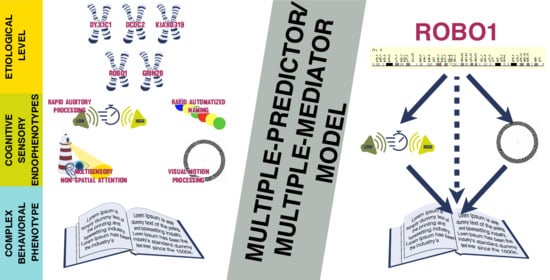
The Mediation Role of Dynamic Multisensory Processing Using Molecular Genetic Data in Dyslexia
Abstract
:1. Introduction
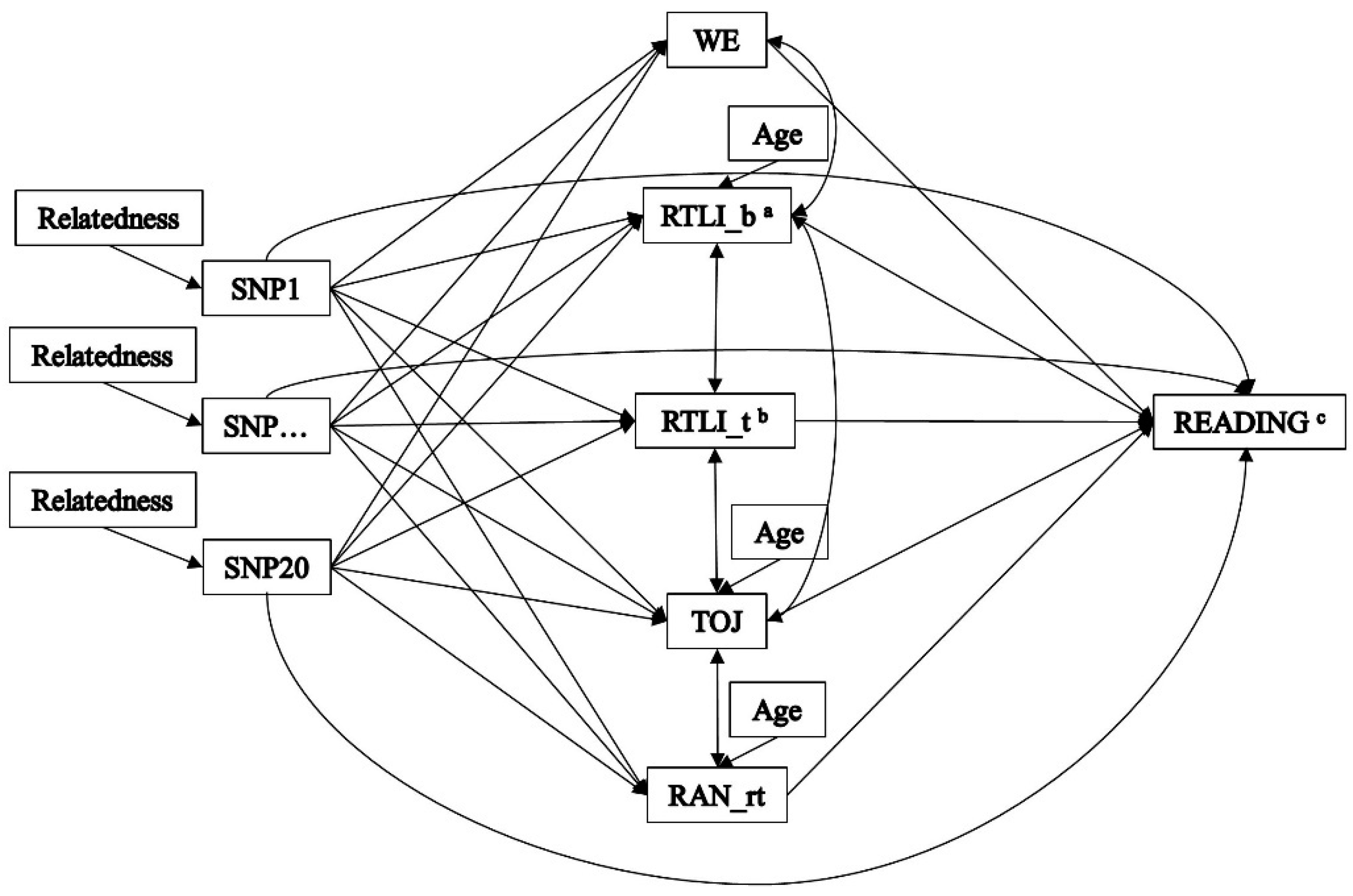
2. Materials and Methods
2.1. Sample
2.2. Genotypic Assessment
2.3. Endophenotypic Assessment
2.3.1. Rapid Auditory Processing: Temporal Order Judgment Task
2.3.2. Rapid Automatized Naming
2.3.3. Multisensory Non-Spatial Attention: Visual and Auditory Attention Tasks
2.3.4. Visual Motion Processing: The Rotating-Tilted-Lines Illusion—RTLI
2.4. Outcome Assessment
2.5. Statistical Analysis
3. Results
3.1. Bivariate Associations between Gene and EPs, Gene and Reading, and EPs and Reading
3.1.1. Bivariate Associations between Gene and EPs
3.1.2. Bivariate Associations between Gene and Reading
3.1.3. Bivariate Associations between EPs and Reading
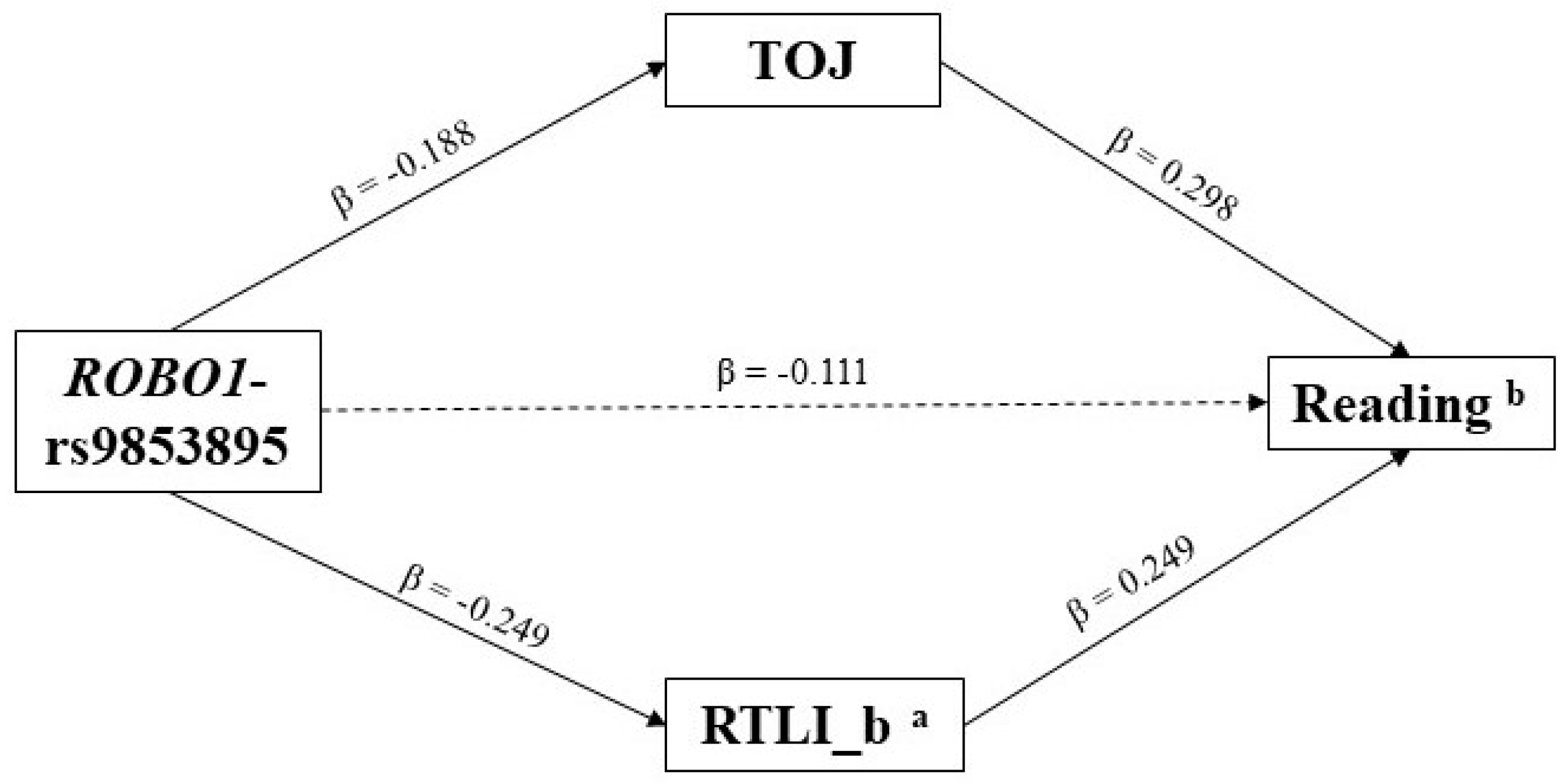
3.2. Indirect Effects—The Multiple-Predictor/Multiple-Mediator Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013. [Google Scholar]
- Peterson, R.L.; Pennington, B.F. Developmental Dyslexia. Annu. Rev. Clin. Psychol. 2015, 11, 283–307. [Google Scholar] [CrossRef]
- Sexton, C.C.; Gelhorn, H.L.; Bell, J.A.; Classi, P.M. The Co-occurrence of Reading Disorder and ADHD. J. Learn. Disabil. 2012, 45, 538–564. [Google Scholar] [CrossRef]
- Hallgren, B. Specific dyslexia (congenital word-blindness); a clinical and genetic study. Acta Psychiatr. Et Neurol. Suppl. 1950, 65, 1–287. [Google Scholar]
- Fisher, S.E.; DeFries, J.C. Developmental dyslexia: Genetic dissection of a complex cognitive trait. Nat. Rev. Neurosci. 2002, 3, 767–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gialluisi, A.; Newbury, D.F.; Wilcutt, E.G.; Olson, R.K.; DeFries, J.C.; Brandler, W.M.; Pennington, B.F.; Smith, S.D.; Scerri, T.S.; Simpson, N.H.; et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014, 13, 686–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gialluisi, A.; Andlauer, T.F.M.; Mirza-Schreiber, N.; Moll, K.; Becker, J.; Hoffmann, P.; Ludwig, K.U.; Czamara, D.; Pourcain, B.S.; Brandler, W.; et al. Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia. Transl. Psychiatry 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, D.T.; Adams, A.K.; Paniagua, S.; Frijters, J.C.; Boada, R.; Hill, D.E.; Lovett, M.W.; Mahone, E.M.; Willcutt, E.G.; Wolf, M.; et al. Multivariate genome-wide association study of rapid automatised naming and rapid alternating stimulus in Hispanic American and African–American youth. J. Med. Genet. 2019, 56, 557–566. [Google Scholar] [CrossRef]
- Gialluisi, A.; Andlauer, T.F.M.; Mirza-Schreiber, N.; Moll, K.; Becker, J.; Hoffmann, P.; Ludwig, K.U.; Czamara, D.; Pourcain, B.S.; Honbolygó, F.; et al. Genome-wide association study reveals new insights into the heritability and genetic correlates of developmental dyslexia. Mol. Psychiatry 2020. [Google Scholar] [CrossRef]
- Becker, J.; Czamara, D.; Scerri, T.S.; Ramus, F.; Csépe, V.; Talcott, J.B.; Stein, J.; Morris, A.; Ludwig, K.U.; Hoffmann, P.; et al. Genetic analysis of dyslexia candidate genes in the European cross-linguistic NeuroDys cohort. Eur. J. Hum. Genet. 2013, 22, 675–680. [Google Scholar] [CrossRef] [Green Version]
- Mascheretti, S.; De Luca, A.; Trezzi, V.; Peruzzo, D.; Nordio, A.; Marino, C.; Arrigoni, F. Neurogenetics of developmental dyslexia: From genes to behavior through brain neuroimaging and cognitive and sensorial mechanisms. Transl. Psychiatry 2017, 7, e987. [Google Scholar] [CrossRef] [Green Version]
- Marino, C.; Citterio, A.; Giorda, R.; Facoetti, A.; Menozzi, G.; Vanzin, L.; Lorusso, M.L.; Nobile, M.; Molteni, M. Association of short-term memory with a variant within DYX1C1 in developmental dyslexia. Genes Brain Behav. 2007, 6, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Marino, C.; Mascheretti, S.; Riva, V.; Cattaneo, F.; Rigoletto, C.; Rusconi, M.; Gruen, J.R.; Giorda, R.; Lazazzera, C.; Molteni, M. Pleiotropic effects of DCDC2 and DYX1C1 genes on language and mathematics traits in nuclear families of developmental dyslexia. Behav. Genet. 2010, 41, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Marino, C.; Meng, H.; Mascheretti, S.; Rusconi, M.; Cope, N.; Giorda, R.; Molteni, M.; Gruen, J.R. DCDC2 genetic variants and susceptibility to developmental dyslexia. Psychiatr. Genet. 2012, 22, 25–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascheretti, S.; Riva, V.; Giorda, R.; Beri, S.; Lanzoni, L.F.E.; Cellino, M.R.; Marino, C. KIAA0319 and ROBO1: Evidence on association with reading and pleiotropic effects on language and mathematics abilities in developmental dyslexia. J. Hum. Genet. 2014, 59, 189–197. [Google Scholar] [CrossRef]
- Mascheretti, S.; Facoetti, A.; Giorda, R.; Beri, S.; Riva, V.; Trezzi, V.; Cellino, M.R.; Marino, C. GRIN2B mediates susceptibility to intelligence quotient and cognitive impairments in developmental dyslexia. Psychiatr. Genet. 2015, 25, 9–20. [Google Scholar] [CrossRef]
