Dynamical Role of Pivotal Brain Regions in Parkinson Symptomatology Uncovered with Deep Learning
Abstract
:1. Introduction
- We implemented an autoencoder neural network for the prediction of future motor/cognitive impairment
- Our best model prediction performed with 85% accuracy, 80% specificity, and 100% sensitivity
- The autoencoder significantly improves upon standard statistical regression results
- Pivotal impaired regions change their relative contributions to Parkinsonian symptomatology over time
2. Material and Methods
3. Results
4. Discussion
4.1. Rationale for Variable Selection
4.2. Pivotal Regions for Cognitive and Motor Decline Prediction
4.3. Dynamical Reordering of Culprits in Parkinsonian Symptomatology
4.4. Limitations and Future Work
4.5. Summary
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Code Availability
Abbreviations
| DBM | Deformation-based Morphometry |
| MCI | Mild Cognitive Impairment |
| MDS-UPDRS-III | Movement Disorder Society-Unified Parkinson’s Disease Rating Scale Part III |
| MoCA | Montreal Cognitive Assessment |
| PD | Parkinson’s Disease |
| PPMI | Parkinson’s Progression Markers Initiative |
References
- Grover, S.; Bhartia, S.; Yadav, A.; Seeja, K.R. Predicting severity of Parkinson’s disease using deep learning. Procedia Comput. Sci. 2018, 132, 1788–1794. [Google Scholar] [CrossRef]
- Hirschauer, T.J.; Adeli, H.; Buford, J.A. Computer-Aided Diagnosis of Parkinson’s Disease Using Enhanced Probabilistic Neural Network. J. Med. Syst. 2015, 39, 179. [Google Scholar] [CrossRef] [PubMed]
- Kamagata, K.; Zalesky, A.; Hatano, T.; Di Biase, M.A.; El Samad, O.; Saiki, S.; Shimoji, K.; Kumamaru, K.K.; Kamiya, K.; Hori, M.; et al. Connectome analysis with diffusion MRI in idiopathic Parkinson’s disease: Evaluation using multi-shell, multi-tissue, constrained spherical deconvolution. Neuroimage Clin. 2018, 17, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, C.; Cerasa, A.; Castiglioni, I.; Gallivanone, F.; Augimeri, A.; Lopez, M.; Arabia, G.; Morelli, M.; Gilardi, M.C.; Quattrone, A. Machine learning on brain MRI data for differential diagnosis of Parkinson’s disease and Progressive Supranuclear Palsy. J. Neurosci. Methods 2014, 222, 230–237. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Bhagwat, N.; Viviano, J.D.; Voineskos, A.N.; Chakravarty, M.M.; Alzheimer’s Disease Neuroimaging, I. Modeling and prediction of clinical symptom trajectories in Alzheimer’s disease using longitudinal data. PLoS Comput. Biol. 2018, 14, e1006376. [Google Scholar] [CrossRef]
