Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions
Abstract
:1. Introduction
2. Material and Methods
2.1. Participants and sMRI Acquisition
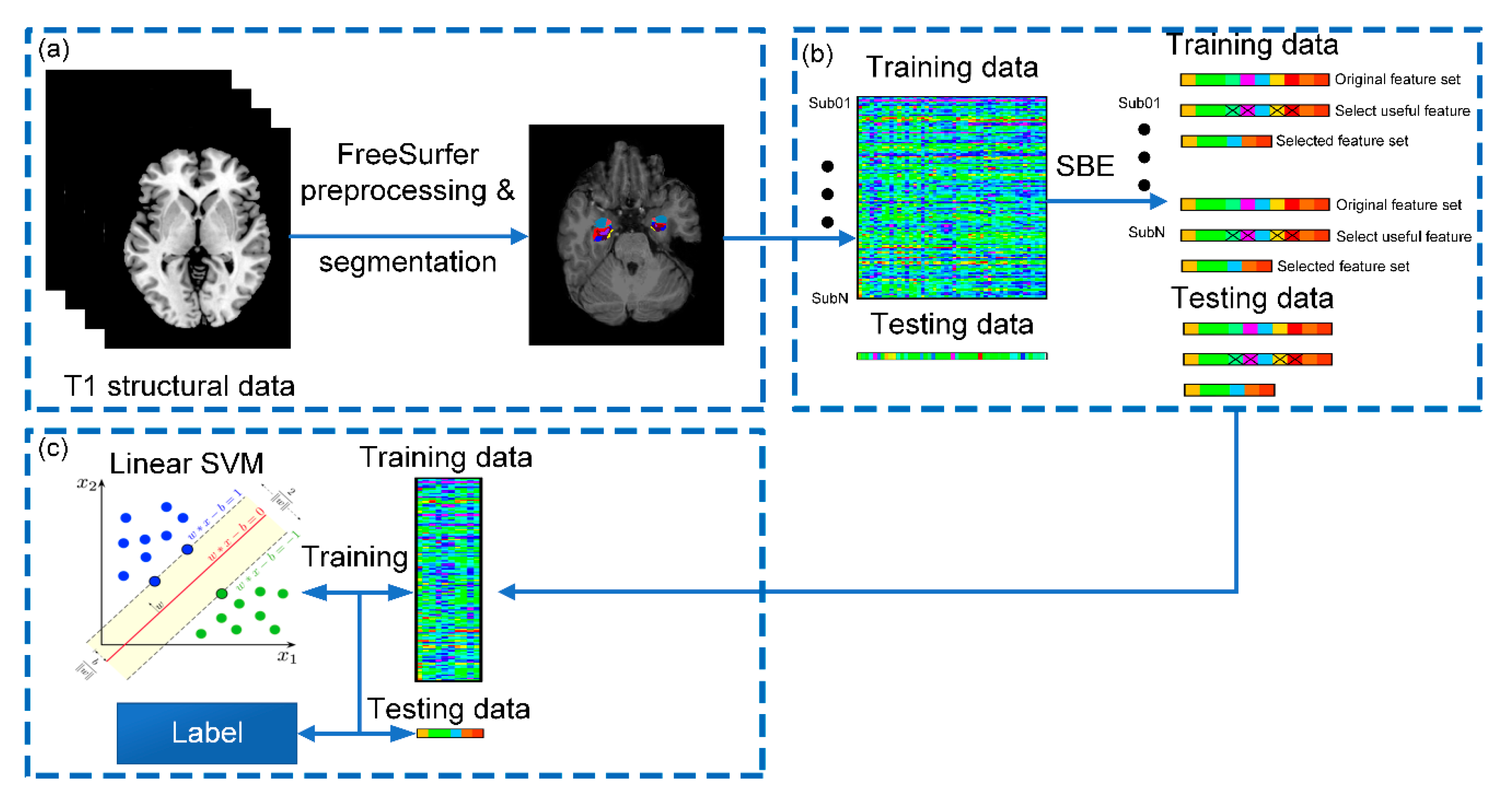
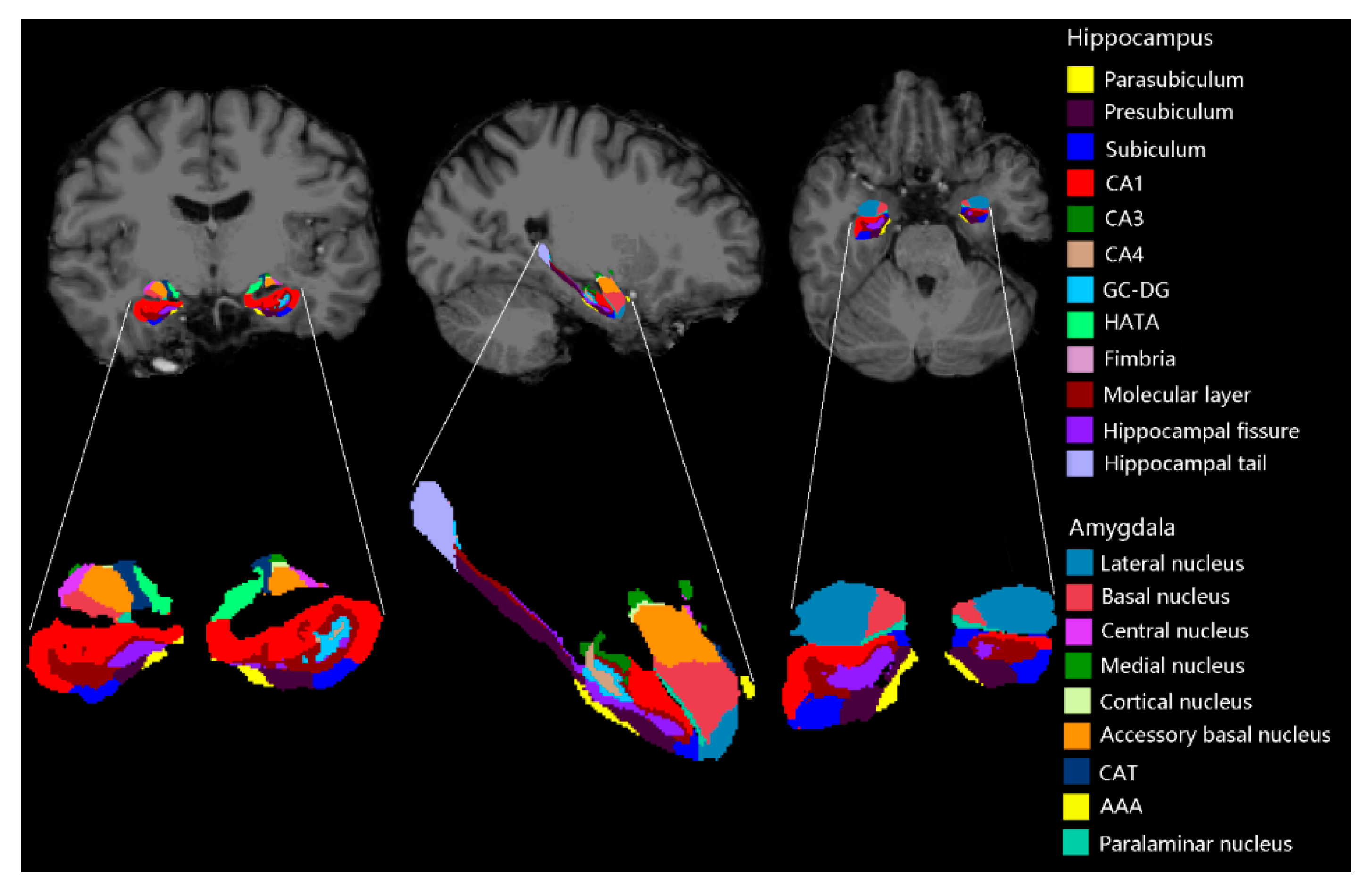
2.2. Imaging Processing
2.3. Feature Selection
2.3.1. Sequential Selection and Its Variants
2.3.2. T-Test
2.3.3. F-Score
2.3.4. Random Forest
2.4. SVM and Evaluating Metrics
2.4.1. Linear SVM
2.4.2. Competing Algorithms
2.4.3. Evaluating Metrics
2.5. Post Hoc Analysis
3. Results
3.1. Demographic Information
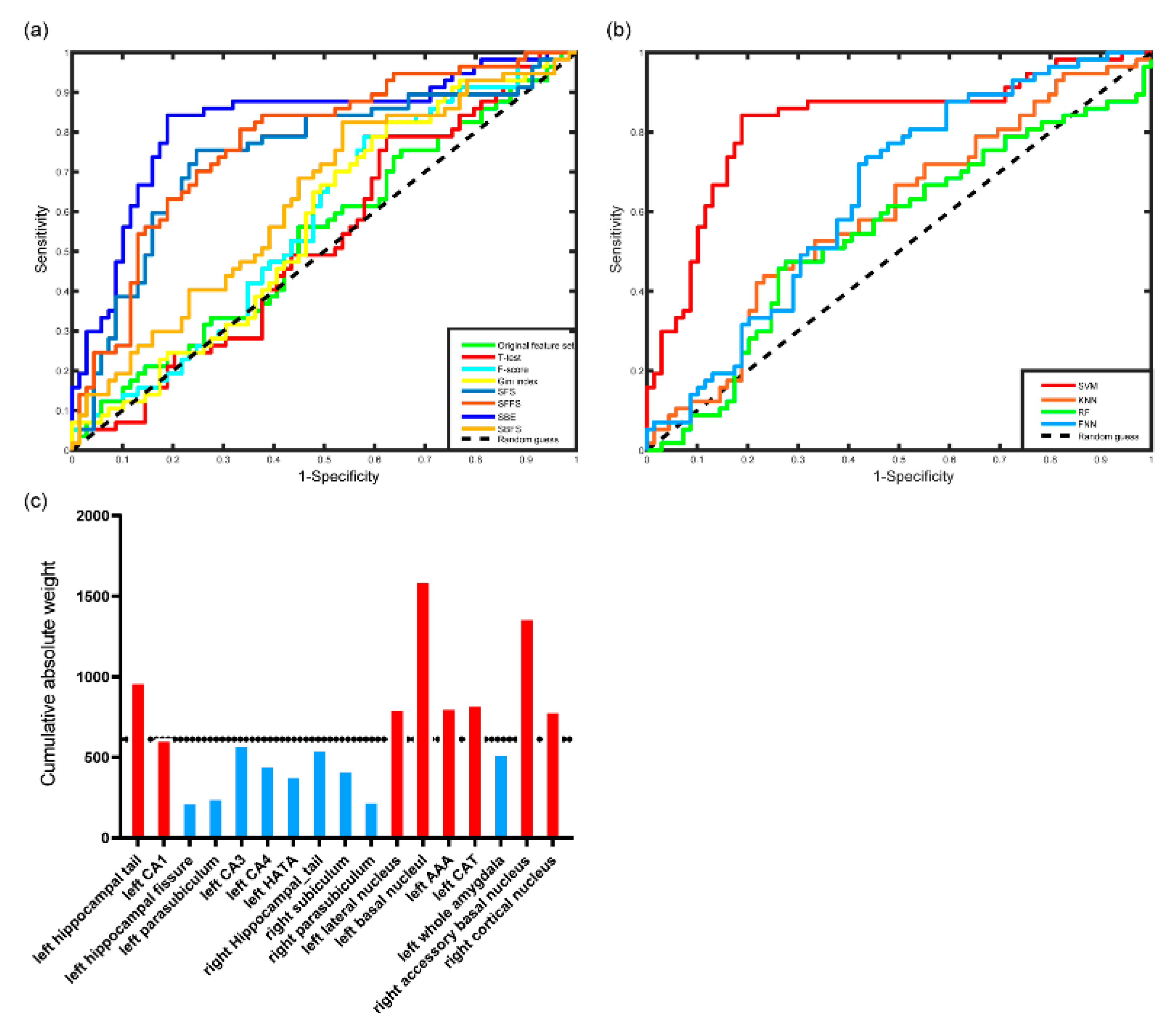
3.2. Performance of the Classifier
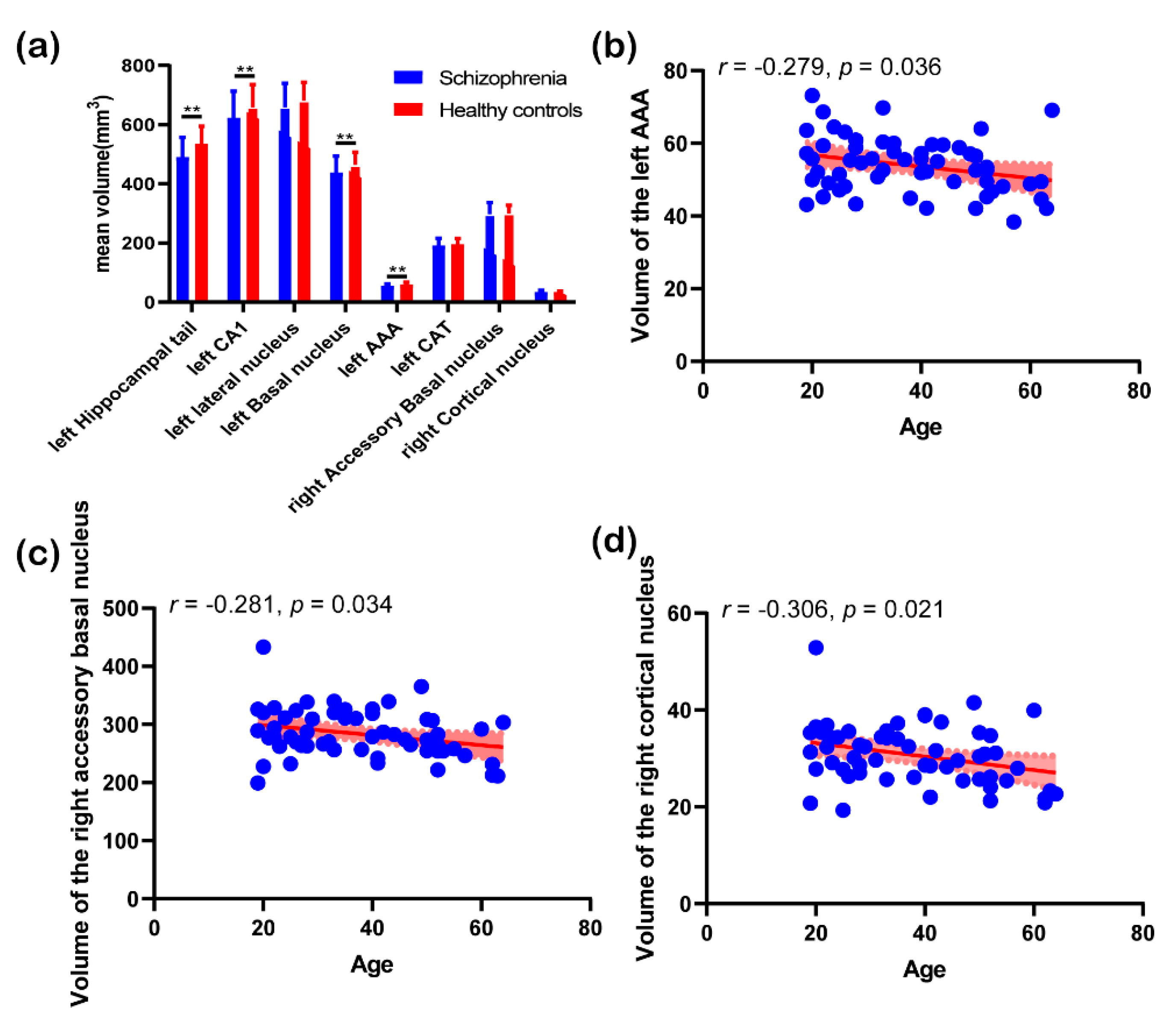
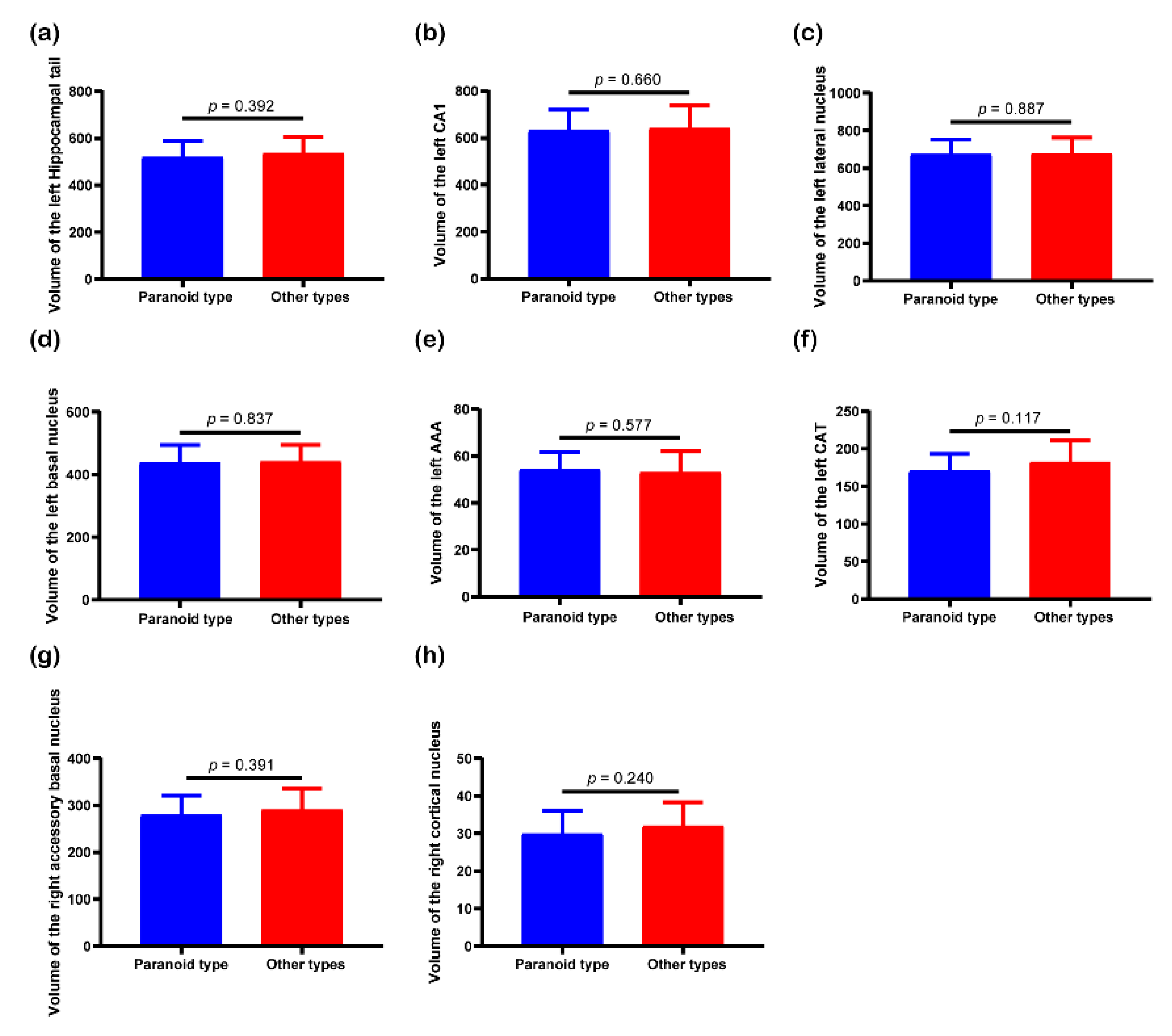
3.3. Post Hoc Analysis Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Krishnan, R.R.; Keefe, R.; Kraus, M. Schizophrenia is a disorder of higher order hierarchical processing. Med. Hypotheses 2009, 72, 740–744. [Google Scholar] [CrossRef]
- Seidman, L.J.; Giuliano, A.J.; Smith, C.W.; Stone, W.S.; Glatt, S.J.; Meyer, E.; Faraone, S.V.; Tsuang, M.T.; Cornblatt, B. Neuropsychological functioning in adolescents and young adults at genetic risk for schizophrenia and affective psychoses: Results from the harvard and hillside adolescent high risk studies. Schizophr. Bull. 2005, 32, 507–524. [Google Scholar] [CrossRef]