- Trezzi, V.; Forni, D.; Giorda, R.; Villa, M.; Molteni, M.; Marino, C.; Mascheretti, S. The role of READ1 and KIAA0319 genetic variations in developmental dyslexia: Testing main and interactive effects. J. Hum. Genet. 2017, 62, 949–955. [Google Scholar] [CrossRef]
- Riva, V.; Mozzi, A.; Forni, D.; Trezzi, V.; Giorda, R.; Riva, S.; Villa, M.; Sironi, M.; Cagliani, R.; Mascheretti, S. The influence of DCDC2 risk genetic variants on reading: Testing main and haplotypic effects. Neuropsychologia 2019, 130, 52–58. [Google Scholar] [CrossRef]
- Gottesman, I.I.; Gould, T.D. The Endophenotype Concept in Psychiatry: Etymology and Strategic Intentions. Am. J. Psychiatry 2003, 160, 636–645. [Google Scholar] [CrossRef]
- Kendler, K.S.; Neale, M.C. Endophenotype: A conceptual analysis. Mol. Psychiatry 2010, 15, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Munafò, M.R. Candidate gene studies in the 21st century: Meta-analysis, mediation, moderation. Genes Brain Behav. 2006, 5, 3–8. [Google Scholar] [CrossRef]
- Kamradt, J.M.; Nigg, J.T.; Friderici, K.H.; Nikolas, M.A. Neuropsychological performance measures as intermediate phenotypes for attention-deficit/hyperactivity disorder: A multiple mediation analysis. Dev. Psychopathol. 2017, 29, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Braff, D.L. The importance of endophenotypes in schizophrenia research. Schizophr. Res. 2015, 163, 1–8. [Google Scholar] [CrossRef]
- Flint, J.; Munafò, M.R. The endophenotype concept in psychiatric genetics. Psychol. Med. 2007, 37, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Flint, J.; Timpson, N.J.; Munafò, M.R. Assessing the utility of intermediate phenotypes for genetic mapping of psychiatric disease. Trends Neurosci. 2014, 37, 733–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szatmari, P.; Maziade, M.; Zwaigenbaum, L.; Mérette, C.; Roy, M.-A.; Joober, R.; Palmour, R. Informative phenotypes for genetic studies of psychiatric disorders. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007, 144B, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Boets, B.; Vandermosten, M.; Cornelissen, P.; Wouters, J.; Ghesquière, P. Coherent Motion Sensitivity and Reading Development in the Transition From Prereading to Reading Stage. Child Dev. 2011, 82, 854–869. [Google Scholar] [CrossRef] [PubMed]
- Cantiani, C.; Riva, V.; Piazza, C.; Bettoni, R.; Molteni, M.; Choudhury, N.; Marino, C.; Benasich, A.A. Auditory discrimination predicts linguistic outcome in Italian infants with and without familial risk for language learning impairment. Dev. Cogn. Neurosci. 2016, 20, 23–34. [Google Scholar] [CrossRef] [Green Version]
- Castles, A.; Coltheart, M. Is there a causal link from phonological awareness to success in learning to read? Cognition 2004, 91, 77–111. [Google Scholar] [CrossRef]
- Clark, K.A.; Helland, T.; Specht, K.; Narr, K.L.; Manis, F.R.; Toga, A.W.; Hugdahl, K. Neuroanatomical precursors of dyslexia identified from pre-reading through to age 11. Brain 2014, 137, 3136–3141. [Google Scholar] [CrossRef]
- Cornelissen, P.; Richardson, A.; Mason, A.; Fowler, S.; Stein, J. Contrast sensitivity and coherent motion detection measured at photopic luminance levels in dyslexics and controls. Vis. Res. 1995, 35, 1483–1494. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, S.; Gori, S.; Ruffino, M.; Pedrolli, K.; Facoetti, A. A Causal Link between Visual Spatial Attention and Reading Acquisition. Curr. Biol. 2012, 22, 814–819. [Google Scholar] [CrossRef] [Green Version]
- Gori, S.; Facoetti, A. Perceptual learning as a possible new approach for remediation and prevention of developmental dyslexia. Vis. Res. 2014, 99, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gori, S.; Facoetti, A. How the visual aspects can be crucial in reading acquisition? The intriguing case of crowding and developmental dyslexia. J. Vis. 2015, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Gori, S.; Molteni, M.; Facoetti, A. Visual Illusions: An Interesting Tool to Investigate Developmental Dyslexia and Autism Spectrum Disorder. Front. Hum. Neurosci. 2016, 10, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gori, S.; Seitz, A.R.; Ronconi, L.; Franceschini, S.; Facoetti, A. Multiple Causal Links Between Magnocellular–Dorsal Pathway Deficit and Developmental Dyslexia. Cereb. Cortex 2016, 26, 4356–4369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hämäläinen, J.A.; Guttorm, T.K.; Richardson, U.; Alku, P.; Lyytinen, H.; Leppänen, P.H.T. Auditory Event-Related Potentials Measured in Kindergarten Predict Later Reading Problems at School Age. Dev. Neuropsychol. 2013, 38, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Hari, R.; Renvall, H. Impaired processing of rapid stimulus sequences in dyslexia. Trends Cogn. Sci. 2001, 5, 525–532. [Google Scholar] [CrossRef]