- Falahati, F.; Westman, E.; Simmons, A. Multivariate data analysis and machine learning in Alzheimer’s disease with a focus on structural magnetic resonance imaging. J. Alzheimers Dis. 2014, 41, 685–708. [Google Scholar] [CrossRef]
- Filipovych, R.; Davatzikos, C.; Alzheimer’s Disease Neuroimaging, I. Semi-supervised pattern classification of medical images: Application to mild cognitive impairment (MCI). Neuroimage 2011, 55, 1109–1119. [Google Scholar] [CrossRef] [Green Version]
- Moradi, E.; Pepe, A.; Gaser, C.; Huttunen, H.; Tohka, J.; Alzheimer’s Disease Neuroimaging, I. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage 2015, 104, 398–412. [Google Scholar] [CrossRef] [Green Version]
- Asgari, M.; Shafran, I. Predicting severity of Parkinson’s disease from speech. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 5201–5204. [Google Scholar]
- Liu, G.; Locascio, J.J.; Corvol, J.C.; Boot, B.; Liao, Z.; Page, K.; Franco, D.; Burke, K.; Jansen, I.E.; Trisini-Lipsanopoulos, A.; et al. Prediction of cognition in Parkinson’s disease with a clinical-genetic score: A longitudinal analysis of nine cohorts. Lancet Neurol. 2017, 16, 620–629. [Google Scholar] [CrossRef]
- Prashanth, R.; Roy, S.D. Novel and improved stage estimation in Parkinson’s disease using clinical scales and machine learning. Neurocomputing 2018, 305, 8–103. [Google Scholar] [CrossRef] [Green Version]
- Abos, A.; Baggio, H.C.; Segura, B.; Garcia-Diaz, A.I.; Compta, Y.; Marti, M.J.; Valldeoriola, F.; Junque, C. Discriminating cognitive status in Parkinson’s disease through functional connectomics and machine learning. Sci. Rep. 2017, 7, 45347. [Google Scholar] [CrossRef] [PubMed]
- Arnaldi, D.; De Carli, F.; Fama, F.; Brugnolo, A.; Girtler, N.; Picco, A.; Pardini, M.; Accardo, J.; Proietti, L.; Massa, F.; et al. Prediction of cognitive worsening in de novo Parkinson’s disease: Clinical use of biomarkers. Mov. Disord. 2017, 32, 1738–1747. [Google Scholar] [CrossRef]
- Choi, H.; Ha, S.; Im, H.J.; Paek, S.H.; Lee, D.S. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef]
- Parkinson Progression Marker, I. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 2011, 95, 629–635. [Google Scholar] [CrossRef]
- Holden, S.K.; Finseth, T.; Sillau, S.H.; Berman, B.D. Progression of MDS-UPDRS Scores Over Five Years in De Novo Parkinson Disease from the Parkinson’s Progression Markers Initiative Cohort. Mov. Disord. Clin. Pract. 2018, 5, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Zeighami, Y.; Ulla, M.; Iturria-Medina, Y.; Dadar, M.; Zhang, Y.; Larcher, K.M.; Fonov, V.; Evans, A.C.; Collins, D.L.; Dagher, A. Network structure of brain atrophy in de novo Parkinson’s disease. Elife 2015, 4. [Google Scholar] [CrossRef]
