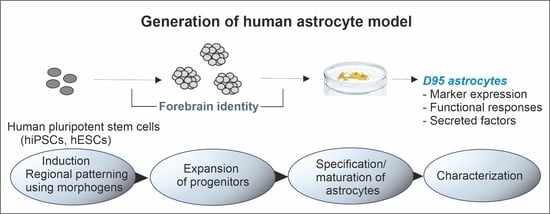
Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity
Abstract
1. Introduction
2. Materials and Methods
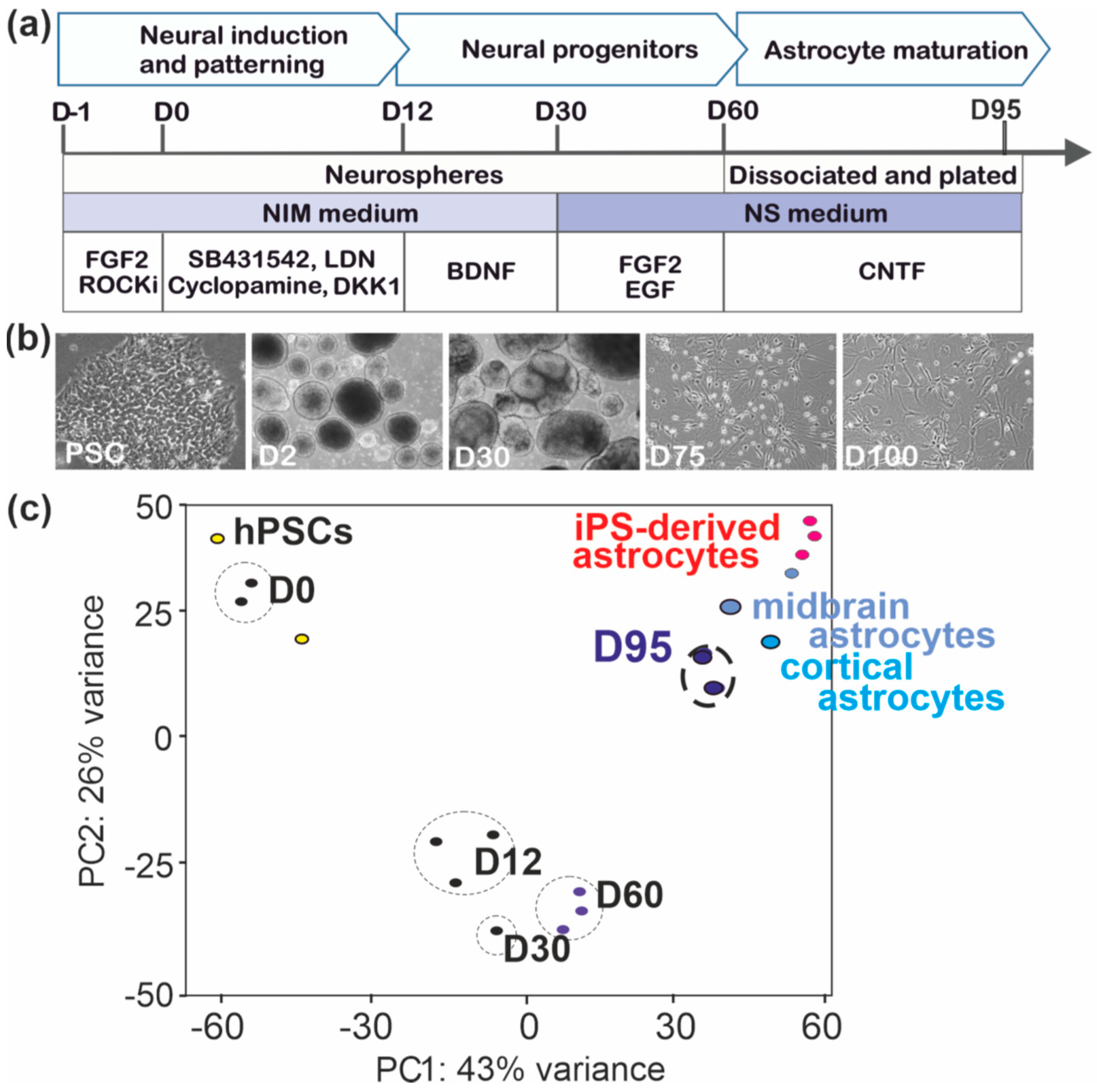
2.1. Generation of hPSC-Derived Astrocytes
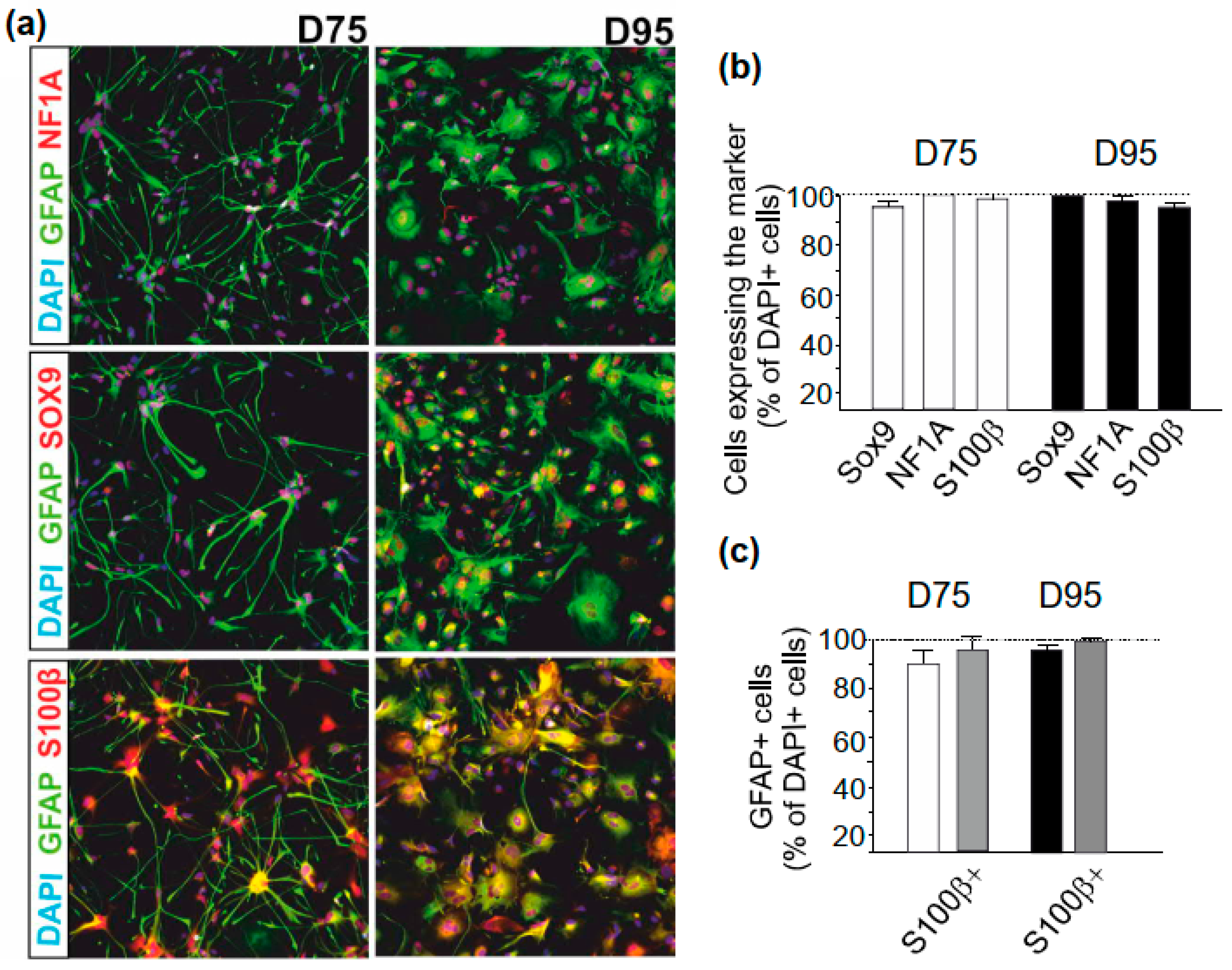
2.2. Immunocytochemistry
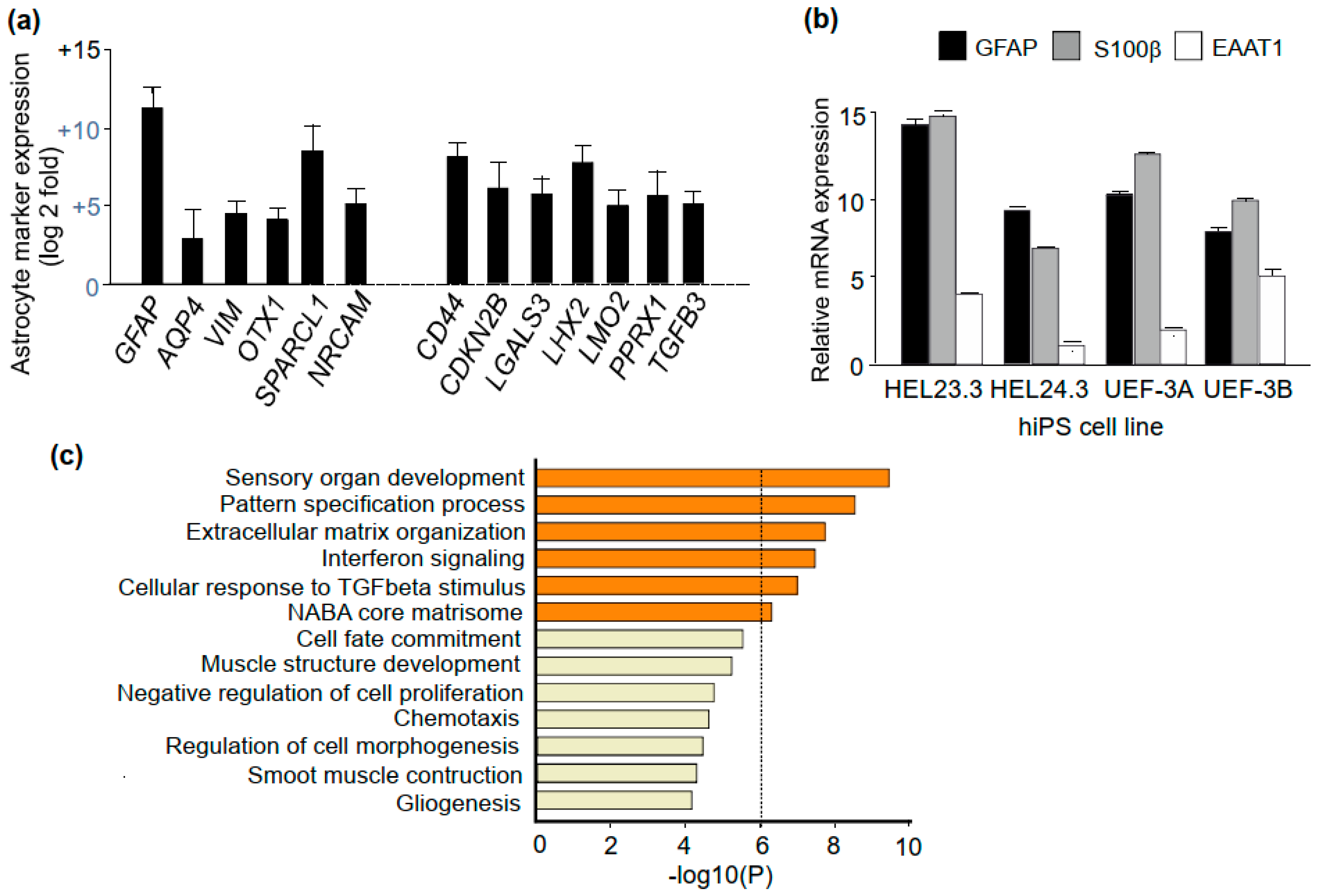
2.3. RNA Sequencing and Analysis
2.4. RNA Expression Analysis with Quantitative Real Time PCR (qRT-PCR)
2.5. Calcium Imaging
2.6. Cytokine Array
3. Results
3.1. Patterning and Differentiation of Neural Progenitors
3.2. Astrocyte Specification
3.3. Properties of hPSC-Derived Astrocytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Sloan, S.; Clarke, L.; Caneda, C.; Plaza, C.; Blumenthal, P.; Vogel, H.; Steinberg, G.; Edwards, M.; Li, G.; et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef]
- Zhao, Z.; Nelson, A.; Betsholtz, C.; Zlokovic, B. Establishment and dysfunction of the blood-brain barrier. Cell 2015, 163, 1064–1078. [Google Scholar] [CrossRef]
