The Application of Complexity Analysis in Brain Blood-Oxygen Signal
Abstract
:1. Introduction
2. Complexity of Brain Blood-Oxygen Signal
2.1. Physiological Complexity of Brain
2.2. Measuring Brain Complexity through Brain Blood-Oxygen Signals
3. Current Studies in Complexity of Brain Blood-Oxygen Signals
3.1. Brain Blood-Oxygen Signal Complexity as a Biomarker
| Measure | Signal Type | Res. Orientation | Participants | Main Findings | Ref |
|---|---|---|---|---|---|
| ApEn | Task-BOLD | Biomarker | Older adults (40) | Cognitive ability was positively correlated with regional brain BOLD complexity. | [22] |
| SampEn | Task-BOLD | Biomarker | ADHD (17); HC (13) | The mean whole brain BOLD complexity of ADHD group was significantly lower than the HC; the mean regional brain complexity values have a significant negative correlation with ADHD score. | [15] |
| MSE | Rest-BOLD | Biomarker | Older adults (99); Younger adults (56) | The mean whole brain BOLD complexity of younger adults was significantly higher than that of older adults; the high cognitive ability group showed significantly higher whole brain BOLD complexity than the low cognitive ability group; regional brain BOLD complexity was significantly correlated with cognitive function. | [17] |
| ApEn | Rest-BOLD | Biomarker | Younger adults (8); Older adults (8); fAD (22) | Brain BOLD complexity decreased with normal aging and cognitive decline. | [18] |
| SampEn | Task-BOLD | Biomarker | SZ (13); HC (16) | Brain BOLD complexity of SZ patients was higher than that of HC when performing Cyberball social exclusion task. | [19] |
| MSE | Rest-BOLD | Biomarker/ Methodology | Older adults (8); Younger adults (8) | Brain BOLD complexity was used to discriminate younger from older participants as well as grey matter from white matter. | [23] |
| SampEn | Rest-BOLD | Biomarker/ Methodology | Older adults (53); Younger adults (53) | SampEn was used to discriminate the younger from the elderly adults with short length data; the suggested value of m was 2. | [16] |
| SampEn | Rest-BOLD | Methodology | 1049 | Using a data-driven clustering method, the entire brain was organized into seven regional brain entropy networks that are consistent with known brain parcellation. | [24] |
| MSE | Rest-BOLD | Biomarker/ Methodology | 20 | Complexity of the BOLD signal showed different patterns from white, pink, and red noises; neural complexity across all networks was negative. | [25] |
| MSE | Rest-BOLD | Biomarker | SZ (105); HC (210) | Complexity of BOLD signals in SZ patients showed two patterns (toward either regularity or randomness), which were respectively associated with positive or negative symptoms of schizophrenia. | [26] |
