Optimization of Neurite Tracing and Further Characterization of Human Monocyte-Derived-Neuronal-like Cells
Abstract
:1. Introduction
2. Methods
2.1. Cell Culture
2.2. Single Cell RNA-Sequencing
2.3. Statistical Analysis
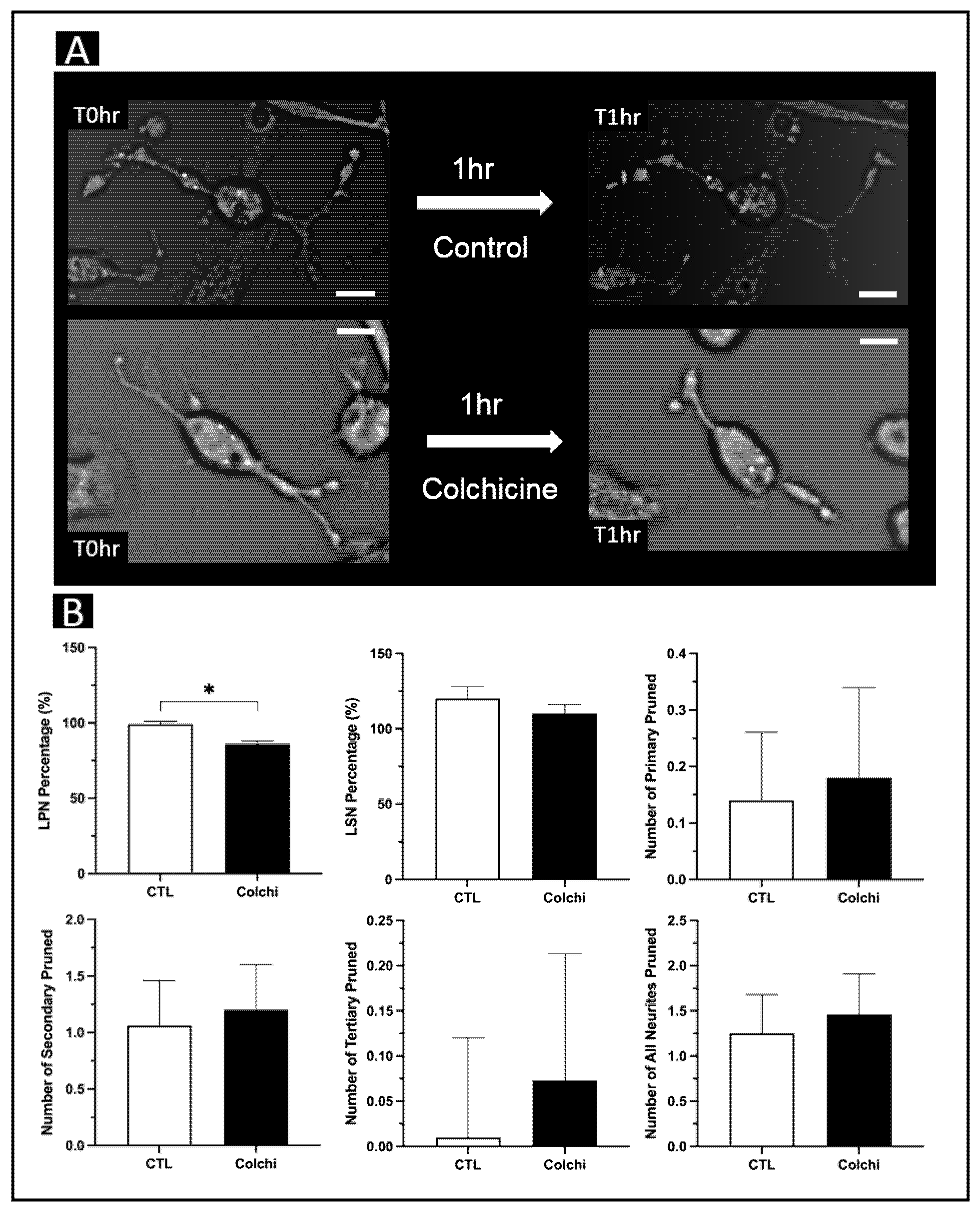
3. Results
3.1. Neuronal and Monocyte Markers in MDNCs, SH-SY5Y and THP-1 Cells
3.2. Whole-Cell Tracing
3.3. Three Neurite Tracing Approaches
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christensen, D.L.; Baio, J.; Braun, K.V.N.; Bilder, D.; Charles, J.; Constantino, J.N.; Daniels, J.; Durkin, M.S.; Fitzgerald, R.T.; Kurzius-Spencer, M.; et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and developmental disabilities monitoring network, 11 Sites, United States, 2012. MMWR Surveill. Summ. 2016, 65, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Chant, D.; Welham, J.; McGrath, J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005, 2, e141. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Tóth, F.; Polyák, H.; Szabó, A.; Mándi, Y.; Vécsei, L. Immune influencers in action: Metabolites and enzymes of the Tryptophan-Kynurenine metabolic pathway. Biomedicines 2021, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.A.; Lin, Y.-K.; Lai, J.-H.; Lo, Y.-C.; Yang, Y.-C.S.H.; Ye, S.-Y.; Lee, C.-J.; Wang, C.-C.; Chiang, Y.-H.; Tseng, S.-H. Maternal immune activation causes social behavior deficits and hypomyelination in male rat offspring with an autism-like microbiota profile. Brain Sci. 2021, 11, 1085. [Google Scholar] [CrossRef] [PubMed]
- Abuaish, S.; Al-Otaibi, N.; Abujamel, T.; Alzahrani, S.; Alotaibi, S.; AlShawakir, Y.; Aabed, K.; El-Ansary, A. Fecal transplant and Bifidobacterium treatments modulate gut Clostridium bacteria and rescue social impairment and hippocampal BDNF expression in a rodent model of autism. Brain Sci. 2021, 11, 1038. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, H.; Watanabe, E.; Fukuchi, M. Psychiatric neural networks and precision therapeutics by machine learning. Biomedicines 2021, 9, 403. [Google Scholar] [CrossRef]
- Correia, B.; Nani, J.; Ricardo, R.W.; Stanisic, D.; Costa, T.; Hayashi, M.; Tasic, L. Effects of psychostimulants and antipsychotics on serum lipids in an animal model for schizophrenia. Biomedicines 2021, 9, 235. [Google Scholar] [CrossRef]
- Rog, J.; Błażewicz, A.; Juchnowicz, D.; Ludwiczuk, A.; Stelmach, E.; Kozioł, M.; Karakula, M.; Niziński, P.; Karakula-Juchnowicz, H. The role of GPR120 receptor in essential fatty acids metabolism in schizophrenia. Biomedicines 2020, 8, 243. [Google Scholar] [CrossRef]
- Kalus, P.; Muller, T.J.; Zuschratter, W.; Senitz, D. The dendritic architecture of prefrontal pyramidal neurons in schizophrenic patients. NeuroReport 2000, 11, 3621–3625. [Google Scholar] [CrossRef]
- Kalus, P.; Bondzio, J.; Federspiel, A.; Müller, T.J.; Zuschratter, W. Cell-type specific alterations of cortical interneurons in schizophrenic patients. NeuroReport 2002, 13, 713–717. [Google Scholar] [CrossRef]
