Sleep Duration in Mouse Models of Neurodevelopmental Disorders
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. TSC
2.3. Oxtr KO
2.4. Shank3 e4-9 KO
2.5. Home-Cage Assessment of Sleep Duration
2.6. Statistical Analysis
3. Results
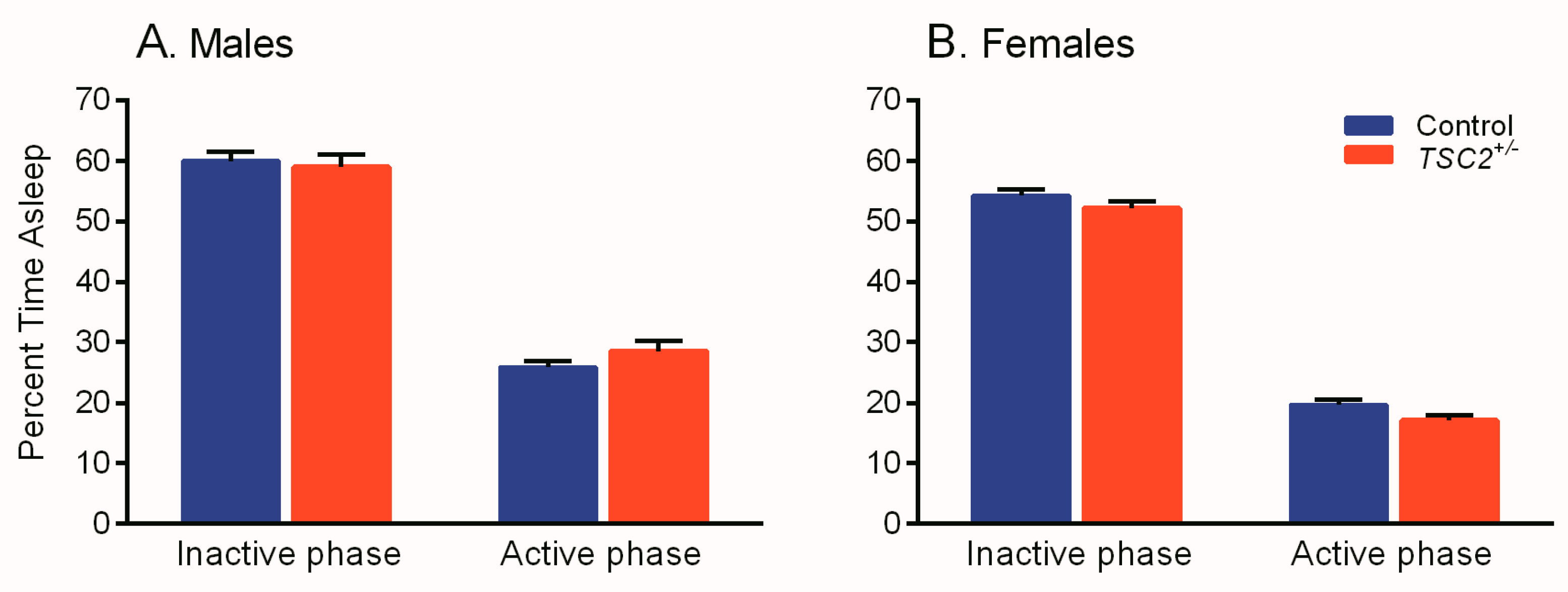
3.1. Sleep Duration in a Mouse Model of TSC
3.2. Sleep Duration in Oxtr KO Mice
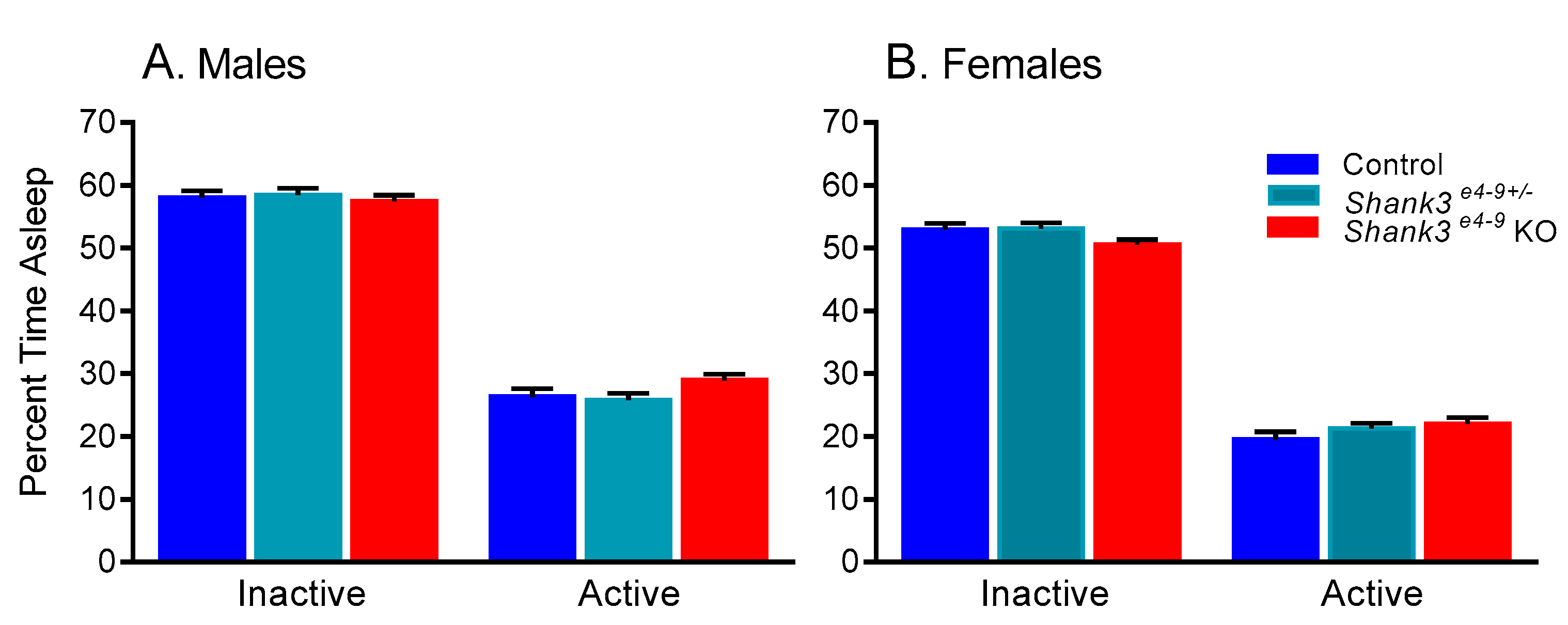
3.3. Sleep Duration in Shank3 e4-9 Mouse Model of Phelan–McDermid Syndrome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Picchioni, D.; Reith, R.M.; Nadel, J.L.; Smith, C.B. Sleep, plasticity and the pathophysiology of neurodevelopmental disorders: The potential roles of protein synthesis and other cellular processes. Brain Sci. 2014, 4, 150–201. [Google Scholar] [CrossRef] [PubMed]
- Colas, D.; Wagstaff, J.; Fort, P.; Salvert, D.; Sarda, N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol. Dis. 2005, 20, 471–478. [Google Scholar] [CrossRef]
- Colas, D.; Valletta, J.S.; Takimoto-Kimura, R.; Nishino, S.; Fujiki, N.; Mobley, W.C.; Mignot, E. Sleep and EEG features in genetic models of Down syndrome. Neurobiol. Dis. 2008, 30, 1–7. [Google Scholar] [CrossRef]
- Sare, R.M.; Harkless, L.; Levine, M.; Torossian, A.; Sheeler, C.A.; Smith, C.B. Deficient Sleep in Mouse Models of Fragile X Syndrome. Front. Mol. Neurosci. 2017, 10, 280. [Google Scholar] [CrossRef]
- Boone, C.E.; Davoudi, H.; Harrold, J.B.; Foster, D.J. Abnormal Sleep Architecture and Hippocampal Circuit Dysfunction in a Mouse Model of Fragile X Syndrome. Neuroscience 2018, 384, 275–289. [Google Scholar] [CrossRef]
- Moretti, P.; Bouwknecht, J.A.; Teague, R.; Paylor, R.; Zoghbi, H.Y. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum. Mol. Genet. 2005, 14, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Horev, G.; Ellegood, J.; Lerch, J.P.; Son, Y.E.; Muthuswamy, L.; Vogel, H.; Krieger, A.M.; Buja, A.; Henkelman, R.M.; Wigler, M.; et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc. Natl. Acad. Sci. USA 2011, 108, 17076–17081. [Google Scholar] [CrossRef] [PubMed]
- Koehl, M.; Battle, S.E.; Turek, F.W. Sleep in female mice: A strain comparison across the estrous cycle. Sleep 2003, 26, 267–272. [Google Scholar] [CrossRef]
- Wisor, J.P.; DeLorey, T.M.; Homanics, G.E.; Edgar, D.M. Sleep states and sleep electroencephalographic spectral power in mice lacking the beta 3 subunit of the GABA(A) receptor. Brain Res. 2002, 955, 221–228. [Google Scholar] [CrossRef]
