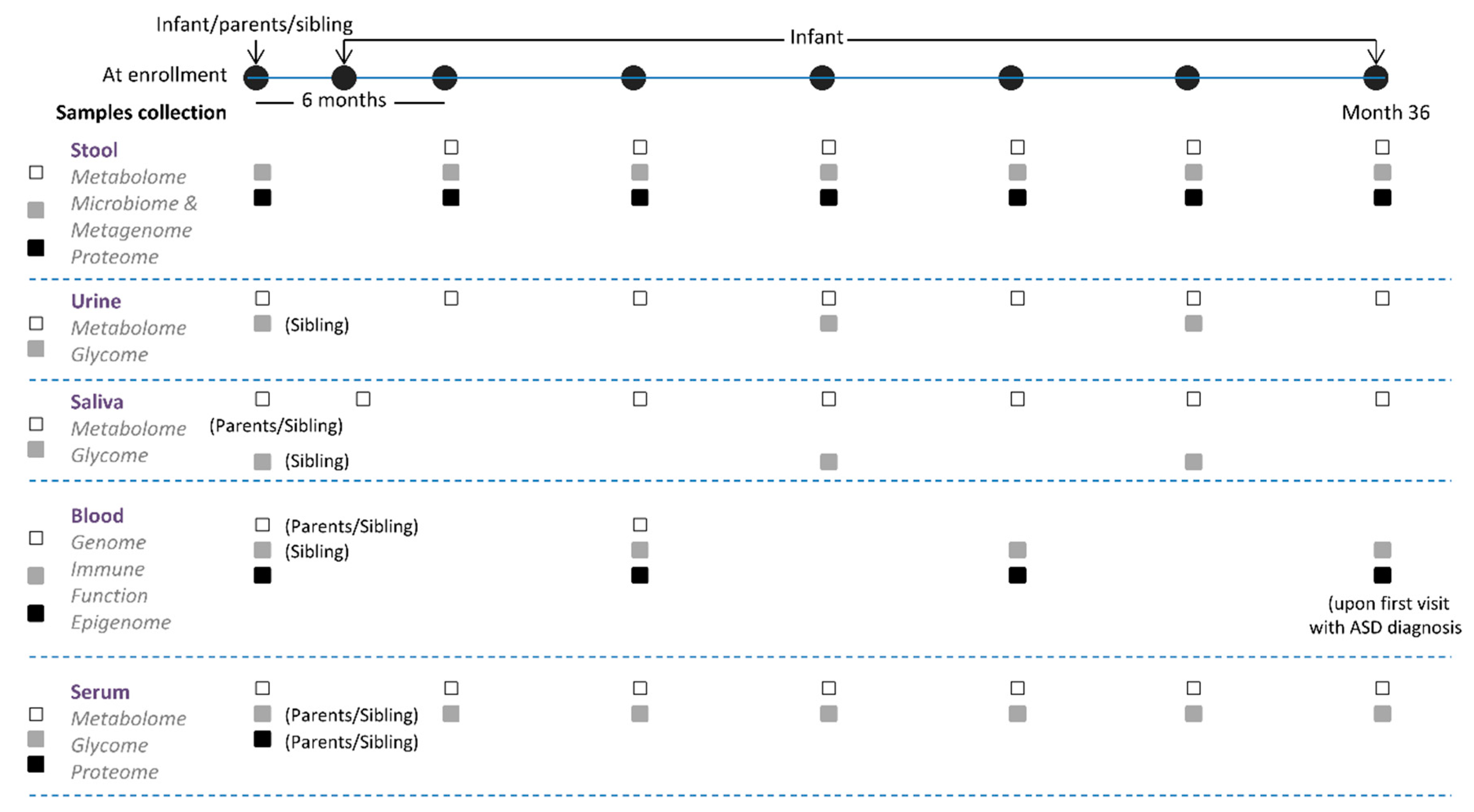
2.2.2. Observational Trial
2.2.3. Interventional Trial
2.3. Data Collection
Parental and Child Questionnaires
2.4. Serological Markers
2.4.1. Whole Blood
2.4.2. Stool, Urine, and Saliva Samples
3. Factor of Interest
3.1. Environmental
3.2. Genetics
3.3. GI Microbiome and Metabolome
3.4. Immune Function
4. Statistical Approach
4.1. Statistical Methods
4.2. Power Analysis
4.3. Developing an Integrative Multilevel Model to Predict ASD
5. Discussions and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evans, B. How autism became autism: The radical transformation of a central concept of child development in Britain. Hist. Hum. Sci. 2013, 26, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Elsabbagh, M.; Divan, G.; Koh, Y.-J.; Kim, Y.S.; Kauchali, S.; Marcín, C.; Montiel-Nava, C.; Patel, V.; Paula, C.S.; Wang, C.; et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012, 5, 160–179. [Google Scholar] [CrossRef] [Green Version]
- Newschaffer, C.J.; Croen, L.A.; Daniels, J.; Giarelli, E.; Grether, J.K.; Levy, S.E.; Mandell, D.S.; Miller, L.A.; Pinto-Martin, J.; Reaven, J.; et al. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health 2007, 28, 235–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wing, L.; Potter, D. The epidemiology of autistic spectrum disorders: Is the prevalence rising? Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 151–161. [Google Scholar] [CrossRef]
- Fombonne, E. The prevalence of autism. JAMA 2003, 289, 87–89. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. (CDC) Prevalence of Autism Spectrum Disorders—Autism and Developmental Disabilities Monitoring Network, 14 Sites USA, 2008; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2012.