- Hornickel, J.; Kraus, N. Unstable Representation of Sound: A Biological Marker of Dyslexia. J. Neurosci. 2013, 33, 3500–3504. [Google Scholar] [CrossRef] [Green Version]
- Kevan, A.; Pammer, K. Predicting early reading skills from pre-reading measures of dorsal stream functioning. Neuropsychologia 2009, 47, 3174–3181. [Google Scholar] [CrossRef]
- Lallier, M.; Donnadieu, S.; Valdois, S. Investigating the role of visual and auditory search in reading and developmental dyslexia. Front. Hum. Neurosci. 2013, 7, 597. [Google Scholar] [CrossRef] [Green Version]
- Leppänen, P.H.T.; Hämäläinen, J.A.; Salminen, H.K.; Eklund, K.M.; Guttorm, T.K.; Lohvansuu, K.; Puolakanaho, A.; Lyytinen, H. Newborn brain event-related potentials revealing atypical processing of sound frequency and the subsequent association with later literacy skills in children with familial dyslexia. Cortex 2010, 46, 1362–1376. [Google Scholar] [CrossRef] [PubMed]
- Lervåg, A.; Hulme, C. Rapid Automatized Naming (RAN) Taps a Mechanism That Places Constraints on the Development of Early Reading Fluency. Psychol. Sci. 2009, 20, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Molfese, D.L. Predicting Dyslexia at 8 Years of Age Using Neonatal Brain Responses. Brain Lang. 2000, 72, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Norton, E.S.; Wolf, M. Rapid Automatized Naming (RAN) and Reading Fluency: Implications for Understanding and Treatment of Reading Disabilities. Annu. Rev. Psychol. 2012, 63, 427–452. [Google Scholar] [CrossRef] [Green Version]
- Plaza, M.; Cohen, H. The contribution of phonological awareness and visual attention in early reading and spelling. Dyslexia 2007, 13, 67–76. [Google Scholar] [CrossRef]
- Stein, J.F. The current status of the magnocellular theory of developmental dyslexia. Neuropsychologia 2019, 130, 66–77. [Google Scholar] [CrossRef]
- Van Der Leij, A.; Van Bergen, E.; Van Zuijen, T.L.; De Jong, P.; Maurits, N.M.; Maassen, B.A.M. Precursors of Developmental Dyslexia: An Overview of the Longitudinal Dutch Dyslexia Programme Study. Dyslexia 2013, 19, 191–213. [Google Scholar] [CrossRef]
- Vandermosten, M.; Boets, B.; Luts, H.; Poelmans, H.; Golestani, N.; Wouters, J.; Ghesquière, P. Adults with dyslexia are impaired in categorizing speech and nonspeech sounds on the basis of temporal cues. Proc. Natl. Acad. Sci. USA 2010, 107, 10389–10394. [Google Scholar] [CrossRef] [Green Version]
- Vidyasagar, T.R.; Pammer, K. Dyslexia: A deficit in visuo-spatial attention, not in phonological processing. Trends Cogn. Sci. 2010, 14, 57–63. [Google Scholar] [CrossRef]
- Mascheretti, S.; Gori, S.; Trezzi, V.; Ruffino, M.; Facoetti, A.; Marino, C. Visual motion and rapid auditory processing are solid endophenotypes of developmental dyslexia. Genes Brain Behav. 2018, 17, 70–81. [Google Scholar] [CrossRef] [Green Version]
- Brewer, C.C.; Zalewski, C.K.; King, K.; Zobay, O.; Riley, A.; Ferguson, M.; Bird, J.E.; McCabe, M.M.; Hood, L.J.; Drayna, D.; et al. Heritability of non-speech auditory processing skills. Eur. J. Hum. Genet. 2016, 24, 1137–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byrne, B.; Wadsworth, S.; Corley, R.; Samuelsson, S.; Quain, P.; DeFries, J.C.; Willcutt, E.; Olson, R.K. Longitudinal Twin Study of Early Literacy Development: Preschool and Kindergarten Phases. Sci. Stud. Read. 2005, 9, 219–235. [Google Scholar] [CrossRef]
- Davis, C.J.; Knopik, V.S.; Olson, R.K.; Wadsworth, S.J.; DeFries, J.C. Genetic and environmental influences on rapid naming and reading ability: A twin study. Ann. Dyslexia 2001, 51, 231–247. [Google Scholar] [CrossRef]
- Olson, R.K.; Hulslander, J.; Christopher, M.; Keenan, J.M.; Wadsworth, S.J.; Willcutt, E.G.; Pennington, B.F.; DeFries, J.C. Genetic and environmental influences on writing and their relations to language and reading. Ann. Dyslexia 2013, 63, 25–43. [Google Scholar] [CrossRef] [Green Version]
- Petrill, S.A.; Deater-Deckard, K.; Thompson, L.A.; De Thorne, L.S.; Schatschneider, C. Reading skills in early readers: Genetic and shared environmental influences. J. Learn. Disabil. 2006, 39, 48–55. [Google Scholar] [CrossRef]
- Willcutt, E.G.; Pennington, B.F.; Duncan, L.; Smith, S.D.; Keenan, J.M.; Wadsworth, S.; DeFries, J.C.; Olson, R.K. Understanding the Complex Etiologies of Developmental Disorders: Behavioral and Molecular Genetic Approaches. J. Dev. Behav. Pediatr. 2010, 31, 533–544. [Google Scholar] [CrossRef]
- Cicchini, G.M.; Marino, C.; Mascheretti, S.; Perani, D.; Morrone, M.C. Strong Motion Deficits in Dyslexia Associated with DCDC2 Gene Alteration. J. Neurosci. 2015, 35, 8059–8064. [Google Scholar] [CrossRef] [Green Version]
- Gori, S.; Mascheretti, S.; Giora, E.; Ronconi, L.; Ruffino, M.; Quadrelli, E.; Facoetti, A.; Marino, C. The DCDC2 Intron 2 Deletion Impairs Illusory Motion Perception Unveiling the Selective Role of Magnocellular-Dorsal Stream in Reading (Dis)ability. Cereb. Cortex 2015, 25, 1685–1695. [Google Scholar] [CrossRef]