| fApEn; SampEN | Rest-BOLD | Biomarker/ Methodology | 86 | Compared to SampEn, fApEn was better at discriminating different age groups and have shown to be a more sensitive method. | [27] |
| SampEn | Rest-BOLD | Biomarker | CPI (29); HC (29) | The BEN map of CPI patients demonstrated significant differences from HC, and altered functional connectivity patterns were associated with abnormal BEN regions. | [28] |
| SampEn | Rest-BOLD | Biomarker | RRMS (34); HC (34) | BOLD complexity of RRMS patients was significantly increased in some regions and was positively correlated with disease severity. | [29] |
| SampEn | Rest-BOLD | Biomarker | seafarers (20); HC (20) | BOLD complexity pattern of seafarers was significant different from HC. | [30] |
| PE | Rest-BOLD | Biomarker | MCI (65); AD (29); HC (30) | The BOLD complexity of AD patients was significantly lower than that of MCI patients and HC; that of AD patients and MCI patients was significantly correlated with ReHo in several brain regions associated with AD. | [31] |
| PE | Task-O2Hb | Biomarker | ADHD (15); HC (16) | BOLD complexity in the right dorsolateral prefrontal cortex of ADHD patients were significantly higher than that of HC. | [20] |
| SampEn; MSE | Rest-BOLD | Methodology | 354 | Proposed a generic strategy to minimize the relative error of SampEn to determine the appropriate complexity measurement parameters. | [32] |
| SampEn | Task-BOLD | Biomarker | CFS (43); HC (26) | Regional brain complexity in CFS patients was lower than that in HC when performing a Stroop task. | [33] |
| SampEn | Rest-BOLD Task-BOLD | Biomarker | CFS (45); HC (27) | BOLD complexity of CFS patients was higher in the default mode network at resting-state or performing a Stroop task. | [21] |
| SampEn | Rest-BOLD | Biomarker | 892 | BOLD complexity was positively associated with intelligence. | [34] |
| SampEn; MSE | Rest-BOLD | Biomarker | MCI (65); AD (29); HC (30) | BOLD complexity of AD and MCI were lower than HC; AD patients showed lower BOLD complexity than MCI. | [35] |
| MSE | Rest-O2Hb | Biomarker | MCI (65); AD (29); HC (30) | O2Hb complexity in AD patients was lower than HC and positive correlated with cognitive ability. | [36] |
| SampEn MSE | Task-O2Hb Task-HHb | Biomarker | AD (11); HC (11) | When performing memory-related tasks, O2Hb complexity of AD was higher than that of HC. | [37] |
| SampEn | Rest-BOLD | Biomarker | 107 | SampEn-CBF and SampEn-fALFF correlations were only observed in a few brain regions, demonstrating that complexity, CBF, and fALFF are independent brain activity measures. | [38] |