- Maenner, M.J. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Autism Spectrum Disorders in the European Union (ASDEU). Consortium Autism Spectrum Disorders in the European Union (ASDEU): Final Report: Main Results of the ASDEU Project, ASDEU, Madrid: 28 August 2018. Available online: http://repositorio.insa.pt/handle/10400.18/6188 (accessed on 15 October 2020).
- Ozonoff, S.; Young, G.S.; Carter, A.; Messinger, D.; Yirmiya, N.; Zwaigenbaum, L.; Bryson, S.; Carver, L.J.; Constantino, J.N.; Dobkins, K.; et al. Recurrence risk for autism spectrum disorders: A Baby Siblings Research Consortium study. Pediatrics 2011, 128, e488–e495. [Google Scholar] [CrossRef] [Green Version]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.-Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e23. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Yip, B.H.K.; Windham, G.C.; Sourander, A.; Francis, R.; Yoffe, R.; Glasson, E.; Mahjani, B.; Suominen, A.; Leonard, H.; et al. Association of Genetic and Environmental Factors With Autism in a 5-Country Cohort. JAMA Psychiatry 2019, 76, 1035–1043. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Larsson, H.; Hultman, C.M.; Reichenberg, A. The familial risk of autism. JAMA 2014, 311, 1770–1777. [Google Scholar] [CrossRef]
- Brucato, M.; Ladd-Acosta, C.; Li, M.; Caruso, D.; Hong, X.; Kaczaniuk, J.; Stuart, E.A.; Fallin, M.D.; Wang, X. Prenatal exposure to fever is associated with autism spectrum disorder in the boston birth cohort. Autism Res. 2017, 10, 1878–1890. [Google Scholar] [CrossRef]
- Croen, L.A.; Qian, Y.; Ashwood, P.; Zerbo, O.; Schendel, D.; Pinto-Martin, J.; Daniele Fallin, M.; Levy, S.; Schieve, L.A.; Yeargin-Allsopp, M.; et al. Infection and Fever in Pregnancy and Autism Spectrum Disorders: Findings from the Study to Explore Early Development. Autism Res. 2019, 12, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- DiStasio, M.M.; Nagakura, I.; Nadler, M.J.; Anderson, M.P. T-lymphocytes and Cytotoxic Astrocyte Blebs Correlate Across Autism Brains. Ann. Neurol. 2019, 86, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Hornig, M.; Bresnahan, M.A.; Che, X.; Schultz, A.F.; Ukaigwe, J.E.; Eddy, M.L.; Hirtz, D.; Gunnes, N.; Lie, K.K.; Magnus, P.; et al. Prenatal fever and autism risk. Mol. Psychiatry 2018, 23, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Nankova, B.B.; Agarwal, R.; MacFabe, D.F.; La Gamma, E.F. Enteric bacterial metabolites propionic and butyric acid modulate gene expression, including CREB-dependent catecholaminergic neurotransmission, in PC12 cells--possible relevance to autism spectrum disorders. PLoS ONE 2014, 9, e103740. [Google Scholar] [CrossRef]
- Macfabe, D. Autism: Metabolism, mitochondria, and the microbiome. Glob. Adv. Health Med. 2013, 2, 52–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scumpia, P.O.; Kelly-Scumpia, K.; Stevens, B.R. Alpha-lipoic acid effects on brain glial functions accompanying double-stranded RNA antiviral and inflammatory signaling. Neurochem. Int. 2014, 64, 55–63. [Google Scholar] [CrossRef]
- Zielinski, B.A.; Prigge, M.B.D.; Nielsen, J.A.; Froehlich, A.L.; Abildskov, T.J.; Anderson, J.S.; Fletcher, P.T.; Zygmunt, K.M.; Travers, B.G.; Lange, N.; et al. Longitudinal changes in cortical thickness in autism and typical development. Brain 2014, 137, 1799–1812. [Google Scholar] [CrossRef] [Green Version]
- Stoner, R.; Chow, M.L.; Boyle, M.P.; Sunkin, S.M.; Mouton, P.R.; Roy, S.; Wynshaw-Boris, A.; Colamarino, S.A.; Lein, E.S.; Courchesne, E. Patches of Disorganization in the Neocortex of Children with Autism. N. Engl. J. Med. 2014, 370, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, D.A.; Frye, R.E. A review of research trends in physiological abnormalities in autism spectrum disorders: Immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol. Psychiatry 2012, 17, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.-M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rook, G.A.; Raison, C.L.; Lowry, C.A. Microbiota, immunoregulatory old friends and psychiatric disorders. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2014; pp. 319–356. [Google Scholar]