- El Helou, J.; Belanger-Nelson, E.; Freyburger, M.; Dorsaz, S.; Curie, T.; La Spada, F.; Gaudreault, P.O.; Beaumont, E.; Pouliot, P.; Lesage, F.; et al. Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. USA 2013, 110, 9974–9979. [Google Scholar] [CrossRef]
- Liu, J.J.; Grace, K.P.; Horner, R.L.; Cortez, M.A.; Shao, Y.; Jia, Z. Neuroligin 3 R451C mutation alters electroencephalography spectral activity in an animal model of autism spectrum disorders. Mol. Brain 2017, 10, 10. [Google Scholar] [CrossRef]
- Osborne, J.P.; Fryer, A.; Webb, D. Epidemiology of tuberous sclerosis. Ann. N. Y. Acad. Sci. 1991, 615, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Smalley, S.L.; Tanguay, P.E.; Smith, M.; Gutierrez, G. Autism and tuberous sclerosis. J. Autism Dev. Disord. 1992, 22, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Lipton, J.O.; Boyle, L.M.; Yuan, E.D.; Hochstrasser, K.J.; Chifamba, F.F.; Nathan, A.; Tsai, P.T.; Davis, F.; Sahin, M. Aberrant Proteostasis of BMAL1 Underlies Circadian Abnormalities in a Paradigmatic mTOR-opathy. Cell Rep. 2017, 20, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Pobbe, R.L.; Pearson, B.L.; Defensor, E.B.; Bolivar, V.J.; Young, W.S., 3rd; Lee, H.J.; Blanchard, D.C.; Blanchard, R.J. Oxytocin receptor knockout mice display deficits in the expression of autism-related behaviors. Horm. Behav. 2012, 61, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Costales, J.L.; Kolevzon, A. Phelan-McDermid Syndrome and SHANK3: Implications for Treatment. Neurother. J. Am. Soc. Exp. Neurother. 2015, 12, 620–630. [Google Scholar] [CrossRef]
- Bro, D.; O’Hara, R.; Primeau, M.; Hanson-Kahn, A.; Hallmayer, J.; Bernstein, J.A. Sleep Disturbances in Individuals With Phelan-McDermid Syndrome: Correlation With Caregivers’ Sleep Quality and Daytime Functioning. Sleep 2017, 40. [Google Scholar] [CrossRef]
- Ingiosi, A.M.; Schoch, H.; Wintler, T.; Singletary, K.G.; Righelli, D.; Roser, L.G.; Medina, E.; Risso, D.; Frank, M.G.; Peixoto, L. Shank3 modulates sleep and expression of circadian transcription factors. Elife 2019, 8. [Google Scholar] [CrossRef]
- Lee, H.J.; Caldwell, H.K.; Macbeth, A.H.; Tolu, S.G.; Young, W.S., 3rd. A conditional knockout mouse line of the oxytocin receptor. Endocrinology 2008, 149, 3256–3263. [Google Scholar] [CrossRef]