| SampEn | Rest-BOLD | Biomarker | ASD (20); HC (17) | BOLD complexity was negatively correlated with severity of ASD behaviors. | [39] |
| SampEn | Rest-BOLD | Biomarker | SZ (53); HC (59) | Compared with HC, SZ showed decreased brain BOLD complexity. | [40] |
| SampEn; MSE | Task-O2Hb | Biomarker | AD (11); HC (11) | AD showed significant differences from HC in O2Hb complexity during VFT and WM tasks. | [41] |
| SampEn | Rest-BOLD | Biomarker | Stroke patients (23); HC (19) | Stroke patients showed reduced BOLD complexity in the motor area. | [42] |
| MSE | Rest-BOLD | Biomarker | MCI (169); HC (176) | BOLD complexity in MCI was significantly lower than that in HC and correlated with severity of MCI. | [43] |
| MSE | Rest-BOLD | Biomarker | BP (125); SZ (107); SAD (98); HC (156) | Significant differences as well as overlaps of brain BOLD signal complexity between different psychotic disorder groups were found. | [12] |
| MSE | Task-O2Hb Task-HHb | Biomarker | 15 | Brain complexity during performing intentional memory task was significantly higher than that during purposefully forgetting. | [44] |
| MSE | Rest-O2Hb Rest-HHb | Biomarker | ASD (25); HC (22) | Brain complexity could be used to distinguish ASD from HC. Compared with HC, altered brain complexity in ASD is seen more in IFG than in TC and in left hemisphere than in right hemisphere. | [45] |
| MSE | Rest-BOLD | Biomarker | LLD (35); HC (22) | LLD patients showed decreased complexity only in the right posterior cingulate gyrus but increased complexity in affective processing, sensory, motor, and temporal nodes. Complexity in the left frontoparietal network partially mediated the relation between depression severity and the mental components of quality of life. | [46] |
3.2. Main Complexity Measures for Brain Blood-Oxygen Signals
3.3. Optimizing Parameters for Complexity Measures
4. Future Directions
4.1. Improving Brain Blood-Oxygen Signal Complexity Measurement
4.2. Accurate Trait Classification Methods
4.3. The Dynamics of Blood Oxygen Signals Complexity
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bassett, D.S.; Gazzaniga, M.S. Understanding complexity in the human brain. Trends Cogn. Sci. 2011, 15, 200–209. [Google Scholar] [CrossRef] [Green Version]
- Freud, S. Introductory Lectures on Psychoanalysis; WW Norton & Company: New York, NY, USA, 1977. [Google Scholar]
- Paulus, M.P.; Geyer, M.A.; Braff, D.L. Use of methods from chaos theory to quantify a fundamental dysfunction in the behavioral organization of schizophrenic patients. Am. J. Psychiatry 1996, 153, 714–717. [Google Scholar] [PubMed] [Green Version]