- Guloksuz, S.; Pries, L.; Delespaul, P.; Kenis, G.; Luykx, J.J.; Lin, B.D.; Richards, A.L.; Akdede, B.; Binbay, T.; Altınyazar, V.; et al. Examining the independent and joint effects of molecular genetic liability and environmental exposures in schizophrenia: Results from the EUGEI study. World Psychiatry 2019, 18, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Saarinen, A.I.L.; Huhtaniska, S.; Pudas, J.; Björnholm, L.; Jukuri, T.; Tohka, J.; Granö, N.; Barnett, J.H.; Kiviniemi, V.; Veijola, J.; et al. Structural and functional alterations in the brain gray matter among first-degree relatives of schizophrenia patients: A multimodal meta-analysis of fMRI and VBM studies. Schizophr. Res. 2020, 216, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Andjus, P.R.; Danijela, B.; Vanhoutte, G.; Mitrecic, D.; Pizzolante, F.; Djogo, N.; Nicaise, C.; Kengne, F.G.; Gangitano, C.; Michetti, F.; et al. In vivo morphological changes in animal models of amyotrophic lateral sclerosis and alzheimer’s-like disease: MRI Approach. Anat. Rec. 2009, 292, 1882–1892. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Zheng, J.; Miao, C.; McKeown, M.J.; Wang, Z.J. 3D CNN based automatic diagnosis of attention deficit hyperactivity disorder using functional and structural MRI. IEEE Access. 2017, 5, 23626–23636. [Google Scholar] [CrossRef]