- Iritani, S.; Kuroki, N.; Niizato, K.; Ikeda, K. Morphological changes in neuropeptide Y-positive fiber in the hippocampal formation of schizophrenics. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2000, 24, 241–249. [Google Scholar] [CrossRef]
- Martínez-Cerdeño, V. Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev. Neurobiol. 2017, 77, 393–404. [Google Scholar] [CrossRef]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.; et al. Autism spectrum disorder: Neuropathology and animal models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef] [PubMed]
- Borgmann-Winter, K.; Rawson, N.; Wang, H.-Y.; MacDonald, M.; Ozdener, M.; Yee, K.; Gomez, G.; Xu, J.; Bryant, B.; Adamek, G.; et al. Human olfactory epithelial cells generated in vitro express diverse neuronal characteristics. Neuroscience 2009, 158, 642–653. [Google Scholar] [CrossRef] [Green Version]
- Borgmann-Winter, K.; Willard, S.L.; Sinclair, D.; Mirza, N.; Turetsky, B.; Berretta, S.; Hahn, C.-G. Translational potential of olfactory mucosa for the study of neuropsychiatric illness. Transl. Psychiatry 2015, 5, e527. [Google Scholar] [CrossRef] [Green Version]
- Féron, F.; Perry, C.; McGrath, J.J.; Mackay-Sim, A. New techniques for biopsy and culture of human olfactory epithelial neurons. Arch. Otolaryngol. Head Neck Surg. 1998, 124, 861–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodbury, D.; Schwarz, E.J.; Prockop, D.J.; Black, I.B. Adult rat and human bone marrow stromal cells differentiate into neurons. J. Neurosci. Res. 2000, 61, 364–370. [Google Scholar] [CrossRef]
- Taran, R.; Mamidi, M.K.; Singh, G.; Dutta, S.; Parhar, I.S.; John, J.P.; Bhonde, R.; Pal, R.; Das, A.K. In vitro and in vivo neurogenic potential of mesenchymal stem cells isolated from different sources. J. Biosci. 2014, 39, 157–169. [Google Scholar] [CrossRef]
- Wen, Z.; Christian, K.M.; Song, H.; Ming, G.-L. Modeling psychiatric disorders with patient-derived iPSCs. Curr. Opin. Neurobiol. 2016, 36, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Urbach, A.; Bar-Nur, O.; Daley, G.Q.; Benvenisty, N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell 2010, 6, 407–411. [Google Scholar] [CrossRef] [Green Version]
- Pera, M.F. Stem cells: The dark side of induced pluripotency. Nature 2011, 471, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Bellin, M.; Marchetto, M.C.; Gage, F.H.; Mummery, C. Induced pluripotent stem cells: The new patient? Nat. Rev. Mol. Cell Biol. 2012, 13, 713–726. [Google Scholar] [CrossRef]
- Hu, B.-Y.; Weick, J.P.; Yu, J.; Ma, L.-X.; Zhang, X.-Q.; Thomson, J.A.; Zhang, S.-C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [Green Version]
- Dolmetsch, R.; Geschwind, D.H. The human brain in a dish: The promise of iPSC-derived neurons. Cell 2011, 145, 831–834. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Ng, Y.H.; Pang, Z.; Südhof, T.C.; Wernig, M. Induced neuronal cells: How to make and define a neuron. Cell Stem Cell 2011, 9, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Su, S.; Zhou, S.; Yang, W.; Deng, X.; Sun, Y.; Li, L.; Li, Y. How to reprogram human fibroblasts to neurons. Cell Biosci. 2020, 10, 116. [Google Scholar] [CrossRef]