- Wigg, K.G.; Couto, J.M.; Feng, Y.; Anderson, B.; Cate-Carter, T.D.; Macciardi, F.; Tannock, R.; Lovett, M.W.; Humphries, T.W.; Barr, C.L. Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol. Psychiatry 2004, 9, 1111–1121. [Google Scholar] [CrossRef]
- Rendall, A.R.; Tarkar, A.; Contreras-Mora, H.M.; LoTurco, J.J.; Fitch, R.H. Deficits in learning and memory in mice with a mutation of the candidate dyslexia susceptibility gene Dyx1c. Brain Lang. 2017, 172, 30–38. [Google Scholar] [CrossRef]
- Szalkowski, C.E.; Booker, A.B.; Truong, N.T.; Threlkeld, S.W.; Rosen, G.D.; Fitch, R.H. Knockdown of the candidate dyslexia susceptibility gene homolog dyx1c1 in rodents: Effects on auditory processing, visual attention, and cortical and thalamic anatomy. Dev. Neurosci. 2013, 35, 50–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Threlkeld, S.W.; McClure, M.M.; Bai, J.; Wang, Y.; LoTurco, J.J.; Rosen, G.D.; Fitch, R.H. Developmental disruptions and behavioral impairments in rats following in utero RNAi of Dyx1c1. Brain Res. Bull. 2007, 71, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centanni, T.M.; Booker, A.B.; Sloan, A.M.; Chen, F.; Maher, B.J.; Carraway, R.S.; Khodaparast, N.; Rennaker, R.; LoTurco, J.J.; Kilgard, M.P. Knockdown of the Dyslexia-Associated Gene Kiaa0319 Impairs Temporal Responses to Speech Stimuli in Rat Primary Auditory Cortex. Cereb. Cortex 2014, 24, 1753–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, D.T.; Che, A.; Rendall, A.R.; Szalkowski, C.E.; LoTurco, J.J.; Galaburda, A.M.; Fitch, R.H. Mutation ofDcdc2in mice leads to impairments in auditory processing and memory ability. Genes Brain Behav. 2014, 13, 802–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centanni, T.M.; Booker, A.B.; Chen, F.; Sloan, A.M.; Carraway, R.S.; Rennaker, R.L.; LoTurco, J.J.; Kilgard, M.P. Knockdown of Dyslexia-Gene Dcdc2 Interferes with Speech Sound Discrimination in Continuous Streams. J. Neurosci. 2017, 36, 4895–4906. [Google Scholar] [CrossRef] [Green Version]
- Che, A.; Girgenti, M.J.; LoTurco, J.J. The dyslexia-associated gene DCDC2 is required for spike-timing precision in mouse neocortex. Biol. Psychiatry 2013, 76, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Che, F.; Zhang, Y.; Wang, G.; Heng, X.; Liu, S.; Du, Y. The role of GRIN2B in Tourette syndrome: Results from a transmission disequilibrium study. J. Affect. Disord. 2015, 187, 62–65. [Google Scholar] [CrossRef]
- Hayes, A.F. Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach; Guilford Press: New York, NY, USA, 2013. [Google Scholar]
- Preacher, K.J.; Hayes, A.F. Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behav. Res. Methods 2008, 40, 879–891. [Google Scholar] [CrossRef]
- Facoetti, A.; Trussardi, A.N.; Ruffino, M.; Lorusso, M.L.; Cattaneo, C.; Galli, R.; Molteni, M.; Zorzi, M. Multisensory Spatial Attention Deficits Are Predictive of Phonological Decoding Skills in Developmental Dyslexia. J. Cogn. Neurosci. 2010, 22, 1011–1025. [Google Scholar] [CrossRef]
- Cornoldi, C.; Colpo, G.; Gruppo, M.T. Prove di Lettura MT per la Scuola Elementare—2; Organizzazioni Speciali: Firenze, Italy, 1998. [Google Scholar]
- Sartori, S.; Job, R.; Tressoldi, P.E. Batteria per la Valutazione Della Dislessia e Della Disortografia Evolutiva; Organizzazioni Speciali: Firenze, Italy, 1995. [Google Scholar]
- Muthén, L.K.; Muthén, B. Mplus User’s Guide, 7th ed.; Muthén & Muthén: Los Angeles, CA, USA, 2014. [Google Scholar]
- Fritz, M.S.; MacKinnon, D.P. Required Sample Size to Detect the Mediated Effect. Psychol. Sci. 2007, 18, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Tofighi, D.; MacKinnon, D.P. RMediation: An R package for mediation analysis confidence intervals. Behav. Res. Methods 2011, 43, 692–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennington, B.F. From single to multiple deficit models of developmental disorders. Cognition 2006, 101, 385–413. [Google Scholar] [CrossRef] [PubMed]
- McGrath, L.M.; Peterson, R.L.; Pennington, B.F. The Multiple Deficit Model: Progress, Problems, and Prospects. Sci. Stud. Read. 2020, 24, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Archer, K.; Pammer, K.; Vidyasagar, T.R. A Temporal Sampling Basis for Visual Processing in Developmental Dyslexia. Front. Hum. Neurosci. 2020, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Boets, B.; Wouters, J.; Van Wieringen, A.; De Smedt, B.; Ghesquière, P. Modelling relations between sensory processing, speech perception, orthographic and phonological ability, and literacy achievement. Brain Lang. 2008, 106, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Lawton, T. Improving Dorsal Stream Function in Dyslexics by Training Figure/Ground Motion Discrimination Improves Attention, Reading Fluency, and Working Memory. Front. Hum. Neurosci. 2016, 10, 397. [Google Scholar] [CrossRef] [Green Version]