- Tong, T.; Wolz, R.; Gao, Q.; Guerrero, R.; Hajnal, J.V.; Rueckert, D. Multiple instance learning for classification of dementia in brain MRI. Med. Image Anal. 2014, 18, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Black, M.; Xia, S.; Zhan, C.; Bertisch, H.C.; Branch, C.A.; DeLisi, L.E. Subcortical structure alterations impact language processing in individuals with schizophrenia and those at high genetic risk. Schizophr. Res. 2015, 169, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Satizabal, C.L.; Adams, H.H.; Hibar, D.P.; White, C.C.; Knol, M.J.; Stein, J.L.; Scholz, M.; Sargurupremraj, M.; Jahanshad, N.; Roshchupkin, G.V.; et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat. Genet. 2019, 51, 1624–1636. [Google Scholar] [CrossRef] [Green Version]
- Zheng, F.; Li, C.; Zhang, N.; Cui, D.; Wang, Z.; Qiu, J. Study on the sub-regions volume of hippocampus and amygdala in schizophrenia. Quant. Imaging Med. Surg. 2019, 9, 1025–1036. [Google Scholar] [CrossRef]
- Okada, N.; Fukunaga, M.; Yamashita, F.; Koshiyama, D.; Yamamori, H.; Ohi, K.; Yasuda, Y.; Fujimoto, M.; Yahata, N.; Nemoto, K.; et al. Abnormal asymmetries in subcortical brain volume in schizophrenia. Mol. Psychiatry 2016, 21, 1460–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallak, J.; De Paula, A.; Chaves, C.; Bressan, R.A.; Machado-De-Sousa, J. An overview on the search for schizophrenia biomarkers. CNS Neurol. Disord. Drug Targets 2015, 14, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Razafsha, M.; Khaku, A.; Azari, H.; Alawieh, A.; Behforuzi, H.; Fadlallah, B.; Kobeissy, F.; Wang, K.K.; Gold, M.S. Biomarker identification in psychiatric disorders. J. Psychiatr. Pr. 2015, 21, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Chyzhyk, D.; Savio, A.; Graña, M. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Networks 2015, 68, 23–33. [Google Scholar] [CrossRef]
- Xiao, Y.; Yan, Z.; Zhao, Y.; Tao, B.; Sun, H.; Li, F.; Yao, L.; Zhang, W.; Chandan, S.; Liu, J.; et al. Support vector machine-based classification of first episode drug-naïve schizophrenia patients and healthy controls using structural MRI. Schizophr. Res. 2017, 214, 11–17. [Google Scholar] [CrossRef]
- Cao, H.; Duan, J.; Lin, D.; Shugart, Y.Y.; Calhoun, V.; Wang, Y.