- Sare, R.M.; Lemons, A.; Torossian, A.; Beebe Smith, C. Noninvasive, High-throughput Determination of Sleep Duration in Rodents. J. Vis. Exp. JoVE 2018. [Google Scholar] [CrossRef]
- Pack, A.I.; Galante, R.J.; Maislin, G.; Cater, J.; Metaxas, D.; Lu, S.; Zhang, L.; Von Smith, R.; Kay, T.; Lian, J.; et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol. Genom. 2007, 28, 232–238. [Google Scholar] [CrossRef]
- Buckley, A.W.; Rodriguez, A.J.; Jennison, K.; Buckley, J.; Thurm, A.; Sato, S.; Swedo, S. Rapid eye movement sleep percentage in children with autism compared with children with developmental delay and typical development. Arch. Pediatr. Adolesc. Med. 2010, 164, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Chilakamarri, P.; Thurm, A.; Farmer, C.; Swedo, S.; Burroughs, S.A.; Holmes, G.L.; Buckley, A.W. Characterizing Sleep Spindles in Children with Autism Spectum Disorder (ASD). Neurology 2017, 88, P3.209. [Google Scholar]
- Borbely, A.A.; Achermann, P. Sleep homeostasis and models of sleep regulation. J. Biol. Rhythm. 1999, 14, 557–568. [Google Scholar]
- Geoffray, M.M.; Nicolas, A.; Speranza, M.; Georgieff, N. Are circadian rhythms new pathways to understand Autism Spectrum Disorder? J. Physiol. Paris 2016, 110, 434–438. [Google Scholar] [CrossRef]
- Gannon, R.L. Non-peptide oxytocin receptor ligands and hamster circadian wheel running rhythms. Brain Res. 2014, 1585, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Angelakos, C.C.; Tudor, J.C.; Ferri, S.L.; Jongens, T.A.; Abel, T. Home-cage hypoactivity in mouse genetic models of autism spectrum disorder. Neurobiol. Learn. Mem. 2019, 165, 107000. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.N.; Dugovic, C.; Turek, F.W.; Laposky, A.D. Diurnal sex differences in the sleep-wake cycle of mice are dependent on gonadal function. Sleep 2006, 29, 1211–1223. [Google Scholar] [CrossRef]
- Koehl, M.; Battle, S.; Meerlo, P. Sex differences in sleep: The response to sleep deprivation and restraint stress in mice. Sleep 2006, 29, 1224–1231. [Google Scholar] [CrossRef]
- Mallampalli, M.P.; Carter, C.L. Exploring sex and gender differences in sleep health: A Society for Women’s Health Research Report. J. Womens Health 2014, 23, 553–562. [Google Scholar] [CrossRef]