- Stein, J. The magnocellular theory of developmental dyslexia. Dyslexia 2001, 7, 12–36. [Google Scholar] [CrossRef]
- Stein, J.; Talcott, J. Impaired neuronal timing in developmental dyslexia—the magnocellular hypothesis. Dyslexia 1999, 5, 59–77. [Google Scholar] [CrossRef]
- Stein, J. To see but not to read; the magnocellular theory of dyslexia. Trends Neurosci. 1997, 20, 147–152. [Google Scholar] [CrossRef]
- Talcott, J.B.; Hansen, P.C.; Assoku, E.L.; Stein, J.F. Visual motion sensitivity in dyslexia: Evidence for temporal and energy integration deficits. Neuropsychologia 2000, 38, 935–943. [Google Scholar] [CrossRef] [Green Version]
- Talcott, J.B.; Witton, C.; Hebb, G.S.; Stoodley, C.J.; Westwood, E.A.; France, S.J.; Hansen, P.C.; Stein, J.F. On the relationship between dynamic visual and auditory processing and literacy skills; results from a large primary-school study. Dyslexia 2002, 8, 204–225. [Google Scholar] [CrossRef] [PubMed]
- Snowling, M.J.; Hulme, C. Annual Research Review: Reading disorders revisited—The critical importance of oral language. J. Child Psychol. Psychiatry 2020. [Google Scholar] [CrossRef] [PubMed]
- Goswami, U. The neural basis of dyslexia may originate in primary auditory cortex. Brain 2014, 137, 3100–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, J.F. Dyslexia: The Role of Vision and Visual Attention. Curr. Dev. Disord. Rep. 2014, 1, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Pammer, K.; Hansen, P.C.; Holliday, I.; Cornelissen, P. Attentional shifting and the role of the dorsal pathway in visual word recognition. Neuropsychologia 2006, 44, 2926–2936. [Google Scholar] [CrossRef]
- Renvall, H.; Hari, R. Auditory Cortical Responses to Speech-Like Stimuli in Dyslexic Adults. J. Cogn. Neurosci. 2002, 14, 757–768. [Google Scholar] [CrossRef] [Green Version]
- Vidyasagar, T.R. Reading into neuronal oscillations in the visual system: Implications for developmental dyslexia. Front. Hum. Neurosci. 2013, 7, 811. [Google Scholar] [CrossRef] [Green Version]
- Vidyasagar, T.R.; Pammer, K. Impaired visual search in dyslexia relates to the role of the magnocellular pathway in attention. NeuroReport 1999, 10, 1283–1287. [Google Scholar] [CrossRef]
- Denison, R.N.; Vu, A.T.; Yacoub, E.; Feinberg, D.A.; Silver, M.A. Functional mapping of the magnocellular and parvocellular subdivisions of human LGN. NeuroImage 2014, 102, 358–369. [Google Scholar] [CrossRef] [Green Version]
- Eden, G.F.; VanMeter, J.W.; Rumsey, J.M.; Maisog, J.M.; Woods, R.P.; Zeffiro, T.A. Abnormal processing of visual motion in dyslexia revealed by functional brain imaging. Nat. Cell Biol. 1996, 382, 66–69. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, H.; Wen, W.; He, S. Layer-specific response properties of the human lateral geniculate nucleus and superior colliculus. NeuroImage 2015, 111, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Belin, P.; Zatorre, R.J. ’What’, ’where’ and ’how’ in auditory cortex. Nat. Neurosci. 2000, 3, 965–966. [Google Scholar] [CrossRef] [PubMed]
- Kaas, J.H.; Hackett, T.A. ’What’ and ’where’ processing in auditory cortex. Nat. Neurosci. 1999, 2, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Maeder, P.P.; Meuli, R.A.; Adriani, M.; Bellmann, A.; Fornari, E.; Thiran, J.-P.; Pittet, A.; Clarke, S. Distinct Pathways Involved in Sound Recognition and Localization: A Human fMRI Study. NeuroImage 2001, 14, 802–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanski, L.M.; Tian, B.; Fritz, J.; Mishkin, M.; Goldman-Rakic, P.S.; Rauschecker, J.P. Dual streams of auditory afferents target multiple domains in the primate prefrontal cortex. Nat. Neurosci. 1999, 2, 1131–1136. [Google Scholar] [CrossRef] [PubMed]
- Zatorre, R.J. Spectral and Temporal Processing in Human Auditory Cortex. Cereb. Cortex 2001, 11, 946–953. [Google Scholar] [CrossRef]
- Frey, A.; François, C.; Chobert, J.; Besson, M.; Ziegler, J.C. Behavioral and electrophysiological investigation of speech perception deficits in silence, noise and envelope conditions in developmental dyslexia. Neuropsychologia 2019, 130, 3–12. [Google Scholar] [CrossRef]
- Geiger, G.; Cattaneo, C.; Galli, R.; Pozzoli, U.; Lorusso, M.L.; Facoetti, A.; Molteni, M. Wide and Diffuse Perceptual Modes Characterize Dyslexics in Vision and Audition. Perception 2008, 37, 1745–1764. [Google Scholar] [CrossRef]
- Hancock, R.; Pugh, K.R.; Hoeft, F. Correction: Neural Noise Hypothesis of Developmental Dyslexia. Trends Cogn. Sci. 2017, 21, 909. [Google Scholar] [CrossRef] [Green Version]
- Sperling, A.J.; Lu, Z.-L.; Manis, F.R.; Seidenberg, M.S. Deficits in perceptual noise exclusion in developmental dyslexia. Nat. Neurosci. 2005, 8, 862–863. [Google Scholar] [CrossRef]