- Zhou, M.; Tao, X.; Sui, M.; Cui, M.; Liu, D.; Wang, B.; Wang, T.; Zheng, Y.; Luo, J.; Mu, Y.; et al. Reprogramming astrocytes to motor neurons by activation of endogenous Ngn2 and Isl1. Stem Cell Rep. 2021, 16, 1777–1791. [Google Scholar] [CrossRef]
- Bellon, A.; Wegener, A.M.-A.; Lescallette, A.R.; Valente, M.; Yang, S.-K.; Gardette, R.; Matricon, J.; Mouaffak, F.; Watts, P.J.; Vimeux, L.; et al. Transdifferentiation of human circulating monocytes into neuronal-like cells in 20 days and without reprograming. Front. Mol. Neurosci. 2018, 11, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosik, K.; Finch, E. MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: An immunocytochemical study of cultured rat cerebrum. J. Neurosci. 1987, 7, 3142–3153. [Google Scholar] [CrossRef] [PubMed]
- Dotti, C.G.; A Sullivan, C.; A Banker, G. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988, 8, 1454–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehmelt, L.; Halpain, S. Actin and microtubules in neurite initiation: Are MAPs the missing link? J. Neurobiol. 2004, 58, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Areal, L.B.; Blakely, R.D. Neurobehavioral changes arising from early life dopamine signaling perturbations. Neurochem. Int. 2020, 137, 104747. [Google Scholar] [CrossRef] [PubMed]
- Sanches, M.; Keshavan, M.S.; Brambilla, P.; Soares, J.C. Neurodevelopmental basis of bipolar disorder: A critical appraisal. Prog. Neuro Psychopharmacol. Biol. Psychiatry 2008, 32, 1617–1627. [Google Scholar] [CrossRef] [PubMed]
- Schnell, U.; Dijk, F.; Sjollema, K.A.; Giepmans, B. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 2012, 9, 152–158. [Google Scholar] [CrossRef]
- Cheng, R.; Zhang, F.; Li, M.; Wo, X.; Su, Y.-W.; Wang, W. Influence of fixation and permeabilization on the mass density of single cells: A surface plasmon resonance imaging study. Front. Chem. 2019, 7, 588. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; A Williams, B.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ma, Z.; Shi, M.; Malty, R.H.; Aoki, H.; Minic, Z.; Phanse, S.; Jin, K.; Wall, D.P.; Zhang, Z.; et al. Identification of Human Neuronal Protein Complexes Reveals Biochemical Activities and Convergent Mechanisms of Action in Autism Spectrum Disorders. Cell Syst. 2015, 1, 361–374. [Google Scholar] [CrossRef] [Green Version]
- Südhof, T.C. The cell biology of synapse formation. J. Cell Biol. 2021, 220, e202103052. [Google Scholar] [CrossRef]
- Antonucci, F.; Corradini, I.; Fossati, G.; Tomasoni, R.; Menna, E.; Matteoli, M. SNAP-25, a known presynaptic protein with emerging postsynaptic functions. Front. Synaptic Neurosci. 2016, 8, 7. [Google Scholar] [CrossRef] [Green Version]