- Andica, C.; Kamagata, K.; Hatano, T.; Saito, A.; Uchida, W.; Ogawa, T.; Takeshige-Amano, H.; Zalesky, A.; Wada, A.; Suzuki, M.; et al. Free-Water Imaging in White and Gray Matter in Parkinson’s Disease. Cells 2019, 8, 839. [Google Scholar] [CrossRef] [Green Version]
- Hoops, S.; Nazem, S.; Siderowf, A.D.; Duda, J.E.; Xie, S.X.; Stern, M.B.; Weintraub, D. Validity of the MoCA and MMSE in the detection of MCI and dementia in Parkinson disease. Neurology 2009, 73, 1738–1745. [Google Scholar] [CrossRef]
- Martinez-Martin, P.; Rodriguez-Blazquez, C.; Mario, A.; Arakaki, T.; Arillo, V.C.; Chana, P.; Fernandez, W.; Garretto, N.; Martinez-Castrillo, J.C.; Rodriguez-Violante, M.; et al. Parkinson’s disease severity levels and MDS-Unified Parkinson’s disease rating scale. Parkinsonism Relat. Disord. 2015, 21, 50–54. [Google Scholar] [CrossRef] [Green Version]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Møller, M.F. A scaled conjugate gradient algorithm for fast supervised learning. Neural Networks 1993, 6, 525–533. [Google Scholar] [CrossRef]
- Luo, G. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw. Model. Anal. Health. Inform. Bioinforma 2016, 5, 18. [Google Scholar] [CrossRef]
- Fengler, S.; Kessler, J.; Timmermann, L.; Zapf, A.; Elben, S.; Wojtecki, L.; Tucha, O.; Kalbe, E. Screening for cognitive impairment in Parkinson’s disease: Improving the diagnostic utility of the MoCA through subtest weighting. PLoS ONE 2016, 11, e0159318. [Google Scholar] [CrossRef] [PubMed]
- Movement Disorder Society Task Force on Rating Scales for Parkinson’s Disease. The Unified Parkinson’s Disease Rating Scale (UPDRS): Status and recommendations. Mov. Disord. 2003, 18, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Greffard, S.; Verny, M.; Bonnet, A.M.; Beinis, J.Y.; Gallinari, C.; Meaume, S.; Piette, F.; Hauw, J.J.; Duyckaerts, C. Motor score of the Unified Parkinson Disease Rating Scale as a good predictor of Lewy body-associated neuronal loss in the substantia nigra. Arch. Neurol. 2006, 63, 584–588. [Google Scholar] [CrossRef] [Green Version]
- Freeze, B.; Pandya, S.; Zeighami, Y.; Raj, A. Regional transcriptional architecture of Parkinson’s disease pathogenesis and network spread. Brain 2019, 142, 3072–3085. [Google Scholar] [CrossRef]
- Pandya, S.; Zeighami, Y.; Freeze, B.; Dadar, M.; Collins, D.L.; Dagher, A.; Raj, A. Predictive model of spread of Parkinson’s pathology using network diffusion. NeuroImage 2019, 192, 178–194. [Google Scholar] [CrossRef]
- Scanlon, C.; Mueller, S.G.; Tosun, D.; Cheong, I.; Garcia, P.; Barakos, J.; Weiner, M.W.; Laxer, K.D. Impact of methodologic choice for automatic detection of different aspects of brain atrophy by using temporal lobe epilepsy as a model. AJNR Am. J. Neuroradiol. 2011, 32, 1669–1676. [Google Scholar] [CrossRef] [Green Version]