- Borre, Y.E.; Moloney, R.D.; Clarke, G.; Dinan, T.G.; Cryan, J.F. The impact of microbiota on brain and behavior: Mechanisms & therapeutic potential. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2014; pp. 373–403. [Google Scholar]
- Shimabukuro, T.T.; Grosse, S.D.; Rice, C. Medical expenditures for children with an autism spectrum disorder in a privately insured population. J. Autism Dev. Disord. 2008, 38, 546–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peacock, G.; Amendah, D.; Ouyang, L.; Grosse, S.D. Autism spectrum disorders and health care expenditures: The effects of co-occurring conditions. J. Dev. Behav. Pediatr. 2012, 33, 2–8. [Google Scholar] [CrossRef]
- Blumberg, S.J.; Bramlett, M.D.; Kogan, M.D.; Schieve, L.A.; Jones, J.R.; Lu, M.C. Changes in prevalence of parent-reported autism spectrum disorder in school-aged U.S. children: 2007 to 2011–2012. Natl. Health Stat. Rep. 2013, 65, 1–11. [Google Scholar]
- Fletcher-Watson, S.; McConnell, F.; Manola, E.; McConachie, H. Interventions based on the Theory of Mind cognitive model for autism spectrum disorder (ASD). Cochrane Database Syst. Rev. 2014, 3, CD008785. [Google Scholar] [CrossRef] [Green Version]
- Sukhodolsky, D.G.; Bloch, M.H.; Panza, K.E.; Reichow, B. Cognitive-behavioral therapy for anxiety in children with high-functioning autism: A meta-analysis. Pediatrics 2013, 132, e1341–e1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millward, C.; Ferriter, M.; Calver, S.; Connell-Jones, G. Gluten- and casein-free diets for autistic spectrum disorder. Cochrane Database Syst. Rev. 2008, 2, CD003498. [Google Scholar] [CrossRef] [PubMed]
- Bolte, E.R. Autism and Clostridium tetani. Med. Hypotheses 1998, 51, 133–144. [Google Scholar] [CrossRef]
- Finegold, S.M.; Molitoris, D.; Song, Y.; Liu, C.; Vaisanen, M.-L.; Bolte, E.; McTeague, M.; Sandler, R.; Wexler, H.; Marlowe, E.M.; et al. Gastrointestinal Microflora Studies in Late-Onset Autism. Clin. Infect. Dis. 2002, 35, S6–S16. [Google Scholar] [CrossRef]
- Pulikkan, J.; Mazumder, A.; Grace, T. Role of the Gut Microbiome in Autism Spectrum Disorders. In Reviews on Biomarker Studies in Psychiatric and Neurodegenerative Disorders; Guest, P.C., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 253–269. ISBN 978-3-030-05542-4. [Google Scholar]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [Green Version]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Masi, A.; DeMayo, M.M.; Glozier, N.; Guastella, A.J. An overview of autism spectrum disorder, heterogeneity and treatment options. Neurosci. Bull. 2017, 33, 183–193. [Google Scholar] [CrossRef] [Green Version]
- de Theije, C.G.M.; Wu, J.; Koelink, P.J.; Korte-Bouws, G.A.H.; Borre, Y.; Kas, M.J.H.; da Silva, S.L.; Korte, S.M.; Olivier, B.; Garssen, J.; et al. Autistic-like behavioural and neurochemical changes in a mouse model of food allergy. Behav. Brain Res. 2014, 261, 265–274. [Google Scholar] [CrossRef] [PubMed]
- de Theije, C.G.M.; van den Elsen, L.W.J.; Willemsen, L.E.M.; Milosevic, V.; Korte-Bouws, G.A.H.; da Silva, S.L.; Broersen, L.M.; Korte, S.M.; Olivier, B.; Garssen, J.; et al. Dietary long chain n-3 polyunsaturated fatty acids prevent impaired social behaviour and normalize brain dopamine levels in food allergic mice. Neuropharmacology 2015, 90, 15–22. [Google Scholar] [CrossRef]