-P. Sparse representation based biomarker selection for schizophrenia with integrated analysis of fMRI and SNPs. NeuroImage 2014, 102, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Bracher-Smith, M.; Crawford, K.; Escott-Price, V. Machine learning for genetic prediction of psychiatric disorders: A systematic review. Mol. Psychiatry. (in press). [CrossRef]
- Steardo, L.; Carbone, E.A.; De Filippis, R.; Pisanu, C.; Segura-Garcia, C.; Squassina, A.; De Fazio, P.; Steardo, L. Application of support vector machine on fMRI data as biomarkers in schizophrenia diagnosis: A systematic review. Front. Psychol. 2020, 11, 588. [Google Scholar] [CrossRef]
- FreeSurfer 6.0. Available online: http://surfer.nmr.mgh.harvard.edu/ (accessed on 1 July 2020).
- FreeSurfer development version. Available online: ftp://surfer.nmr.mgh.harvard.edu/pub/dist/freesurfer/dev (accessed on 1 July 2020).
- Iglesias, J.E.; Augustinack, J.C.; Nguyen, K.; Player, C.M.; Player, A.; Wright, M.; Roy, N.; Frosch, M.P.; McKee, A.C.; Wald, L.L.; et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage 2015, 115, 117–137. [Google Scholar] [CrossRef]
- Saygin, Z.; Kliemann, D.; Iglesias, J.E.; Van Der Kouwe, A.; Boyd, E.; Reuter, M.; Stevens, A.; Van Leemput, K.; McKee, A.; Frosch, M.; et al. High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: Manual segmentation to automatic atlas. NeuroImage 2017, 155, 370–382. [Google Scholar] [CrossRef]
- Koutsouleris, N.; Kambeitz-Ilankovic, L.; Ruhrmann, S.; Rosen, M.; Ruef, A.; Dwyer, D.B.; Paolini, M.; Chisholm, K.; Kambeitz, J.; Haidl, T.; et al. Prediction models of functional outcomes for individuals in the clinical high-risk state for psychosis or with recent-onset depression: A multimodal, multisite machine learning analysis. JAMA Psychiatry 2018, 75, 1156–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Zhang, Q.C.; Chen, Z.; Meng, Y.; Guan, J.; Zhou, S. PredHS: A web server for predicting protein–Protein interaction hot spots by using structural neighborhood properties. Nucleic Acids Res. 2014, 42, W290–W295. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Guo, W.; Fouche, J.-P.; Wang, Y.; Wang, W.; Ding, J.; Zeng, L.; Qiu, C.; Gong, Q.; Zhang, W.; et al. Multivariate classification of social anxiety disorder using whole brain functional connectivity. Brain Struct. Funct. 2013, 220, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.C.; Sheridan, R.P.; Feuston, B.P. Random forest: A Classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intel. Syst. Tec. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- LIBSVM. Available online: https://www.csie.ntu.edu.tw/~cjlin/libsvm/ (accessed on 1 July 2020).