- Hausdorff, J.M.; Mitchell, S.L.; Firtion, R.; Peng, C.K.; Cudkowicz, M.E.; Wei, J.Y.; Goldberger, A.L. Altered fractal dynamics of gait: Reduced stride-interval correlations with aging and Huntington’s disease. J. Appl. Physiol. 1997, 82, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Lipsitz, L.A.; Goldberger, A.L. Loss of ‘complexity’ and aging: Potential applications of fractals and chaos theory to senescence. JAMA 1992, 267, 1806–1809. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Peng, C.K.; Lipsitz, L.A. What is physiologic complexity and how does it change with aging and disease? Neurobiol. Aging 2002, 23, 23–26. [Google Scholar] [CrossRef]
- Simon, H.A. The architecture of complexity. In Facets of Systems Science; Springer: Boston, MA, USA, 1991; pp. 457–476. [Google Scholar]
- Singer, W. The Brain, a Complex Self-organizing System. Eur. Rev. 2009, 17, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Başar, E. Oscillations in “brain-body-mind”—A holistic view including the autonomous system. Brain Res. 2008, 1235, 2–11. [Google Scholar] [CrossRef]
- Sporns, O. Neural Complexity. In Networks of the Brain; MIT Press: Cambridge, MA, USA, 2011; pp. 277–304. [Google Scholar]
- Hager, B.; Yang, A.C.; Brady, R.; Meda, S.; Clementz, B.; Pearlson, G.D.; Sweeney, J.A.; Tamminga, C.; Keshavan, M. Neural complexity as a potential translational biomarker for psychosis. J. Affect. Disord. 2017, 216, 89–99. [Google Scholar] [CrossRef]
- Goldberger, A.L. Non-linear dynamics for clinicians: Chaos theory, fractals, and complexity at the bedside. Lancet 1996, 347, 1312–1314. [Google Scholar] [CrossRef]
- Goldberger, A.L. Fractal variability versus pathologic periodicity: Complexity loss and stereotypy in disease. Perspect. Biol. Med. 1997, 40, 543–561. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Hausdorff, J.M.; Ivanov, P.C.; Peng, C.K.; Stanley, H.E. Fractal dynamics in physiology: Alterations with disease and aging. Proc. Natl. Acad. Sci. USA 2002, 99, 2466–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokunbi, M.O.; Fung, W.; Sawlani, V.; Choppin, S.; Linden, D.E.J.; Thome, J. Resting state fMRI entropy probes complexity of brain activity in adults with ADHD. Psychiatry Res. 2013, 214, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokunbi, M.O. Sample entropy reveals high discriminative power between young and elderly adults in short fMRI data sets. Front. Neuroinform. 2014, 8, 69. [Google Scholar] [CrossRef]
- Yang, A.C.; Huang, C.-C.; Yeh, H.-L.; Liu, M.-E.; Hong, C.-J.; Tu, P.-C.; Chen, J.-F.; Huang, N.E.; Peng, C.-K.; Lin, C.-P.; et al. Complexity of spontaneous BOLD activity in default mode network is correlated with cognitive function in normal male elderly: A multiscale entropy analysis. Neurobiol. Aging 2013, 34, 428–438. [Google Scholar] [CrossRef]