- Nowack, A.; Yao, J.; Custer, K.L.; Bajjalieh, S.M. SV2 regulates neurotransmitter release via multiple mechanisms. Am. J. Physiol. Physiol. 2010, 299, C960–C967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, S.; Stewart, B.; Niemeyer, B.A.; Burgess, R.; McCabe, B.; Lin, P.; Boulianne, G.; O’Kane, C.; Schwarz, T.L. Members of the synaptobrevin/vesicle-associated membrane protein (VAMP) family in Drosophila are functionally interchangeable in vivo for neurotransmitter release and cell viability. Proc. Natl. Acad. Sci. USA 2002, 99, 13867–13872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Naisbitt, S.; Yoon, J.; Hwang, J.-I.; Suh, P.-G.; Sheng, M.; Kim, E. Characterization of the shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J. Biol. Chem. 1999, 274, 29510–29518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burré, J. The Synaptic Function of α-Synuclein. J. Park. Dis. 2015, 5, 699–713. [Google Scholar] [CrossRef] [Green Version]
- Bennett, M.K.; Calakos, N.; Scheller, R.H. Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 1992, 257, 255–259. [Google Scholar] [CrossRef]
- Schumacher, N.; Borawski, J.M.; Leberfinger, C.B.; Gessler, M.; Kerkhoff, E. Overlapping expression pattern of the actin organizers Spir-1 and formin-2 in the developing mouse nervous system and the adult brain. Gene Expr. Patterns 2004, 4, 249–255. [Google Scholar] [CrossRef]
- Barbier, P.; Zejneli, O.; Martinho, M.; Lasorsa, A.; Belle, V.; Smet-Nocca, C.; Tsvetkov, P.O.; Devred, F.; Landrieu, I. Role of Tau as a microtubule-associated protein: Structural and Functional aspects. Front. Aging Neurosci. 2019, 11, 204. [Google Scholar] [CrossRef] [Green Version]
- Toriyama, M.; Shimada, T.; Kim, K.B.; Mitsuba, M.; Nomura, E.; Katsuta, K.; Sakumura, Y.; Roepstorff, P.; Inagaki, N. Shootin1: A protein involved in the organization of an asymmetric signal for neuronal polarization. J. Cell Biol. 2006, 175, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Meiri, K.F.; Pfenninger, K.H.; Willard, M.B. Growth-associated protein, GAP-43, a polypeptide that is induced when neurons extend axons, is a component of growth cones and corresponds to pp46, a major polypeptide of a subcellular fraction enriched in growth cones. Proc. Natl. Acad. Sci. USA 1986, 83, 3537–3541. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, M.; Ebert, B.; Wafford, K.; Smart, T.G. Distinct activities of GABA agonists at synaptic- and extrasynaptic-type GABAA receptors. J. Physiol. 2010, 588, 1251–1268. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.C.; Khan, S.Q.; Kaneda, M.M.; Pathria, P.; Shepard, R.; Louis, T.L.; Anand, S.; Woo, G.; Leem, C.; Faridi, M.H.; et al. Integrin CD11b activation drives anti-tumor innate immunity. Nat. Commun. 2018, 9, 5379. [Google Scholar] [CrossRef]
- Tu, M.M.; Abdel-Hafiz, H.A.; Jones, R.T.; Jean, A.; Hoff, K.J.; Duex, J.E.; Chauca-Diaz, A.; Costello, J.C.; Dancik, G.M.; Tamburini, B.A.J.; et al. Inhibition of the CCL2 receptor, CCR2, enhances tumor response to immune checkpoint therapy. Commun. Biol. 2020, 3, 720. [Google Scholar] [CrossRef]