- Sperling, A.J.; Lu, Z.-L.; Manis, F.R.; Seidenberg, M.S. Motion-Perception Deficits and Reading Impairment. Psychol. Sci. 2006, 17, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W.; Liapi, A.; Plachez, C.; Camurri, L.; Zhang, J.; Mori, S.; Murakami, F.; Parnavelas, J.G.; Sundaresan, V.; Richards, L.J. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development 2006, 133, 2243–2252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Okanoya, K.; Koike, T.; Sasaki, E.; Okano, H.; Watanabe, S.; Iriki, A. Human speech- and reading-related genes display partially overlapping expression patterns in the marmoset brain. Brain Lang. 2014, 133, 26–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannula-Jouppi, K.; Kaminen-Ahola, N.; Taipale, M.; Eklund, R.; Nopola-Hemmi, J.; Kääriäinen, H.; Kere, J. The Axon Guidance Receptor Gene ROBO1 Is a Candidate Gene for Developmental Dyslexia. PLoS Genet. 2005, 1, e50. [Google Scholar] [CrossRef] [PubMed]
- Kidd, T.; Bland, K.S.; Goodman, C.S. Slit Is the Midline Repellent for the Robo Receptor in Drosophila. Cell 1999, 96, 785–794. [Google Scholar] [CrossRef] [Green Version]
- Kidd, T.; Brose, K.; Mitchell, K.J.; Fetter, R.D.; Tessier-Lavigne, M.; Goodman, C.S.; Tear, G. Roundabout Controls Axon Crossing of the CNS Midline and Defines a Novel Subfamily of Evolutionarily Conserved Guidance Receptors. Cell 1998, 92, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Massinen, S.; Wang, J.; Laivuori, K.; Bieder, A.; Tapia-Páez, I.; Jiao, H.; Kere, J. Genomic sequencing of a dyslexia susceptibility haplotype encompassing ROBO1. J. Neurodev. Disord. 2016, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Nguyen-Ba-Charvet, K.T.; Brose, K.; Marillat, V.; Kidd, T.; Goodman, C.S.; Tessier-Lavigne, M.; Sotelo, C.; Chédotal, A. Slit2-Mediated Chemorepulsion and Collapse of Developing Forebrain Axons. Neuron 1999, 22, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Seeger, M.; Tear, G.; Ferres-Marco, D.; Goodman, C.S. Mutations affecting growth cone guidance in drosophila: Genes necessary for guidance toward or away from the midline. Neuron 1993, 10, 409–426. [Google Scholar] [CrossRef]
- Whitford, K.L.; Marillat, V.; Stein, E.; Goodman, C.S.; Tessier-Lavigne, M.; Chédotal, A.; Ghosh, A. Regulation of Cortical Dendrite Development by Slit-Robo Interactions. Neuron 2002, 33, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Andrews, W.D.; Barber, M.; Hernadez-Miranda, L.R.; Xian, J.; Rakic, S.; Sundaresan, V.; Rabbitts, T.H.; Pannell, R.; Rabbitts, P.; Thompson, H.; et al. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 2008, 313, 648–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Meglio, T.; Nguyen-Ba-Charvet, K.T.; Tessier-Lavigne, M.; Sotelo, C.; Chédotal, A. Molecular Mechanisms Controlling Midline Crossing by Precerebellar Neurons. J. Neurosci. 2008, 28, 6285–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Miranda, L.R.; Cariboni, A.; Faux, C.; Ruhrberg, C.; Cho, J.H.; Cloutier, J.-F.; Eickholt, B.J.; Parnavelas, J.G.; Andrews, W.D. Robo1 Regulates Semaphorin Signaling to Guide the Migration of Cortical Interneurons through the Ventral Forebrain. J. Neurosci. 2011, 31, 6174–6187. [Google Scholar] [CrossRef] [PubMed]
- Gonda, Y.; Andrews, W.D.; Tabata, H.; Namba, T.; Parnavelas, J.G.; Nakajima, K.; Kohsaka, S.; Hanashima, C.; Uchino, S. Robo1 Regulates the Migration and Laminar Distribution of Upper-Layer Pyramidal Neurons of the Cerebral Cortex. Cereb. Cortex 2013, 23, 1495–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominici, C.; Rappeneau, Q.; Zelina, P.; Fouquet, S.; Chédotal, A. Non-cell autonomous control of precerebellar neuron migration by Slits and Robos. Development 2018, 145, 150375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darki, F.; Peyrard-Janvid, M.; Matsson, H.; Kere, J.; Klingberg, T. Three Dyslexia Susceptibility Genes, DYX1C1, DCDC2, and KIAA0319, Affect Temporo-Parietal White Matter Structure. Biol. Psychiatry 2012, 72, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Darki, F.; Peyrard-Janvid, M.; Matsson, H.; Kere, J.; Klingberg, T. DCDC2 Polymorphism Is Associated with Left Temporoparietal Gray and White Matter Structures during Development. J. Neurosci. 2014, 34, 14455–14462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascheretti, S.; Perdue, M.V.; Feng, B.; Andreola, C.; Dionne, G.; Jasińska, K.K.; Pugh, K.R.; Grigorenko, E.L.; Landi, N. From BDNF to Reading: Neural Activation and Phonological Processing as Multiple Mediators. Behav. Brain Res. 2020, 396, 112859. [Google Scholar] [CrossRef]
- Perdue, M.V.; Mascheretti, S.; Kornilov, S.A.; Jasińska, K.K.; Ryherd, K.; Mencl, W.E.; Frost, S.J.; Grigorenko, E.L.; Pugh, K.R.; Landi, N. Common variation within the SETBP1 gene is associated with reading-related skills and patterns of functional neural activation. Neuropsychologia 2019, 130, 44–51. [Google Scholar] [CrossRef]
- Gabrieli, J.D.E. Dyslexia: A New Synergy Between Education and Cognitive Neuroscience. Science 2009, 325, 280–283. [Google Scholar] [CrossRef] [Green Version]