- Gaser, C.; Nenadic, I.; Buchsbaum, B.R.; Hazlett, E.A.; Buchsbaum, M.S. Deformation-based morphometry and its relation to conventional volumetry of brain lateral ventricles in MRI. Neuroimage 2001, 13, 1140–1145. [Google Scholar] [CrossRef] [Green Version]
- Melzer, T.R.; Watts, R.; MacAskill, M.R.; Pitcher, T.L.; Livingston, L.; Keenan, R.J.; Dalrymple-Alford, J.C.; Anderson, T.J. Grey matter atrophy in cognitively impaired Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 2012, 83, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Kehagia, A.A.; Barker, R.A.; Robbins, T.W. Neuropsychological and clinical heterogeneity of cognitive impairment and dementia in patients with Parkinson’s disease. Lancet Neurol. 2010, 9, 1200–1213. [Google Scholar] [CrossRef]
- Hirata, K.; Hattori, N.; Takeuchi, W.; Shiga, T.; Morimoto, Y.; Umegaki, K.; Kobayashi, K.; Manabe, O.; Okamoto, S.; Tamaki, N. Metabolic Activity of Red Nucleus and Its Correlation with Cerebral Cortex and Cerebellum: A Study Using a High-Resolution Semiconductor PET System. J. Nucl. Med. 2015, 56, 1206–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, V.; Josien, E.; Pasquier, F.; Steinling, M.; Petit, H. Infarction of the red nucleus and crossed cerebellar diaschisis. Rev. Neurol. (Paris) 1993, 149, 294–296. [Google Scholar] [PubMed]
- Weintraub, D.B.; Zaghloul, K.A. The role of the subthalamic nucleus in cognition. Rev. Neurosci. 2013, 24, 125–138. [Google Scholar] [CrossRef] [Green Version]
- Beyer, M.K.; Janvin, C.C.; Larsen, J.P.; Aarsland, D. A magnetic resonance imaging study of patients with Parkinson’s disease with mild cognitive impairment and dementia using voxel-based morphometry. J. Neurol. Neurosurg. Psychiatry 2007, 78, 254–259. [Google Scholar] [CrossRef] [Green Version]
- Vriend, C.; Pattij, T.; van der Werf, Y.D.; Voorn, P.; Booij, J.; Rutten, S.; Berendse, H.W.; van den Heuvel, O.A. Depression and impulse control disorders in Parkinson’s disease: Two sides of the same coin? Neurosci. Biobehav. Rev. 2014, 38, 60–71. [Google Scholar] [CrossRef]
- Muralidharan, A.; Jensen, A.L.; Connolly, A.; Hendrix, C.M.; Johnson, M.D.; Baker, K.B.; Vitek, J.L. Physiological changes in the pallidum in a progressive model of Parkinson’s disease: Are oscillations enough? Exp. Neurol. 2016, 279, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Abosch, A.; Hutchison, W.D.; Saint-Cyr, J.A.; Dostrovsky, J.O.; Lozano, A.M. Movement-related neurons of the subthalamic nucleus in patients with Parkinson disease. J. Neurosurg. 2002, 97, 1167–1172. [Google Scholar] [CrossRef]