- Daniels, M.P. Colchicine inhibition of nerve fiber formation in vitro. J. Cell Biol. 1972, 53, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Brat, D.J.; Brimijoin, S. A paradigm for examining toxicant effects on viability, structure, and axonal transport of neurons in culture. Mol. Neurobiol. 1992, 6, 125–135. [Google Scholar] [CrossRef]
- Drubin, D.; Kobayashi, S.; Kellogg, D.; Kirschner, M. Regulation of microtubule protein levels during cellular morphogenesis in nerve growth factor-treated PC12 cells. J. Cell Biol. 1988, 106, 1583–1591. [Google Scholar] [CrossRef] [Green Version]
- Vaags, A.K.; Lionel, A.C.; Sato, D.; Goodenberger, M.; Stein, Q.; Curran, S.; Ogilvie, C.; Ahn, J.W.; Drmic, I.; Senman, L.; et al. Rare Deletions at the Neurexin 3 Locus in Autism Spectrum Disorder. Am. J. Hum. Genet. 2012, 90, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Onwordi, E.C.; Halff, E.F.; Whitehurst, T.; Mansur, A.; Cotel, M.-C.; Wells, L.; Creeney, H.; Bonsall, D.; Rogdaki, M.; Shatalina, E.; et al. Synaptic density marker SV2A is reduced in schizophrenia patients and unaffected by antipsychotics in rats. Nat. Commun. 2020, 11, 246. [Google Scholar] [CrossRef]
- Halim, N.D.; Weickert, C.S.; McClintock, B.W.; Hyde, T.M.; Weinberger, D.R.; E Kleinman, J.; Lipska, B.K. Presynaptic proteins in the prefrontal cortex of patients with schizophrenia and rats with abnormal prefrontal development. Mol. Psychiatry 2003, 8, 797–810. [Google Scholar] [CrossRef]
- Braida, D.; Guerini, F.R.; Ponzoni, L.; Corradini, I.; De Astis, S.; Pattini, L.; Bolognesi, E.; Benfante, R.; Fornasari, D.; Chiappedi, M.; et al. Association between SNAP-25 gene polymorphisms and cognition in autism: Functional consequences and potential therapeutic strategies. Transl. Psychiatry 2015, 5, e500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corradini, I.; Verderio, C.; Sala, M.; Wilson, M.C.; Matteoli, M. SNAP-25 in neuropsychiatric disorders. Ann. N. Y. Acad. Sci. 2009, 1152, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demirel, F.; Cetin, I.; Turan, S.; Yıldız, N.; Sağlam, T.; Duran, A. Total Tau and Phosphorylated Tau protein serum levels in patients with schizophrenia compared with controls. Psychiatr. Q. 2017, 88, 921–928. [Google Scholar] [CrossRef]
- Weickert, C.S.; Webster, M.J.; Hyde, T.M.; Herman, M.M.; Bachus, S.E.; Bali, G.; Weinberger, D.R.; Kleinman, J.E. Reduced GAP-43 mRNA in dorsolateral prefrontal cortex of patients with schizophrenia. Cereb. Cortex 2001, 11, 136–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zikopoulos, V.; Barbas, H. Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. 2010, 30, 14595–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, S.E.; Lee, V.M.; Gur, R.E.; Trojanowski, J.Q. Abnormal expression of two microtubule-associated proteins (MAP2 and MAP5) in specific subfields of the hippocampal formation in schizophrenia. Proc. Natl. Acad. Sci. USA 1991, 88, 10850–10854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, L.B.; Johnson, N.; Byne, W. Alterations in MAP2 immunocytochemistry in areas 9 and 32 of schizophrenic prefrontal cortex. Psychiatry Res. Neuroimaging 2002, 114, 137–148. [Google Scholar] [CrossRef]