- Goswami, U. Why theories about developmental dyslexia require developmental designs. Trends Cogn. Sci. 2003, 7, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Melby-Lervåg, M.; Lyster, S.-A.H.; Hulme, C. Phonological skills and their role in learning to read: A meta-analytic review. Psychol. Bull. 2012, 138, 322–352. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.L.; Pennington, B.F. Developmental dyslexia. Lancet 2012, 379, 1997–2007. [Google Scholar] [CrossRef] [Green Version]
- Shaywitz, B.; Shaywitz, S.; Blachman, B.; Pugh, K.R.; Fulbright, R.K.; Skudlarski, P.; Mencl, W.; Constable, R.; Holahan, J.M.; Marchione, K.E.; et al. Development of left occipitotemporal systems for skilled reading in children after a phonologically- based intervention. Biol. Psychiatry 2004, 55, 926–933. [Google Scholar] [CrossRef]
- Snowling, M.J. From language to reading and dyslexia. Dyslexia 2001, 7, 37–46. [Google Scholar] [CrossRef]
- Vellutino, F.R.; Fletcher, J.M.; Snowling, M.J.; Scanlon, D.M. Specific reading disability (dyslexia): What have we learned in the past four decades? J. Child Psychol. Psychiatry 2004, 45, 2–40. [Google Scholar] [CrossRef]
- De Vos, A.; Vanvooren, S.; Vanderauwera, J.; Ghesquière, P.; Wouters, J. Atypical neural synchronization to speech envelope modulations in dyslexia. Brain Lang. 2017, 164, 106–117. [Google Scholar] [CrossRef]
- De Vos, A.; Vanvooren, S.; Vanderauwera, J.; Ghesquière, P.; Wouters, J. A longitudinal study investigating neural processing of speech envelope modulation rates in children with (a family risk for) dyslexia. Cortex 2017, 93, 206–219. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Montag, C.; Ebstein, R.P.; Jawinski, P.; Markett, S. Molecular genetics in psychology and personality neuroscience: On candidate genes, genome wide scans, and new research strategies. Neurosci. Biobehav. Rev. 2020, 118, 163–174. [Google Scholar] [CrossRef]
- Moore, S.R. Commentary: What is the case for candidate gene approaches in the era of high-throughput genomics? A response to Border and Keller (2017). J. Child Psychol. Psychiatry 2017, 58, 331–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKinnon, D.P.; Lockwood, C.M.; Williams, J. Confidence Limits for the Indirect Effect: Distribution of the Product and Resampling Methods. Multivar. Behav. Res. 2004, 39, 99–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacKinnon, D.P.; Fritz, M.S.; Williams, J.; Lockwood, C.M. Distribution of the product confidence limits for the indirect effect: Program PRODCLIN. Behav. Res. Methods 2007, 39, 384–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoemmes, F.; MacKinnon, D.P.; Reiser, M.R. Power Analysis for Complex Mediational Designs Using Monte Carlo Methods. Struct. Equ. Model. A Multidiscip. J. 2010, 17, 510–534. [Google Scholar] [CrossRef]
- Gaab, N.; Gabrieli, J.D.; Deutsch, G.K.; Tallal, P.; Temple, E. Neural correlates of rapid auditory processing are disrupted in children with developmental dyslexia and ameliorated with training: An fMRI study. Restor. Neurol. Neurosci. 2007, 25, 295–310. [Google Scholar]
- Tallal, P. Improving neural response to sound improves reading. Proc. Natl. Acad. Sci. USA 2012, 109, 16406–16407. [Google Scholar] [CrossRef] [Green Version]
- Temple, E.; Deutsch, G.K.; Poldrack, R.A.; Miller, S.L.; Tallal, P.; Merzenich, M.M.; Gabrieli, J.D.E. Neural deficits in children with dyslexia ameliorated by behavioral remediation: Evidence from functional MRI. Proc. Natl. Acad. Sci. USA 2003, 100, 2860–2865. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, S.; Bertoni, S. Improving action video games abilities increases the phonological decoding speed and phonological short-term memory in children with developmental dyslexia. Neuropsychologia 2019, 130, 100–106. [Google Scholar] [CrossRef]
- Franceschini, S.; Gori, S.; Ruffino, M.; Viola, S.; Molteni, M.; Facoetti, A. Action Video Games Make Dyslexic Children Read Better. Curr. Biol. 2013, 23, 462–466. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, S.; Trevisan, P.; Ronconi, L.; Bertoni, S.; Colmar, S.; Double, K.; Facoetti, A.; Gori, S. Action video games improve reading abilities and visual-to-auditory attentional shifting in English-speaking children with dyslexia. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
| Allele | Frequency in Unrelated Subjects * | Hardy-Weinberg Equilibrium | ||
|---|---|---|---|---|
| DYX1C1 | rs3743205 | G | 0.927 | 0.018 |
| A | 0.073 | |||
| rs57809907 | G | 0.891 | 0.984 | |
| T | 0.109 | |||
| rs189983504 | C | 0.899 | 0.546 | |
| G | 0.101 | |||
| DCDC2 | rs793842 | C | 0.584 | 0.196 |
| T | 0.416 | |||
| READ1 | Deletion ° | 0.078 | 0.286 | |
| rs793862 | G | 0.758 | 0.581 | |
| A | 0.242 | |||
| KIAA0319 | rs4504469 | C | 0.634 | 0.527 |
| T | 0.366 | |||
| rs2038137 | G | 0.642 | 0.556 | |
| T | 0.358 | |||
| rs9461045 | C | 0.792 | 0.306 | |
| T | 0.208 | |||
| rs2143340 § | A | 0.839 | 0.181 | |
| G | 0.161 | |||