- Liu, C.Y.; Krishnan, A.P.; Yan, L.; Smith, R.X.; Kilroy, E.; Alger, J.R.; Ringman, J.M.; Wang, D.J.J. Complexity and synchronicity of resting state blood oxygenation level-dependent (BOLD) functional MRI in normal aging and cognitive decline. J. Magn. Reson. Imaging 2013, 38, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Sokunbi, M.O.; Gradin, V.B.; Waiter, G.D.; Cameron, G.G.; Ahearn, T.S.; Murray, A.D.; Steele, D.J.; Staff, R.T. Nonlinear complexity analysis of brain FMRI signals in schizophrenia. PLoS ONE 2014, 9, e95146. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Miao, S.; Han, J.; Zeng, K.; Ouyang, G.; Yang, J.; Li, X. Complexity analysis of fNIRS signals in ADHD children during working memory task. Sci. Rep. 2017, 7, 829. [Google Scholar] [CrossRef] [Green Version]
- Shan, Z.Y.; Finegan, K.; Bhuta, S.; Ireland, T.; Staines, D.R.; Marshall-Gradisnik, S.M.; Barnden, L.R. Decreased Connectivity and Increased Blood Oxygenation Level Dependent Complexity in the Default Mode Network in Individuals with Chronic Fatigue Syndrome. Brain Connect. 2018, 8, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Sokunbi, M.O.; Staff, R.T.; Waiter, G.D.; Ahearn, T.S.; Fox, H.C.; Deary, I.J.; Starr, J.M.; Whalley, L.J.; Murray, A.D. Inter-individual differences in fMRI entropy measurements in old age. IEEE Trans. Biomed. Eng. 2011, 58, 3206–3214. [Google Scholar] [CrossRef]
- Smith, R.X.; Yan, L.; Wang, D.J.J. Multiple time scale complexity analysis of resting state FMRI. Brain Imaging Behav. 2014, 8, 284–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Li, Y.; Childress, A.R.; Detre, J.A. Brain entropy mapping using fMRI. PLoS ONE 2014, 9, e89948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonough, I.M.; Nashiro, K. Network complexity as a measure of information processing across resting-state networks: Evidence from the Human Connectome Project. Front. Hum. Neurosci. 2014, 8, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, A.C.; Hong, C.-J.; Liou, Y.-J.; Huang, K.-L.; Huang, C.-C.; Liu, M.-E.; Lo, M.-T.; Huang, N.E.; Peng, C.-K.; Lin, C.-P.; et al. Decreased resting-state brain activity complexity in schizophrenia characterized by both increased regularity and randomness. Hum. Brain Mapp. 2015, 36, 2174–2186. [Google Scholar] [CrossRef]
- Sokunbi, M.O.; Cameron, G.G.; Ahearn, T.S.; Murray, A.D.; Staff, R.T. Fuzzy approximate entropy analysis of resting state fMRI signal complexity across the adult life span. Med. Eng. Phys. 2015, 37, 1082–1090. [Google Scholar] [CrossRef]