- Saunders, J.M.; Moreno, J.L.; Ibi, D.; Sikaroodi, M.; Kang, D.J.; Muñoz-Moreno, R.; Dalmet, S.S.; García-Sastre, A.; Gillevet, P.M.; Dozmorov, M.G.; et al. Gut microbiota manipulation during the prepubertal period shapes behavioral abnormalities in a mouse neurodevelopmental disorder model. Sci. Rep. 2020, 10, 4697. [Google Scholar] [CrossRef]
- Wu, J.; de Theije, C.G.M.; da Silva, S.L.; Abbring, S.; van der Horst, H.; Broersen, L.M.; Willemsen, L.; Kas, M.; Garssen, J.; Kraneveld, A.D. Dietary interventions that reduce mTOR activity rescue autistic-like behavioral deficits in mice. Brain Behav. Immun. 2017, 59, 273–287. [Google Scholar] [CrossRef]
- Le Roy, T.; Debédat, J.; Marquet, F.; Da-Cunha, C.; Ichou, F.; Guerre-Millo, M.; Kapel, N.; Aron-Wisnewsky, J.; Clément, K. Comparative Evaluation of Microbiota Engraftment Following Fecal Microbiota Transfer in Mice Models: Age, Kinetic and Microbial Status Matter. Front. Microbiol. 2019, 9, 3289. [Google Scholar] [CrossRef]
- Stilling, R.M.; Moloney, G.M.; Ryan, F.J.; Hoban, A.E.; Bastiaanssen, T.F.; Shanahan, F.; Clarke, G.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Social interaction-induced activation of RNA splicing in the amygdala of microbiome-deficient mice. eLife 2018, 7, e33070. [Google Scholar] [CrossRef] [Green Version]
- Hoban, A.E.; Stilling, R.M.; Moloney, G.; Shanahan, F.; Dinan, T.G.; Clarke, G.; Cryan, J.F. The microbiome regulates amygdala-dependent fear recall. Mol. Psychiatry 2018, 23, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Luczynski, P.; Whelan, S.O.; O’Sullivan, C.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: Differential effects in the amygdala and hippocampus. Eur. J. Neurosci. 2016, 44, 2654–2666. [Google Scholar] [CrossRef] [Green Version]
- Desbonnet, L.; Clarke, G.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. Microbiota is essential for social development in the mouse. Mol. Psychiatry 2014, 19, 146–148. [Google Scholar] [CrossRef]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, K.M.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011, 23, 255–264.e119. [Google Scholar] [CrossRef] [PubMed]
- Arentsen, T.; Raith, H.; Qian, Y.; Forssberg, H.; Diaz Heijtz, R. Host microbiota modulates development of social preference in mice. Microb. Ecol. Health Dis. 2015, 26, 29719. [Google Scholar] [CrossRef]
- Lionetti, E.; Castellaneta, S.; Francavilla, R.; Pulvirenti, A.; Tonutti, E.; Amarri, S.; Barbato, M.; Barbera, C.; Barera, G.; Bellantoni, A. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014, 371, 1295–1303. [Google Scholar] [CrossRef] [Green Version]
- Leonard, M.M.; Camhi, S.; Huedo-Medina, T.B.; Fasano, A. Celiac Disease Genomic, Environmental, Microbiome, and Metabolomic (CDGEMM) Study Design: Approach to the Future of Personalized Prevention of Celiac Disease. Nutrients 2015, 7, 9325–9336. [Google Scholar] [CrossRef] [Green Version]
- Gotham, K.; Pickles, A.; Lord, C. Standardizing ADOS Scores for a Measure of Severity in Autism Spectrum Disorders. J. Autism Dev. Disord. 2009, 39, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Guthrie, W.; Swineford, L.B.; Nottke, C.; Wetherby, A.M. Early diagnosis of autism spectrum disorder: Stability and change in clinical diagnosis and symptom presentation. J. Child. Psychol. Psychiatry 2013, 54, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Wetherby, A.; Lord, C.; Woods, J.; Guthrie, W.; Pierce, K.; Shumway, S.; Thurm, A.; Ozonoff, S. The Early Screening for Autism and Communication Disorders (ESAC): Preliminary Field-Testing of An. Autism-Specific Screening Tool for Children 12 to 36 Months of Age. In Proceedings of the International Meeting for Autism Research, Chicago, IL, USA, 7–9 May 2009; Available online: https://www.researchgate.net/publication/268144139_The_Early_Screening_for_Autism_and_Communication_Disorders_ESAC_Preliminary_Field-Testing_of_An_Autism-Specific_Screening_Tool_for_Children_12_to_36_Months_of_Age (accessed on 15 October 2020).