| ROBO1 | rs333491 | A | 0.545 | 0.378 |
| G | 0.455 | |||
| rs6803202 | C | 0.494 | 0.361 | |
| T | 0.506 | |||
| rs9853895 | C | 0.587 | 0.232 | |
| T | 0.413 | |||
| rs7644521 | T | 0.836 | 0.744 | |
| C | 0.164 | |||
| GRIN2B | rs5796555 | - | 0.694 | 0.498 |
| A | 0.306 | |||
| rs1012586 | G | 0.695 | 0.126 | |
| C | 0.305 | |||
| rs2268119 | A | 0.768 | 0.729 | |
| T | 0.232 | |||
| rs2216128 | A | 0.784 | 0.593 | |
| G | 0.216 | |||
| rs11609779 | C | 0.817 | 0.310 | |
| T | 0.183 | |||
| rs2192973 | G | 0.775 | 0.410 | |
| A | 0.225 | |||
| GENE | SNP | COGNITIVE ENDOPHENOTYPES | READING # | ||||
|---|---|---|---|---|---|---|---|
| ATTENTION | VISUAL MOTION PROCESSING | RAP | RAN | ||||
| WE | RTLI_b a | RTLI_t b | RAN_rt | ||||
| DYX1C1 | rs3743205G/A | −0.012 | 0.147 * | −0.065 | 0.071 | 0.044 | −0.007 |
| rs57809907G/T | 0.016 | 0.100 | −0.082 | 0.124 * | 0.030 | −0.091 | |
| rs189983504C/G | 0.044 | 0.113 | −0.074 | 0.031 | 0.173 ** | 0.066 | |
| DCDC2 | rs793842C/T | 0.058 | −0.019 | 0.001 | −0.080 | 0.108 | −0.127 * |
| READ1-Deletion ° | 0.004 | 0.052 | −0.057 | −0.010 | −0.025 | 0.090 | |
| rs793862G/A | 0.045 | −0.048 | 0.032 | −0.008 | 0.056 | −0.083 | |
| KIAA0319 | rs4504469C/T | 0.032 | −0.104 | 0.080 | −0.016 | 0.073 | −0.014 |
| rs2038137G/T | 0.028 | −0.008 | −0.016 | 0.010 | 0.026 | 0.065 | |
| rs9461045C/T | −0.063 | −0.043 | 0.034 | 0.030 | 0.021 | −0.071 | |
| rs2143340A/G § | −0.013 | −0.022 | 0.029 | 0.061 | −0.006 | 0.000 | |
| ROBO1 | rs333491A/G | −0.031 | −0.115 | 0.141 * | −0.017 | −0.117 * | 0.136 * |
| rs6803202C/T | −0.119 * | −0.029 | 0.026 | −0.040 | −0.084 | 0.080 | |
| rs9853895C/T | 0.088 | −0.131 | 0.158 * | −0.107 | 0.168 ** | −0.195 ** | |
| rs7644521T/C | −0.029 | −0.013 | 0.005 | 0.014 | 0.018 | 0.097 | |
| GRIN2B | rs5796555-/A | 0.030 | 0.018 | −0.110 | −0.059 | 0.082 | −0.033 |
| rs1012586G/C | 0.052 | 0.048 | −0.060 | −0.027 | 0.086 | 0.014 | |
| rs2268119A/T | −0.053 | −0.008 | −0.055 | −0.037 | 0.061 | −0.007 | |
| rs2216128A/G | −0.089 | −0.035 | 0.023 | −0.135 * | −0.010 | −0.023 | |
| rs11609779C/T | −0.035 | 0.039 | −0.059 | 0.087 | 0.093 | −0.069 | |
| rs2192973G/A | −0.084 | −0.055 | 0.071 | −0.165 * | 0.004 | −0.068 | |
| β | SE | 95% CI * | |
|---|---|---|---|
| DYX1C1-rs3743205 | −0.004 | 0.039 | −0.442/0.436 |
| DYX1C1-rs57809907 | 0.055 | 0.038 | −0.154/0.585 |
| DYX1C1-rs189983504 | −0.010 | 0.030 | −0.293/0.243 |
| DCDC2-rs793842 | −0.015 | 0.035 | −0.147/0.107 |
| DCDC2-READ1d ° | −0.020 | 0.031 | −0.432/0.191 |
| DCDC2-rs793862 | 0.018 | 0.033 | −0.172/0.311 |
| KIAA0319-rs4504469 | −0.018 | 0.035 | −0.142/0.100 |
| KIAA0319-rs2038137 | 0.001 | 0.035 | −0.127/0.118 |
| KIAA0319-rs9461045 | −0.046 | 0.050 | −0.491/0.220 |
| KIAA0319-rs2143340 § | 0.060 | 0.045 | −0.143/0.554 |
| ROBO1-rs333491 | −0.018 | 0.029 | −0.132/0.078 |
| ROBO1-rs6803202 | −0.057 | 0.038 | −0.215/0.042 |
| ROBO1-rs9853895 | −0.099 | 0.042 | −0.306/−0.007 |
| ROBO1-rs7644521 | 0.013 | 0.028 | −0.169/0.260 |
| GRIN2B-rs5796555 | −0.058 | 0.045 | −0.497/0.127 |
| GRIN2B-rs1012586 | 0.019 | 0.046 | −0.275/0.356 |
| GRIN2B-rs2268119 | 0.015 | 0.041 | −0.248/0.331 |
| GRIN2B-rs2216128 | 0.065 | 0.083 | −0.492/0.881 |
| GRIN2B-rs11609779 | −0.002 | 0.030 | −0.225/0.199 |
| GRIN2B-rs2192973 | −0.100 | 0.085 | −0.996/0.382 |
| β | SE | 95% CI * | ||
|---|---|---|---|---|
| ATTENTION | WE | −0.002 | 0.015 | −0.062/0.048 |
| VISUAL MOTION PROCESSING | RTLI_b ° | −0.062 | 0.033 | −0.231/−0.006 |
| RTLI_t § | 0.038 | 0.031 | −0.020/0.192 | |
| RAP | −0.056 | 0.027 | −0.194/−0.003 | |
| RAN | RAN_rt | −0.017 | 0.012 | −0.076/0.007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mascheretti, S.; Riva, V.; Feng, B.; Trezzi, V.; Andreola, C.; Giorda, R.; Villa, M.; Dionne, G.; Gori, S.; Marino, C.; et al. The Mediation Role of Dynamic Multisensory Processing Using Molecular Genetic Data in Dyslexia. Brain Sci. 2020, 10, 993. https://doi.org/10.3390/brainsci10120993
Mascheretti S, Riva V, Feng B, Trezzi V, Andreola C, Giorda R, Villa M, Dionne G, Gori S, Marino C, et al. The Mediation Role of Dynamic Multisensory Processing Using Molecular Genetic Data in Dyslexia. Brain Sciences. 2020; 10(12):993. https://doi.org/10.3390/brainsci10120993
Chicago/Turabian StyleMascheretti, Sara, Valentina Riva, Bei Feng, Vittoria Trezzi, Chiara Andreola, Roberto Giorda, Marco Villa, Ginette Dionne, Simone Gori, Cecilia Marino, and et al. 2020. "The Mediation Role of Dynamic Multisensory Processing Using Molecular Genetic Data in Dyslexia" Brain Sciences 10, no. 12: 993. https://doi.org/10.3390/brainsci10120993
APA StyleMascheretti, S., Riva, V., Feng, B., Trezzi, V., Andreola, C., Giorda, R., Villa, M., Dionne, G., Gori, S., Marino, C., & Facoetti, A. (2020). The Mediation Role of Dynamic Multisensory Processing Using Molecular Genetic Data in Dyslexia. Brain Sciences, 10(12), 993. https://doi.org/10.3390/brainsci10120993