- Zhou, F.; Huang, S.; Gao, L.; Zhuang, Y.; Ding, S.; Gong, H. Temporal regularity of intrinsic cerebral activity in patients with chronic primary insomnia: A brain entropy study using resting-state fMRI. Brain Behav. 2016, 6, e00529. [Google Scholar] [CrossRef]
- Zhou, F.; Zhuang, Y.; Gong, H.; Zhan, J.; Grossman, M.; Wang, Z. Resting State Brain Entropy Alterations in Relapsing Remitting Multiple Sclerosis. PLoS ONE 2016, 11, e0146080. [Google Scholar] [CrossRef]
- Wang, N.; Wu, H.; Xu, M.; Yang, Y.; Chang, C.; Zeng, W.; Yan, H. Occupational functional plasticity revealed by brain entropy: A resting-state fMRI study of seafarers. Hum. Brain Mapp. 2018, 39, 2997–3004. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Niu, Y.; Miao, L.; Cao, R.; Yan, P.; Guo, H.; Li, D.; Guo, Y.; Yan, T.; Wu, J.; et al. Decreased Complexity in Alzheimer’s Disease: Resting-State fMRI Evidence of Brain Entropy Mapping. Front. Aging Neurosci. 2017, 9, 378. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.C.; Tsai, S.-J.; Lin, C.-P.; Peng, C.-K. A Strategy to Reduce Bias of Entropy Estimates in Resting-State fMRI Signals. Front. Neurosci. 2018, 12, 398. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.Y.; Finegan, K.; Bhuta, S.; Ireland, T.; Staines, D.R.; Marshall-Gradisnik, S.M.; Barnden, L.R. Brain function characteristics of chronic fatigue syndrome: A task fMRI study. Neuroimage Clin. 2018, 19, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Saxe, G.N.; Calderone, D.; Morales, L.J. Brain entropy and human intelligence: A resting-state fMRI study. PLoS ONE 2018, 13, e0191582. [Google Scholar] [CrossRef]
- Niu, Y.; Wang, B.; Zhou, M.; Xue, J.; Shapour, H.; Cao, R.; Cui, X.; Wu, J.; Xiang, J. Dynamic Complexity of Spontaneous BOLD Activity in Alzheimer’s Disease and Mild Cognitive Impairment Using Multiscale Entropy Analysis. Front. Neurosci. 2018, 12, 677. [Google Scholar] [CrossRef]
- Li, X.; Zhu, Z.; Zhao, W.; Sun, Y.; Wen, D.; Xie, Y.; Liu, X.; Niu, H.; Han, Y. Decreased resting-state brain signal complexity in patients with mild cognitive impairment and Alzheimer’s disease: A multiscale entropy analysis. Biomed. Opt. Express 2018, 9, 1916–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perpetuini, D.; Bucco, R.; Zito, M.; Merla, A. Study of memory deficit in Alzheimer’s disease by means of complexity analysis of fNIRS signal. Neurophotonics 2018, 5, 011010. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Chang, D.; Zhang, J.; Ge, Q.; Zang, Y.-F.; Wang, Z. Associations of brain entropy (BEN) to cerebral blood flow and fractional amplitude of low-frequency fluctuations in the resting brain. Brain Imaging Behav. 2019, 13, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Easson, A.K.; McIntosh, A.R. BOLD signal variability and complexity in children and adolescents with and without autism spectrum disorder. Dev. Cogn. Neurosci. 2019, 36, 100630. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.-W.; Yu, Q.; Guo, Y.; Song, D.; Wang, Z. Resting-state brain entropy in schizophrenia. Compr. Psychiatry 2019, 89, 16–21. [Google Scholar] [CrossRef]