- Bellon, A. New genes associated with schizophrenia in neurite formation: A review of cell culture experiments. Mol. Psychiatry 2007, 12, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Bellon, A.; Krebs, M.-O.; Jay, T.M. Factoring neurotrophins into a neurite-based pathophysiological model of schizophrenia. Prog. Neurobiol. 2011, 94, 77–90. [Google Scholar] [CrossRef]
- Emerelo, V.; Edurand, D.; Lescallette, A.R.; Vrana, K.; Hong, L.E.; Faghihi, M.A.; Ebellon, A. Associating schizophrenia, long non-coding RNAs and neurostructural dynamics. Front. Mol. Neurosci. 2015, 8, 57. [Google Scholar] [CrossRef]
- Donohue, D.E.; Ascoli, G.A. Automated reconstruction of neuronal morphology: An overview. Brain Res. Rev. 2011, 67, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C.; Thompson-Peer, K.L. Comparing automated morphology quantification software on dendrites of uninjured and injured drosophila neurons. Neuroinformatics 2021, 1–15. [Google Scholar] [CrossRef]


| Protein | Gene | Identifier | Function | Reference |
|---|---|---|---|---|
| Neurexin 3 | NRXN3 | ENSG00000021645 | Synapsis | Sudhof 2021 [39] |
| Synaptosome-associated protein 25 | SNAP25 | ENSG00000132639 | Synapsis | Antonucci et al. 2016 [40] |
| Synaptic vesicle glycoprotein 2A | SV2A | ENSG00000159164 | Synapsis | Nowack et al. 2010 [41] |
| Vesicle-associated membrane protein 1 | VAMP1 | ENSG00000139190 | Synapsis | Bhattacharya et al. 2002 [42] |
| SH3 and multiple ankyrin repeat domains 2 | SHANK2 | ENSG00000162105 | Synapsis | Lim et al. 1999 [43] |
| Synuclein alpha | SNCA | ENSG00000145335 | Synapsis | Burre 2015 [44] |
| Syntaxin 1A | STX1A | ENSG00000106089 | Synapsis | Bennett et al. 1992 [45] |
| Spire type actin nucleation factor 1 | SPIRE1 | ENSG00000134278 | Neuronal structure | Schumacher et al, 2004 [46] |
| Microtubule-associated protein tau | MAPT | ENSG00000186868 | Neuronal structure | Barbier et al. 2019 [47] |
| Shootin 1 | SHTN1 (KIAA1598) | ENSG00000187164 | Neuronal structure | Toriyama et al. 2006 [48] |
| Growth-associated protein 43 | GAP43 | ENSG00000172020 | Neuronal structure | Meiri et al. 1986 [49] |
| Gamma-aminobutyric acid (GABA) type A receptor subunit beta 3 | GABRB3 | ENSG00000166206 | GABA receptor | Mortensen et al. 2010 [50] |
| Integrin subunit alpha M CD11B | ITGAM | ENSG00000169896 | Immune system | Schmid et al. 2018 [51] |
| Monocyte chemoattractant protein 1 peceptor | CCR2 | ENSG00000121807 | Immune system | Tu et al. 2020 [52] |
| Gene | THP-1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | SH-SY5Y 1 * | SH-SY5Y 2 * |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NRXN3 | 0 | 0 | 0 | 0.101 | 3.106 | 0.04 | 0.012 | 0 | 0.009 | 0 | 0.288 | 0 | 0 | 0.062 | 0.061 | 0.022 | 0 | 0.98 | 0.01 | 0.135 |