- Hamani, C.; Saint-Cyr, J.A.; Fraser, J.; Kaplitt, M.; Lozano, A.M. The subthalamic nucleus in the context of movement disorders. Brain 2004, 127, 4–20. [Google Scholar] [CrossRef] [Green Version]
- Lanotte, M.M.; Rizzone, M.; Bergamasco, B.; Faccani, G.; Melcarne, A.; Lopiano, L. Deep brain stimulation of the subthalamic nucleus: Anatomical, neurophysiological, and outcome correlations with the effects of stimulation. J. Neurol. Neurosurg. Psychiatry 2002, 72, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Cyr, J.A.; Hoque, T.; Pereira, L.C.; Dostrovsky, J.O.; Hutchison, W.D.; Mikulis, D.J.; Abosch, A.; Sime, E.; Lang, A.E.; Lozano, A.M. Localization of clinically effective stimulating electrodes in the human subthalamic nucleus on magnetic resonance imaging. J. Neurosurg. 2002, 97, 1152–1166. [Google Scholar] [CrossRef] [PubMed]
- Seger, C.A.; Cincotta, C.M. The roles of the caudate nucleus in human classification learning. J. Neurosci. 2005, 25, 2941–2951. [Google Scholar] [CrossRef] [PubMed]
- Starr, P.A.; Christine, C.W.; Theodosopoulos, P.V.; Lindsey, N.; Byrd, D.; Mosley, A.; Marks, W.J., Jr. Implantation of deep brain stimulators into the subthalamic nucleus: Technical approach and magnetic resonance imaging-verified lead locations. J. Neurosurg. 2002, 97, 370–387. [Google Scholar] [CrossRef]
- Grahn, J.A.; Parkinson, J.A.; Owen, A.M. The cognitive functions of the caudate nucleus. Prog. Neurobiol. 2008, 86, 141–155. [Google Scholar] [CrossRef]
- Blumen, H.M.; Brown, L.L.; Habeck, C.; Allali, G.; Ayers, E.; Beauchet, O.; Callisaya, M.; Lipton, R.B.; Mathuranath, P.S.; Phan, T.G.; et al. Gray matter volume covariance patterns associated with gait speed in older adults: A multi-cohort MRI study. Brain Imaging Behav. 2019, 13, 446–460. [Google Scholar] [CrossRef]
- McDonald, A.J.; Mott, D.D. Functional neuroanatomy of amygdalohippocampal interconnections and their role in learning and memory. J. Neurosci. Res. 2017, 95, 797–820. [Google Scholar] [CrossRef] [Green Version]
- Bouhrara, M.; Cortina, L.E.; Rejimon, A.C.; Khattar, N.; Bergeron, C.; Bergeron, J.; Melvin, D.; Zukley, L.; Spencer, R.G. Quantitative age-dependent differences in human brainstem myelination assessed using high-resolution magnetic resonance mapping. NeuroImage 2019, 206, 116307. [Google Scholar] [CrossRef]

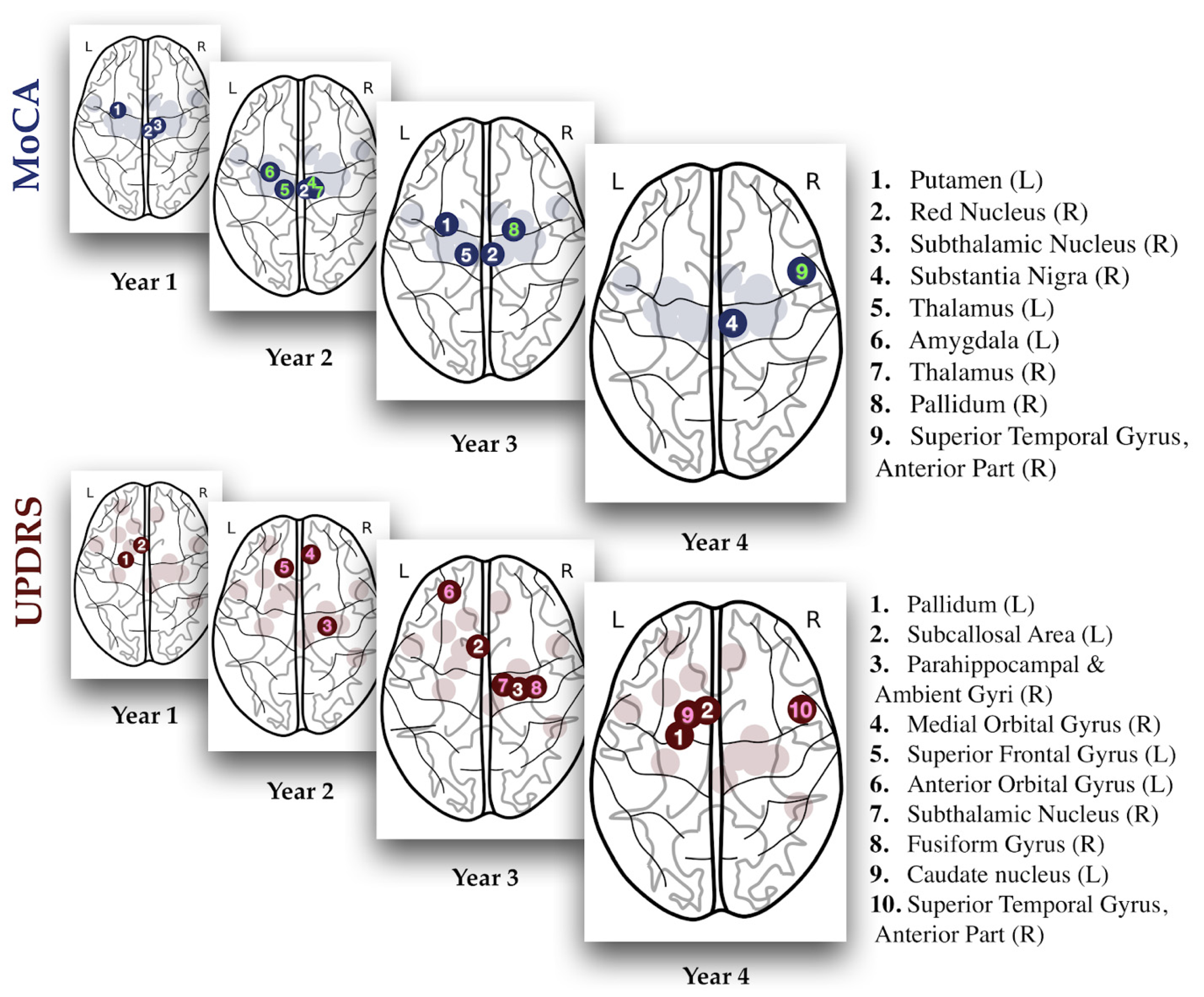

| Year | Predictor | Type | Side | R Value | p Value |
|---|---|---|---|---|---|
| 1 | MDS-UPDRS-III | 0.425 | <0.001 | ||
| Caudate | SBR | R | −0.209 | 0.044 | |
| Putamen | SBR | R | −0.351 | <0.001 | |