- Perpetuini, D.; Chiarelli, A.M.; Cardone, D.; Filippini, C.; Bucco, R.; Zito, M.; Merla, A. Complexity of Frontal Cortex fNIRS Can Support Alzheimer Disease Diagnosis in Memory and Visuo-Spatial Tests. Entropy 2019, 21, 26. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Hu, R.; Luo, X.; Feng, B.; Long, W.; Song, R. Reduced Complexity in Stroke with Motor Deficits: A Resting-State fMRI Study. Neuroscience 2020, 434, 35–43. [Google Scholar] [CrossRef]
- Zheng, H.; Onoda, K.; Nagai, A.; Yamaguchi, S. Reduced Dynamic Complexity of BOLD Signals Differentiates Mild Cognitive Impairment from Normal Aging. Front. Aging Neurosci. 2020, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Angsuwatanakul, T.; O’Reilly, J.; Ounjai, K.; Kaewkamnerdpong, B.; Iramina, K. Multiscale Entropy as a New Feature for EEG and fNIRS Analysis. Entropy 2020, 22, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Hua, Q.; Yu, J.; Li, J. Classification of autism spectrum disorder based on sample entropy of spontaneous functional near infra-red spectroscopy signal. Clin. Neurophysiol. 2020, 131, 1365–1374. [Google Scholar] [CrossRef]
- Lin, C.; Lee, S.-H.; Huang, C.-M.; Chen, G.-Y.; Ho, P.-S.; Liu, H.-L.; Chen, Y.-L.; Lee, T.M.-C.; Wu, S.-C. Increased brain entropy of resting-state fMRI mediates the relationship between depression severity and mental health-related quality of life in late-life depressed elderly. J. Affect. Disord. 2019, 250, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.-H.; Zhu, C.-Z.; Yang, Y.; Zuo, X.-N.; Long, X.-Y.; Cao, Q.-J.; Wang, Y.-F.; Zang, Y.-F. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J. Neurosci. Methods 2008, 172, 137–141. [Google Scholar] [CrossRef] [Green Version]
- Zang, Y.F.; Jiang, T.Z.; Lu, Y.L.; He, Y.; Tian, L.X. Regional homogeneity approach to fMRI data analysis. NeuroImage 2004, 22, 394–400. [Google Scholar] [CrossRef]
- Yang, A.C.; Tsai, S.-J. Is mental illness complex? From behavior to brain. Prog. Neuropsychopharmacol. Biol. Psychiatry 2013, 45, 253–257. [Google Scholar] [CrossRef]
- Welvaert, M.; Durnez, J.; Moerkerke, B.; Verdoolaege, G.; Rosseel, Y. neuRosim: An R package for generating fMRI data. J. Stat. Softw. 2011, 44, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Bonal, A.; Marshak, A. Approximate Entropy and Sample Entropy: A Comprehensive Tutorial. Entropy 2019, 21, 541. [Google Scholar] [CrossRef] [Green Version]