- Akshoomoff, N. Use of the Mullen Scales of Early Learning for the assessment of young children with Autism Spectrum Disorders. Child. Neuropsychol. 2006, 12, 269–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, B.J.; Del’Homme, M.; Guthrie, D.; Zhang, F. Vineland Adaptive Behavior Scale Scores as a Function of Age and Initial IQ in 210 Autistic Children. J. Autism Dev. Disord. 1999, 29, 379–384. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research Electronic Data Capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houser, B. Bio-Rad’s Bio-Plex® suspension array system, xMAP technology overview. Arch. Physiol. Biochem. 2012, 118, 192–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wetie, A.G.N.; Wormwood, K.L.; Russell, S.; Ryan, J.P.; Darie, C.C.; Woods, A.G. A Pilot Proteomic Analysis of Salivary Biomarkers in Autism Spectrum Disorder. Autism Res. 2015, 8, 338–350. [Google Scholar] [CrossRef]
- Curran, E.A.; Dalman, C.; Kearney, P.M.; Kenny, L.C.; Cryan, J.F.; Dinan, T.G.; Khashan, A.S. Association Between Obstetric Mode of Delivery and Autism Spectrum Disorder: A Population-Based Sibling Design Study. JAMA Psychiatry 2015, 72, 935–942. [Google Scholar] [CrossRef] [Green Version]
- Tseng, P.-T.; Chen, Y.-W.; Stubbs, B.; Carvalho, A.F.; Whiteley, P.; Tang, C.-H.; Yang, W.-C.; Chen, T.-Y.; Li, D.-J.; Chu, C.-S.; et al. Maternal breastfeeding and autism spectrum disorder in children: A systematic review and meta-analysis. Nutr. Neurosci. 2019, 22, 354–362. [Google Scholar] [CrossRef]
- Bailey, A.; Le Couteur, A.; Gottesman, I.; Bolton, P.; Simonoff, E.; Yuzda, E.; Rutter, M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol. Med. 1995, 25, 63–77. [Google Scholar] [CrossRef]
- Rosenberg, R.E.; Law, J.K.; Yenokyan, G.; McGready, J.; Kaufmann, W.E.; Law, P.A. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch. Pediatrics Adolesc. Med. 2009, 163, 907–914. [Google Scholar] [CrossRef] [Green Version]
- Losh, M.; Adolphs, R.; Poe, M.D.; Couture, S.; Penn, D.; Baranek, G.T.; Piven, J. Neuropsychological profile of autism and the broad autism phenotype. Arch. Gen. Psychiatry 2009, 66, 518–526. [Google Scholar] [CrossRef] [Green Version]
- Warren, Z.E.; Foss-Feig, J.H.; Malesa, E.E.; Lee, E.B.; Taylor, J.L.; Newsom, C.R.; Crittendon, J.; Stone, W.L. Neurocognitive and behavioral outcomes of younger siblings of children with autism spectrum disorder at age five. J. Autism Dev. Disord. 2012, 42, 409–418. [Google Scholar] [CrossRef]
- Constantino, J.N. The quantitative nature of autistic social impairment. Pediatric Res. 2011, 69, 55R–62R. [Google Scholar] [CrossRef] [PubMed]
- Gamliel, I.; Yirmiya, N.; Jaffe, D.H.; Manor, O.; Sigman, M. Developmental trajectories in siblings of children with autism: Cognition and language from 4 months to 7 years. J. Autism Dev. Disord. 2009, 39, 1131–1144. [Google Scholar] [CrossRef] [Green Version]
- Pickles, A.; Starr, E.; Kazak, S.; Bolton, P.; Papanikolaou, K.; Bailey, A.; Goodman, R.; Rutter, M. Variable expression of the autism broader phenotype: Findings from extended pedigrees. J. Child. Psychol. Psychiatry Allied Discip. 2000, 41, 491–502. [Google Scholar] [CrossRef]
- Constantino, J.N.; Todd, R.D. Intergenerational transmission of subthreshold autistic traits in the general population. Biol. Psychiatry 2005, 57, 655–660. [Google Scholar] [CrossRef]