- Seney, M.L.; Sibille, E. Sex differences in mood disorders: Perspectives from humans and rodent models. Biol. Sex Differ. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Vina, J.; Lloret, A. Why women have more Alzheimer’s disease than men: Gender and mitochondrial toxicity of amyloid-beta peptide. J. Alzheimers Dis. 2010, 20 (Suppl. 2), S527–S533. [Google Scholar] [CrossRef]

| Model | Interaction | Main Effect | F(df, Error) Value | p-Value |
|---|---|---|---|---|
| Tsc2+/− | ||||
| Sex × genotype × phase | F(1, 84) = 1.325 | 0.253 | ||
| Sex × phase | F(1, 84) = 2.361 | 0.128 | ||
| Genotype × phase | F(1, 84) = 1.210 | 0.274 | ||
| Sex × genotype | F(1, 84) = 1.921 | 0.169 | ||
| Sex | F(1, 84) = 49.048 | <0.001 * | ||
| Genotype | F(1, 84) = 0.312 | 0.578 | ||
| Phase | F(1, 84) = 1806.221 | <0.001 * | ||
| Oxtr KO | ||||
| Sex × genotype × phase | F(2, 126) = 1.022 | 0.363 | ||
| Sex × phase | F(1, 126) =0.005 | 0.945 | ||
| Genotype × phase | F(2, 126) = 0.983 | 0.377 | ||
| Sex × genotype | F(2, 126) = 0.396 | 0.674 | ||
| Sex | F(1, 126) = 63.486 | <0.001 * | ||
| Genotype | F(2, 126) = 0.987 | 0.375 | ||
| Phase | F(1, 126) = 2481.879 | <0.001 * | ||
| Shank3 e4-9 KO | ||||
| Sex × genotype × phase | F(2, 137) = 0.537 | 0.586 | ||
| Sex × phase | F(1, 137) =0.125 | 0.725 | ||
| Genotype × phase | F(2, 137) = 6.480 | 0.002 * | ||
| Sex × genotype | F(2, 137) = 0.708 | 0.494 | ||
| Sex | F(1, 137) = 74.197 | <0.001 * | ||
| Genotype | F(2, 137) = 0.230 | 0.795 | ||
| Phase | F(1, 137) = 3771.461 | <0.001 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saré, R.M.; Lemons, A.; Song, A.; Smith, C.B. Sleep Duration in Mouse Models of Neurodevelopmental Disorders. Brain Sci. 2021, 11, 31. https://doi.org/10.3390/brainsci11010031
Saré RM, Lemons A, Song A, Smith CB. Sleep Duration in Mouse Models of Neurodevelopmental Disorders. Brain Sciences. 2021; 11(1):31. https://doi.org/10.3390/brainsci11010031
Chicago/Turabian StyleSaré, Rachel Michelle, Abigail Lemons, Alex Song, and Carolyn Beebe Smith. 2021. "Sleep Duration in Mouse Models of Neurodevelopmental Disorders" Brain Sciences 11, no. 1: 31. https://doi.org/10.3390/brainsci11010031
APA StyleSaré, R. M., Lemons, A., Song, A., & Smith, C. B. (2021). Sleep Duration in Mouse Models of Neurodevelopmental Disorders. Brain Sciences, 11(1), 31. https://doi.org/10.3390/brainsci11010031