| Hoehn and Yahr | 0.292 | 0.005 | |||
| Symbol Digit Modalities Score | −0.28 | 0.007 | |||
| MDS-UPDRS Total Score | 0.401 | <0.001 | |||
| MDS-UPDRS-II PQ | 0.293 | 0.005 | |||
| Modified Schwab England ADL | −0.205 | 0.049 | |||
| UPSIT Total Score | −0.243 | 0.019 | |||
| Superior temporal gyrus, anterior part | DBM | R | 0.204 | 0.049 | |
| Subthalamic Nucleus | DBM | R | −0.23 | 0.026 | |
| 2 | MDS-UPDRS-III | 0.494 | <0.001 | ||
| Hoehn and Yahr | 0.357 | <0.001 | |||
| HVLT Immediate Recall | −0.314 | 0.004 | |||
| HVLT Delayed Recognition Hits | −0.274 | 0.012 | |||
| Symbol Digit Modalities Score | -0.229 | 0.036 | |||
| MDS-UPDRS Total Score | 0.402 | <0.001 | |||
| MDS-UPDRS-II PQ | 0.291 | 0.007 | |||
| Modified Schwab England ADL | −0.333 | 0.002 | |||
| UPSIT Total Score | −0.224 | 0.041 | |||
| Hippocampus | DBM | L | −0.244 | 0.022 | |
| Amygdala | DBM | R | −0.12 | 0.025 | |
| Parahippocampal and ambient gyri | DBM | R | −0.218 | 0.047 | |
| Inferolateral remainder of parietal lobe | DBM | R | 0.22 | 0.045 | |
| Medial orbital gyrus | DBM | R | 0.221 | 0.044 | |
| Superior temporal gyrus, anterior part | DBM | L | −0.221 | 0.044 | |
| 3 | MDS-UPDRS-III | 0.38 | <0.001 | ||
| Caudate | SBR | R | −0.312 | 0.004 | |
| Putamen | SBR | R | −0.326 | 0.002 | |
| Alpha-synuclein | −0.237 | 0.03 | |||
| Hoehn and Yahr | 0.353 | 0.001 | |||
| QUIP Positive Buying | −0.223 | 0.041 | |||
| QUIP Positive Hobbies | −0.223 | 0.041 | |||
| MDS-UPDRS Total Score | 0.42 | <0.001 | |||
| MDS-UPDRS-I PQ | 0.253 | 0.02 | |||
| MDS-UPDRS-II PQ | 0.382 | <0.001 | |||
| Modified Schwab England ADL | −0.314 | 0.004 | |||
| Fusiform (lateral occipitotemporal) gyrus | DBM | R | 0.216 | 0.048 | |
| Pallidum | DBM | L | 0.23 | 0.035 | |
| Superior frontal gyrus | DBM | L | −0.251 | 0.021 | |
| 4 | Age | 0.281 | 0.014 | ||
| MDS-UPDRS-III | 0.51 | <0.001 | |||
| Caudate | SBR | L | −0.293 | 0.01 | |
| Caudate | SBR | R | −0.334 | 0.003 | |
| Putamen | SBR | L | −0.227 | 0.049 | |
| Putamen | SBR | R | −0.282 | 0.014 | |
| Hoehn and Yahr | 0.248 | 0.031 | |||
| HVLT Delayed Recognition Hits | −0.325 | 0.012 | |||
| Benton Judgement of Line Orientation | −0.302 | 0.008 | |||
| QUIP Positive Eating | 0.227 | 0.049 | |||
| MDS-UPDRS Total Score | 0.524 | <0.001 | |||
| MDS-UPDRS-I PQ | 0.296 | 0.01 | |||
| MDS-UPDRS-II PQ | 0.348 | 0.002 | |||
| SCOPA-AUT | 0.255 | 0.026 | |||
| Amygdala | DBM | R | −0.249 | 0.03 | |
| Gyrus cinguli, posterior part | DBM | R | −0.302 | 0.008 | |
| Caudate nucleus | DBM | L | −0.309 | 0.007 | |
| Anterior orbital gyrus | DBM | L | −0.257 | 0.025 | |