- Constantino, J.N.; Lajonchere, C.; Lutz, M.; Gray, T.; Abbacchi, A.; McKenna, K.; Singh, D.; Todd, R.D. Autistic social impairment in the siblings of children with pervasive developmental disorders. Am. J. Psychiatry 2006, 163, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Steer, C.D.; Golding, J.; Bolton, P.F. Traits contributing to the autistic spectrum. PLoS ONE 2010, 5, e12633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronald, A.; Happé, F.; Bolton, P.; Butcher, L.M.; Price, T.S.; Wheelwright, S.; Baron-Cohen, S.; Plomin, R. Genetic heterogeneity between the three components of the autism spectrum: A twin study. J. Am. Acad. Child Adolesc. Psychiatry 2006, 45, 691–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronald, A.; Larsson, H.; Anckarsäter, H.; Lichtenstein, P. A twin study of autism symptoms in Sweden. Mol. Psychiatry 2011, 16, 1039. [Google Scholar] [CrossRef]
- Geschwind, D.H. Autism: Many genes, common pathways? Cell 2008, 135, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Coleman, M.; Gillberg, C. The Biology of the Autistic Syndromes; Praeger: Westport, NY, USA, 1985; ISBN 0-275-91309-0. [Google Scholar]
- Abrahams, B.S.; Geschwind, D.H. Advances in autism genetics: On the threshold of a new neurobiology. Nat. Rev. Genet. 2008, 9, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, G.W.; Copeland, R.J. Glycomics hits the big time. Cell 2010, 143, 672–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasilewska, J.; Klukowski, M. Gastrointestinal symptoms and autism spectrum disorder: Links and risks-a possible new overlap syndrome. Pediatric Health Med. Ther. 2015, 6, 153–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Inclusion Criteria (All Must be Met) | Exclusion Criteria (Participant Will be Excluded from the Study if Any of the Criteria are Met) |
|---|---|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Troisi, J.; Autio, R.; Beopoulos, T.; Bravaccio, C.; Carraturo, F.; Corrivetti, G.; Cunningham, S.; Devane, S.; Fallin, D.; Fetissov, S.; et al. Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach. Brain Sci. 2020, 10, 743. https://doi.org/10.3390/brainsci10100743
Troisi J, Autio R, Beopoulos T, Bravaccio C, Carraturo F, Corrivetti G, Cunningham S, Devane S, Fallin D, Fetissov S, et al. Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach. Brain Sciences. 2020; 10(10):743. https://doi.org/10.3390/brainsci10100743
Chicago/Turabian StyleTroisi, Jacopo, Reija Autio, Thanos Beopoulos, Carmela Bravaccio, Federica Carraturo, Giulio Corrivetti, Stephen Cunningham, Samantha Devane, Daniele Fallin, Serguei Fetissov, and et al. 2020. "Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach" Brain Sciences 10, no. 10: 743. https://doi.org/10.3390/brainsci10100743
APA StyleTroisi, J., Autio, R., Beopoulos, T., Bravaccio, C., Carraturo, F., Corrivetti, G., Cunningham, S., Devane, S., Fallin, D., Fetissov, S., Gea, M., Giorgi, A., Iris, F., Joshi, L., Kadzielski, S., Kraneveld, A., Kumar, H., Ladd-Acosta, C., Leader, G., ... Fasano, A. (2020). Genome, Environment, Microbiome and Metabolome in Autism (GEMMA) Study Design: Biomarkers Identification for Precision Treatment and Primary Prevention of Autism Spectrum Disorders by an Integrated Multi-Omics Systems Biology Approach. Brain Sciences, 10(10), 743. https://doi.org/10.3390/brainsci10100743