- De Filippis, R.; Carbone, E.A.; Gaetano, R.; Bruni, A.; Pugliese, V.; Garcia, C.S.; De Fazio, P. Machine learning techniques in a structural and functional MRI diagnostic approach in schizophrenia: A systematic review. Neuropsychiatr. Dis. Treat. 2019, 15, 1605–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvador, R.; Radua, J.; Canales-Rodríguez, E.J.; Solanes, A.; Sarró, S.; Goikolea, J.M.; Valiente-Gómez, A.; Monté, G.C.; Natividad, M.D.C.; Guerrero-Pedraza, A.; et al. Evaluation of machine learning algorithms and structural features for optimal MRI-based diagnostic prediction in psychosis. PLoS ONE 2017, 12, e0175683. [Google Scholar] [CrossRef] [Green Version]
- Castellani, U.; Rossato, E.; Murino, V.; Bellani, M.; Rambaldelli, G.; Perlini, C.; Tomelleri, L.; Tansella, M.; Brambilla, P. Classification of schizophrenia using feature-based morphometry. J. Neural Transm. 2011, 119, 395–404. [Google Scholar] [CrossRef]
- Rajarethinam, R.; Dequardo, J.R.; Miedler, J.; Arndt, S.; Kirbat, R.; Brunberg, J.A.; Tandon, R. Hippocampus and amygdala in schizophrenia: Assessment of the relationship of neuroanatomy to psychopathology. Psychiatry Res. Neuroimaging 2001, 108, 79–87. [Google Scholar] [CrossRef]
- Joca, S.R.L.; Padovan, C.M.; Guimaraes, F.S. Activation of post-synaptic 5-HT(1A) receptors in the dorsal hippocampus prevents learned helplessness development. Brain Res. 2003, 978, 177–184. [Google Scholar] [CrossRef]
- Kesner, R.P.; Lee, I.; Gilbert, P. A behavioral assessment of hippocampal function based on a subregional analysis. Rev. Neurosci. 2004, 15, 333–352. [Google Scholar] [CrossRef] [PubMed]
- Schobel, S.A.; Lewandowski, N.M.; Corcoran, C.; Moore, H.; Brown, T.; Malaspina, D.; Small, S.A. Differential targeting of the CA1 subfield of the hippocampal formation by schizophrenia and related psychotic disorders. Arch. Gen. Psychiatry 2009, 66, 938–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eastwood, S.; Falkai, P.; Bogerts, B.; Harrison, P. Poly(A)+mRNA as a marker of metabolic activity in schizophrenia: Differential changes in parahippocampal gyrus and CA. Schizophr. Res. 1995, 15, 57. [Google Scholar] [CrossRef]
- Fudge, J.L.; Decampo, D.M.; Becoats, K.T. Revisiting the hippocampal–amygdala pathway in primates: Association with immature-appearing neurons. Neuroscience 2012, 212, 104–119. [Google Scholar] [CrossRef] [Green Version]
- Fudge, J.L.; Tucker, T. Amygdala projections to central amygdaloid nucleus subdivisions and transition zones in the primate. Neuroscience 2009, 159, 819–841. [Google Scholar] [CrossRef] [Green Version]
- Pawel, K.; Helmut, H.; Valentina, M.; Woltersdorf, F.; Masson, T.; Ulfig, N.; Schmidt-Kastner, R.; Korr, H.; Steinbusch, H.W.M.; Hof, P.R.; et al. Volume, neuron density and total neuron number in five subcortical regions in schizophrenia. Brain 2007, 130, 678–692. [Google Scholar]
- Romanski, L.M.; LeDoux, J.E. Information cascade from primary auditory cortex to the amygdala: Corticocortical and corticoamygdaloid projections of temporal cortex in the rat. Cereb. Cortex 1993, 3, 515–532. [Google Scholar] [CrossRef]
- Brown, S.; Rutland, J.W.; Verma, G.; Feldman, R.E.; Alper, J.; Schneider, M.; Delman, B.N.; Murrough, J.M.; Balchandani, P. Structural MRI at 7T reveals amygdala nuclei and hippocampal subfield volumetric association with Major Depressive Disorder symptom severity. Sci. Rep. 2019, 9, 10166. [Google Scholar] [CrossRef] [Green Version]
- McMahon, F.; Lee, D.S.; Trinh, H.; Tward, D.; Miller, M.I.; Younes, L.; Barta, P.E.; Ratnanather, J.T. Morphometry of the amygdala in schizophrenia and psychotic bipolar disorder. Schizophr. Res. 2015, 164, 199–202. [Google Scholar] [CrossRef] [Green Version]