| Posterior orbital gyrus | DBM | L | −0.232 | 0.043 | |
| Subcallosal area | DBM | L | 0.25 | 0.03 | |
| Subthalamic Nucleus | DBM | R | −0.266 | 0.02 |
| Year | Predictor | Type | Side | R Value | p Value |
|---|---|---|---|---|---|
| 1 | Age | 0.373 | <0.001 | ||
| MoCA | −0.402 | <0.001 | |||
| Amyloid Beta (1–42) | −0.212 | 0.034 | |||
| HVLT Immediate Recall | −0.422 | <0.001 | |||
| HVLT Delayed Recognition Hits | −0.252 | 0.011 | |||
| Benton Judgement of Line Orientation | −0.243 | 0.015 | |||
| Semantic Fluency Score | −0.436 | <0.001 | |||
| Symbol Digit Modalities Score | −0.459 | <0.001 | |||
| MDS-UPDRS-I | 0.214 | 0.032 | |||
| Hippocampus | DBM | R | −0.212 | 0.034 | |
| Amygdala | DBM | L | −0.209 | 0.037 | |
| Putamen | DBM | R | −0.221 | 0.027 | |
| Precentral gyrus | DBM | R | 0.289 | 0.004 | |
| Subthalamic Nucleus | DBM | L | −0.235 | 0.019 | |
| Subthalamic Nucleus | DBM | R | −0.228 | 0.022 | |
| 2 | Age | 0.29 | 0.003 | ||
| MoCA | −0.368 | <0.001 | |||
| Amyloid Beta (1–42) | −0.269 | 0.007 | |||
| HVLT Immediate Recall | −0.379 | <0.001 | |||
| HVLT Delayed Recognition Hits | −0.229 | 0.022 | |||
| HVLT Delayed Recognition False Alarms | 0.306 | 0.002 | |||
| Semantic Fluency Score | −0.296 | 0.003 | |||
| Symbol Digit Modalities Score | −0.375 | <0.001 | |||
| MDS-UPDRS Total | 0.217 | 0.029 | |||
| MDS-UPDRS-I | 0.259 | 0.009 | |||
| MDS-UPDRS-II PQ | 0.245 | 0.014 | |||
| Modified Schwab England ADL | −0.26 | 0.009 | |||
| Hippocampus | DBM | R | −0.2 | 0.045 | |
| Amygdala | DBM | R | −0.215 | 0.031 | |
| Amygdala | DBM | L | −0.205 | 0.039 | |
| Putamen | DBM | L | −0.282 | 0.004 | |
| Putamen | DBM | R | −0.285 | 0.004 | |
| Thalamus | DBM | R | −0.225 | 0.024 | |
| Precentral gyrus | DBM | L | −0.21 | 0.035 | |
| Superior temporal gyrus, anterior part | DBM | R | −0.198 | 0.047 | |
| Red Nucleus | DBM | L | −0.203 | 0.042 | |
| Substantia Nigra | DBM | L | −0.236 | 0.017 | |
| Substantia Nigra | DBM | R | −0.255 | 0.01 | |
| Subthalamic Nucleus | DBM | L | −0.208 | 0.037 | |
| Subthalamic Nucleus | DBM | R | −0.28 | 0.005 | |
| 3 | Age | 0.44 | <0.001 | ||
| MDS-UPDRS-III | 0.286 | 0.003 | |||
| MoCA | −0.442 | <0.001 | |||
| SBR Right Putamen | −0.205 | 0.038 | |||
| Hoehn and Yahr | 0.206 | 0.037 | |||
| HVLT Immediate Recall | −0.471 | <0.001 | |||
| HVLT Delayed Recognition Hits | −0.41 | <0.001 | |||
| Benton Judgement of Line Orientation | −0.235 | 0.017 | |||
| Semantic Fluency Score | −0.51 | <0.001 | |||
| Symbol Digit Modalities Score | −0.518 | <0.001 | |||
| MDS-UPDRS Total | 0.255 | 0.009 | |||
| UPSIT Total Score | −0.243 | 0.013 | |||
| Hippocampus | DBM | R | −0.197 | 0.046 | |