| SNAP25 | 0 | 0 | 0 | 0.779 | 0.167 | 0.13 | 0.219 | 0.401 | 0 | 0 | 0 | 0 | 1.19 | 0.288 | 0 | 0 | 0.171 | 0 | 25.06 | 42.23 |
| SV2A | 0 | 0 | 0 | 0.089 | 0 | 0 | 0 | 0 | 0 | 0.127 | 0.044 | 0 | 0 | 0 | 0.086 | 0.171 | 0 | 0.055 | 10.37 | 6.810 |
| VAMP1 | 0 | 0 | 0 | 0 | 0 | 0.256 | 0 | 0 | 0 | 0 | 0.692 | 0 | 0 | 0 | 0.054 | 0 | 0 | 0 | 0.942 | 1.472 |
| SHANK2 | 0 | 0 | 0 | 0 | 0 | 0.132 | 0.031 | 0 | 0.76 | 0.068 | 0.929 | 0 | 0.059 | 0 | 0.223 | 0.552 | 0 | 0.199 | 0.219 | 0.229 |
| SNCA | 0 | 5.35 | 8.08 | 0 | 0 | 0.811 | 1.12 | 0 | 0 | 0 | 0 | 0 | 276.9 | 0 | 0 | 136.3 | 1.63 | 0 | 3.776 | 7.513 |
| STX1A | 0 | 0 | 0 | 0 | 0 | 0 | 0.028 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.742 | 9.759 |
| SPIRE1 | 0 | 27.2 | 58.02 | 0.292 | 5.19 | 0 | 0.01 | 0 | 16.34 | 0.223 | 3.37 | 0 | 0 | 17.32 | 28.61 | 110.5 | 2.85 | 23.25 | 9.341 | 6.986 |
| MAPT | 0 | 0.089 | 0.023 | 0 | 0 | 0.075 | 0.192 | 0.024 | 0.128 | 0 | 0.045 | 0.042 | 0.028 | 0 | 0 | 0.019 | 0.032 | 0.073 | 1.535 | 4.176 |
| SHTN1 | 0 | 45.73 | 25.8 | 4.66 | 0.27 | 4.62 | 2.47 | 43.59 | 4.53 | 16.44 | 20.04 | 0 | 0 | 1.36 | 12.95 | 0.154 | 0 | 83.34 | 2.038 | 3.827 |
| GAP43 | 0 | 0 | 0 | 0 | 0.211 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62.16 | 86.84 |
| GABRB3 | 0 | 0.324 | 0.458 | 0.065 | 0.254 | 0.247 | 0.079 | 0.095 | 0.21 | 0.13 | 0.06 | 0.225 | 0 | 1.06 | 0.093 | 0.901 | 0.173 | 0.182 | 10.25 | 9.763 |
| ITGAM | 14.91 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.034 | 0 | 0 | 0 | 0 | 0 | 0 | 0.014 |
| CCR2 | 24.62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.078 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellon, A.; Hasoglu, T.; Peterson, M.; Gao, K.; Chen, M.; Blandin, E.; Cortez-Resendiz, A.; Clawson, G.A.; Hong, L.E. Optimization of Neurite Tracing and Further Characterization of Human Monocyte-Derived-Neuronal-like Cells. Brain Sci. 2021, 11, 1372. https://doi.org/10.3390/brainsci11111372
Bellon A, Hasoglu T, Peterson M, Gao K, Chen M, Blandin E, Cortez-Resendiz A, Clawson GA, Hong LE. Optimization of Neurite Tracing and Further Characterization of Human Monocyte-Derived-Neuronal-like Cells. Brain Sciences. 2021; 11(11):1372. https://doi.org/10.3390/brainsci11111372
Chicago/Turabian StyleBellon, Alfredo, Tuna Hasoglu, Mallory Peterson, Katherine Gao, Michael Chen, Elisabeta Blandin, Alonso Cortez-Resendiz, Gary A. Clawson, and Liyi Elliot Hong. 2021. "Optimization of Neurite Tracing and Further Characterization of Human Monocyte-Derived-Neuronal-like Cells" Brain Sciences 11, no. 11: 1372. https://doi.org/10.3390/brainsci11111372
APA StyleBellon, A., Hasoglu, T., Peterson, M., Gao, K., Chen, M., Blandin, E., Cortez-Resendiz, A., Clawson, G. A., & Hong, L. E. (2021). Optimization of Neurite Tracing and Further Characterization of Human Monocyte-Derived-Neuronal-like Cells. Brain Sciences, 11(11), 1372. https://doi.org/10.3390/brainsci11111372