- Decampo, D.M.; Fudge, J.L. Amygdala projections to the lateral bed nucleus of the stria terminalis in the macaque: Comparison with ventral striatal afferents. J. Comp. Neurol. 2013, 521, 3191–3216. [Google Scholar] [CrossRef] [Green Version]
- Epstein, J.; Stern, E.; Silbersweig, D. Mesolimbic activity associated with psychosis in schizophrenia: Symptom-specific PET studies. Ann. N. Y. Acad. Sci. 1999, 877, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Prestia, A.; Boccardi, M.; Galluzzi, S.; Cavedoa, E.; Adorniab, A.; Soricellic, A.; Bonettid, M.; Geroldiab, C.; Giannakopoulosef, P.; Thompson, P.; et al. Hippocampal and amygdalar volume changes in elderly patients with Alzheimer’s disease and schizophrenia. Psychiatry Res. 2011, 192, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Van Haren, N.E.M.; Schnack, H.G.; Koevoets, M.G.; Cahn, W.; Pol, H.H.; Kahn, R.S.; Information, P.E.K.F.C. Trajectories of subcortical volume change in schizophrenia: A 5-year follow-up. Schizophr. Res. 2016, 173, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Barth, C.; Jørgensen, K.N.; Wortinger, L.A.; Nerland, S.; Jönsson, E.G.; Agartz, I. Trajectories of brain volume change over 13 years in chronic schizophrenia. Schizophr. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lutz, O.; Lizano, P.; Mothi, S.S.; Zeng, V.; Hegde, R.R.; Hoang, D.T.; Henson, P.; Brady, R.; Tamminga, C.A.; Pearlson, G.; et al. Do neurobiological differences exist between paranoid and non-paranoid schizophrenia? Findings from the bipolar schizophrenia network on intermediate phenotypes study. Schizophr. Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chand, G.B.; Dwyer, D.B.; Erus, G.; Sotiras, A.; Varol, E.; Srinivasan, D.; Doshi, J.; Pomponio, R.; Pigoni, A.; Dazzan, P.; et al. Two distinct neuroanatomical subtypes of schizophrenia revealed using machine learning. Brain 2020, 143, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
| Index | Patients (n = 57) | Controls (n = 69) | T/χ2 | p |
|---|---|---|---|---|
| Age | 37.85 ± 13.63 | 35.32 ± 11.17 | 1.15 1 | 0.252 |
| Gender (F/M) | 9/48 | 20/49 | 3.068 2 | 0.08 |
| Method | Accuracy | Sensitivity | Specificity | AUC | Feature number | p |
|---|---|---|---|---|---|---|
| Full feature set | 53.97% | 56.14% | 55.07% | 0.5362 | 46 | 0.226 |
| F score | 55.56% | 70.17% | 47.83% | 0.5751 | 11 | 0.062 |
| T-test | 56.35% | 78.95% | 37.68% | 0.5238 | 11 | 0.329 |
| Gini Index | 57.14% | 64.91% | 52.17% | 0.5700 | 13 | 0.088 |
| SFS | 74.60% | 75.44% | 75.36% | 0.7442 | 5 | <0.001 |
| SFFS | 71.43% | 80.70% | 66.67% | 0.7732 | 6 | <0.001 |
| SBFS | 58.73% | 82.46% | 46.38% | 0.6293 | 30 | 0.007 |
| SBE | 81.75% | 84.21% | 81.15% | 0.8241 | 17 | <0.001 |
| Classifier | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| SVM | 81.75% | 84.21% | 81.15% | 0.8241 |
| KNN | 61.90% | 52.63% | 66.67% | 0.5960 |
| RF | 58.73% | 47.37% | 72.46% | 0.5489 |
| FNN | 57.14% | 63.16% | 59.42% | 0.5934 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Qiu, J.; Lu, W. Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions. Brain Sci. 2020, 10, 562. https://doi.org/10.3390/brainsci10080562
Guo Y, Qiu J, Lu W. Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions. Brain Sciences. 2020; 10(8):562. https://doi.org/10.3390/brainsci10080562
Chicago/Turabian StyleGuo, Yingying, Jianfeng Qiu, and Weizhao Lu. 2020. "Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions" Brain Sciences 10, no. 8: 562. https://doi.org/10.3390/brainsci10080562
APA StyleGuo, Y., Qiu, J., & Lu, W. (2020). Support Vector Machine-Based Schizophrenia Classification Using Morphological Information from Amygdaloid and Hippocampal Subregions. Brain Sciences, 10(8), 562. https://doi.org/10.3390/brainsci10080562