| Hippocampus | DBM | L | −0.248 | 0.012 | |
| Amygdala | DBM | L | −0.24 | 0.015 | |
| Anterior temporal lobe | DBM | L | −0.217 | 0.028 | |
| Putamen | DBM | L | −0.324 | <0.001 | |
| Putamen | DBM | R | −0.307 | 0.002 | |
| Thalamus | DBM | L | −0.223 | 0.023 | |
| Thalamus | DBM | R | −0.272 | 0.006 | |
| Pallidum | DBM | R | −0.269 | 0.006 | |
| Red Nucleus | DBM | L | −0.304 | 0.002 | |
| Red Nucleus | DBM | R | −0.231 | 0.019 | |
| Substantia Nigra | DBM | L | −0.258 | 0.009 | |
| Substantia Nigra | DBM | R | −0.252 | 0.01 | |
| Subthalamic Nucleus | DBM | L | −0.254 | 0.01 | |
| Subthalamic Nucleus | DBM | R | −0.276 | 0.005 | |
| 4 | Age | 0.452 | <0.001 | ||
| MoCA | −0.438 | <0.001 | |||
| HVLT Immediate Recall | −0.505 | <0.001 | |||
| HVLT Delayed Recognition Hits | −0.344 | <0.001 | |||
| HVLT Delayed Recognition False Alarms | 0.219 | 0.033 | |||
| Semantic Fluency Score | −0.424 | <0.001 | |||
| Symbol Digit Modalities Score | −0.492 | <0.001 | |||
| MDS-UPDRS Total | 0.276 | 0.007 | |||
| MDS-UPDRS-I PQ | 0.305 | 0.003 | |||
| UPSIT Total Score | −0.222 | 0.03 | |||
| SCOPA-AUT | 0.367 | <0.001 | |||
| Nucleus accumbens | DBM | R | −0.26 | 0.012 | |
| Putamen | DBM | L | −0.35 | <0.001 | |
| Putamen | DBM | R | −0.322 | 0.002 | |
| Thalamus | DBM | L | −0.227 | 0.027 | |
| Thalamus | DBM | R | −0.27 | 0.009 | |
| Pallidum | DBM | L | −0.207 | 0.044 | |
| Pallidum | DBM | R | −0.24 | 0.019 | |
| Red Nucleus | DBM | L | −0.219 | 0.033 | |
| Red Nucleus | DBM | R | −0.213 | 0.038 | |
| Substantia Nigra | DBM | L | −0.202 | 0.049 | |
| Subthalamic Nucleus | DBM | L | −0.252 | 0.014 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, A.A.; Maia, P.D.; Gao, X.; Damasceno, P.F.; Raj, A. Dynamical Role of Pivotal Brain Regions in Parkinson Symptomatology Uncovered with Deep Learning. Brain Sci. 2020, 10, 73. https://doi.org/10.3390/brainsci10020073
Nguyen AA, Maia PD, Gao X, Damasceno PF, Raj A. Dynamical Role of Pivotal Brain Regions in Parkinson Symptomatology Uncovered with Deep Learning. Brain Sciences. 2020; 10(2):73. https://doi.org/10.3390/brainsci10020073
Chicago/Turabian StyleNguyen, Alex A., Pedro D. Maia, Xiao Gao, Pablo F. Damasceno, and Ashish Raj. 2020. "Dynamical Role of Pivotal Brain Regions in Parkinson Symptomatology Uncovered with Deep Learning" Brain Sciences 10, no. 2: 73. https://doi.org/10.3390/brainsci10020073
APA StyleNguyen, A. A., Maia, P. D., Gao, X., Damasceno, P. F., & Raj, A. (2020). Dynamical Role of Pivotal Brain Regions in Parkinson Symptomatology Uncovered with Deep Learning. Brain Sciences, 10(2), 73. https://doi.org/10.3390/brainsci10020073