- Richman, J.S.; Moorman, J.R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H2039–H2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanin, M.; Zunino, L.; Rosso, O.A.; Papo, D. Permutation entropy and its main biomedical and econophysics applications: A review. Entropy 2012, 14, 1553–1577. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.-K. Multiscale entropy analysis of complex physiologic time series. Phys. Rev. Lett. 2002, 89, 068102. [Google Scholar] [CrossRef] [Green Version]
- Bandt, C.; Pompe, B. Permutation entropy: A natural complexity measure for time series. Phys. Rev. Lett. 2002, 88, 174102. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Yang, Z.; Zhan, M.Y.; Li, H.; Weng, X.C. Decoding the Representation of Cognition: The principles and applications of MVPA. Adv. Psychol. Sci. 2010, 18, 1934–1941. [Google Scholar]
- Cooper, S.R.; Jackson, J.J.; Barch, D.M.; Braver, T.S. Neuroimaging of individual differences: A latent variable modeling perspective. Neurosci. Biobehav. Rev. 2019, 98, 29–46. [Google Scholar] [CrossRef]
- Hutchison, R.M.; Womelsdorf, T.; Allen, E.A.; Bandettini, P.A.; Calhoun, V.D.; Corbetta, M.; Della Penna, S.; Duyn, J.H.; Glover, G.H.; Gonzalez-Castillo, J.; et al. Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage 2013, 80, 360–378. [Google Scholar] [CrossRef] [Green Version]

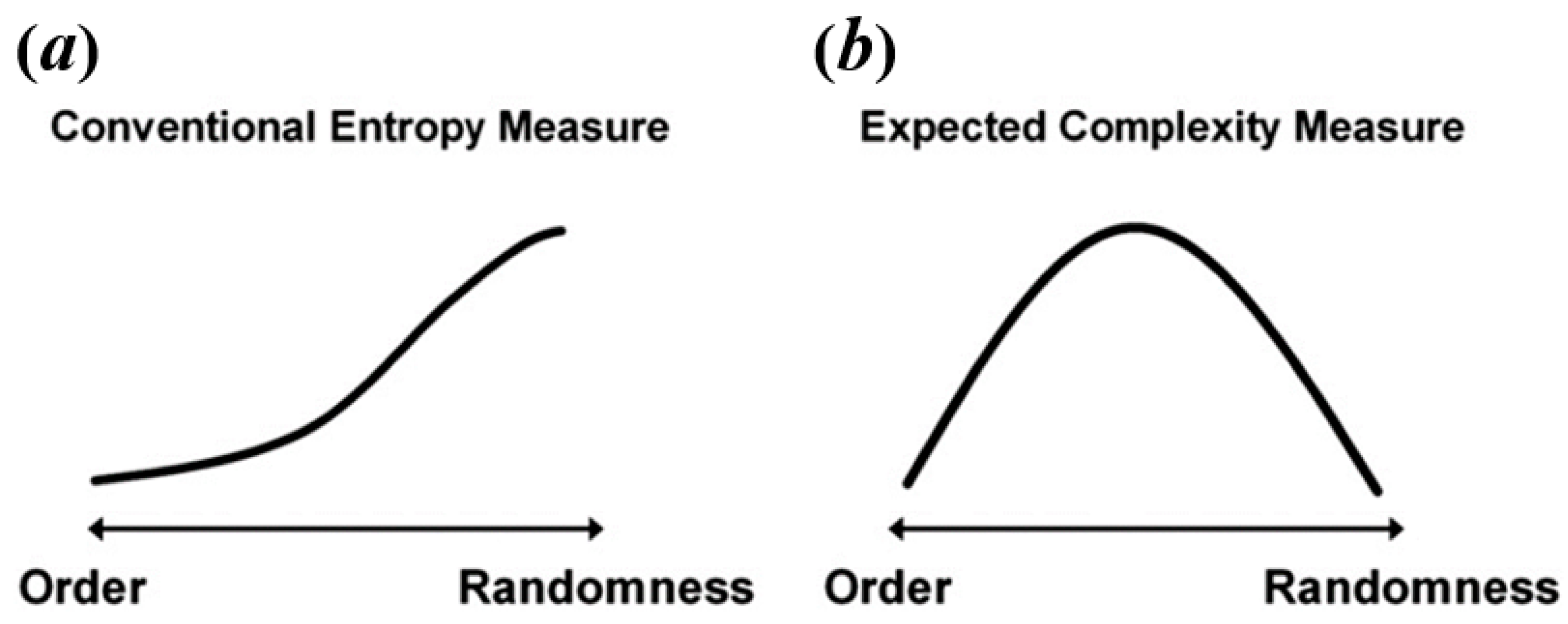

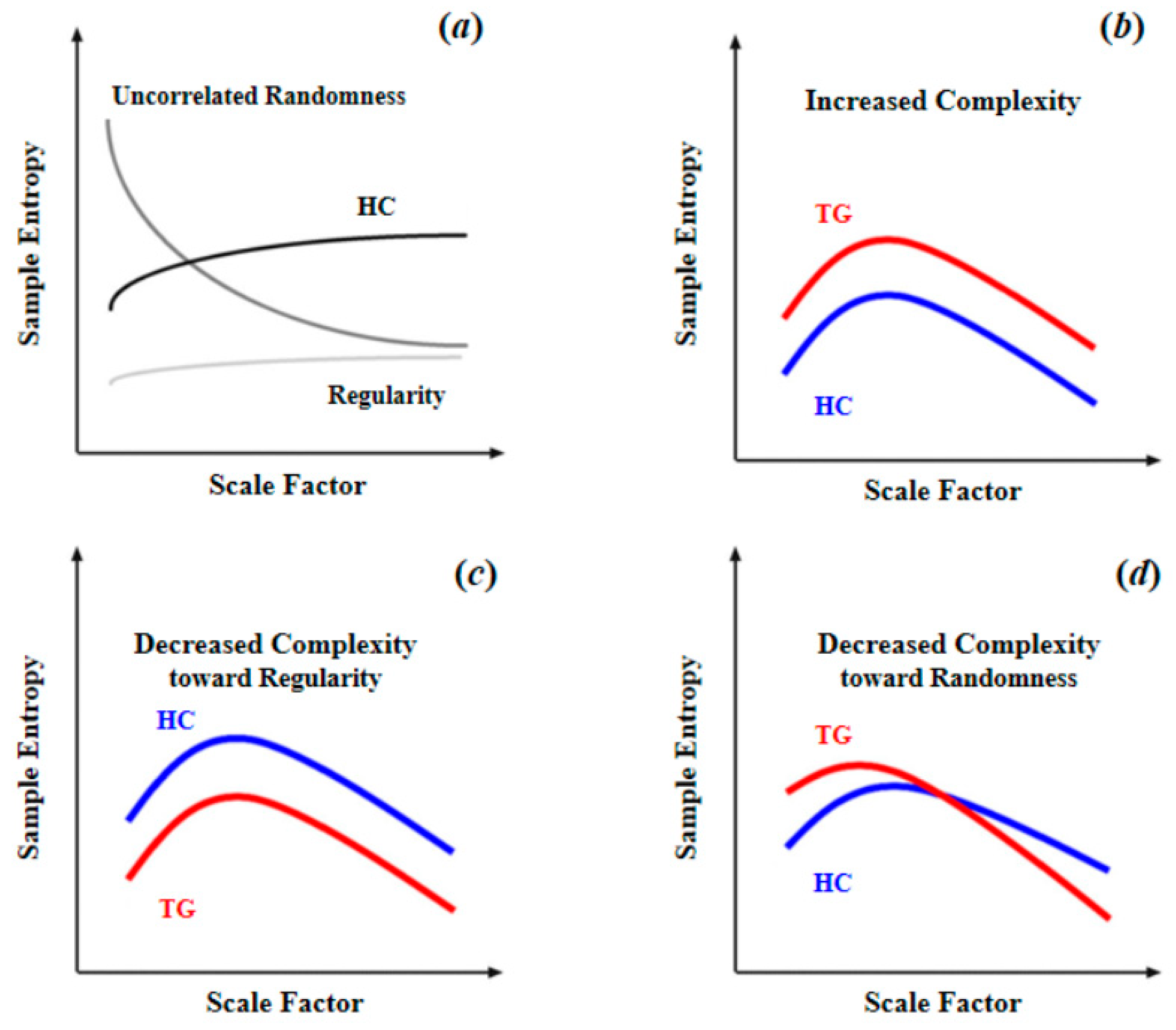
| Category | Ref | Parameters | Recommended Range |
|---|---|---|---|
| ApEn | [51] | m, pattern length; r, tolerance value | |
| SampEn | [53] | m, pattern length; r, tolerance value | |
| MSE | [55] | m, pattern length; r, tolerance value; l, scale factor | |
| PE | [56] | m, pattern length |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, X.; Long, S.; Sun, M.; Gao, X. The Application of Complexity Analysis in Brain Blood-Oxygen Signal. Brain Sci. 2021, 11, 1415. https://doi.org/10.3390/brainsci11111415
Xin X, Long S, Sun M, Gao X. The Application of Complexity Analysis in Brain Blood-Oxygen Signal. Brain Sciences. 2021; 11(11):1415. https://doi.org/10.3390/brainsci11111415
Chicago/Turabian StyleXin, Xiaoyang, Shuyang Long, Mengdan Sun, and Xiaoqing Gao. 2021. "The Application of Complexity Analysis in Brain Blood-Oxygen Signal" Brain Sciences 11, no. 11: 1415. https://doi.org/10.3390/brainsci11111415
APA StyleXin, X., Long, S., Sun, M., & Gao, X. (2021). The Application of Complexity Analysis in Brain Blood-Oxygen Signal. Brain Sciences, 11(11), 1415. https://doi.org/10.3390/brainsci11111415