- Allen, N.; Lyons, D. Glia as architects of central nervous system formation and function. Science 2018, 362, 6181–6185. [Google Scholar] [CrossRef] [PubMed]
- Marina, N.; Turovsky, E.; Christie, I.; Hosford, P.; Hadjihambi, A.; Korsak, A.; Ang, R.; Mastitskaya, S.; Sheikhbahaei, S.; Theparambil, S.; et al. Brain metabolic sensing and metabolic signaling at the level of an astrocyte. Glia 2018, 66, 185–1199. [Google Scholar] [CrossRef] [PubMed]
- Bandeira, F.; Lent, R.; Herculano-Houzel, S. Changing numbers of neuronal and non-neuronal cells underlie postnatal brain growth in the rat. Proc. Natl. Acad. Sci. USA 2009, 106, 14108–14113. [Google Scholar] [CrossRef] [PubMed]
- Barnabé-Heider, F.; Wasylnka, J.; Fernandes, K.; Porsche, C.; Sendtner, M.; Kaplan, D.; Miller, F. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron 2005, 48, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Takouda, J.; Katada, S.; Nakashima, K. Emerging mechanisms underlying astrogenesis in the developing mammalian brain. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Molofsky, A.; Krenick, R.; Ullian, E.; Tsai, H.; Deneen, B.; Richardson, W.; Barres, B.; Rowitch, D. Astrocytes and disease: A neurodevelopmental perspective. Genes Dev. 2012, 26, 891–907. [Google Scholar] [CrossRef]
- Rowitch, D.; Kriegstein, A. Developmental genetics of vertebrate glial-cell specification. Nature 2010, 468, 214–222. [Google Scholar] [CrossRef]
- Chambers, C.; Peng, Y.; Nguyen, H.; Gaiano, N.; Fishell, G.; Nye, J. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development 2001, 128, 689–702. [Google Scholar] [PubMed]
- Li, J.; Khankan, R.; Caneda, C.; Godoy, M.; Haney, M.; Krawczyk, M.; Bassik, M.; Sloan, S.; Zhang, Y. Astrocyte-to-astrocyte contact and a positive feedback loop of growth factor signaling regulate astrocyte maturation. Glia 2019, 67, 1571–1597. [Google Scholar] [CrossRef] [PubMed]
- Duran, R.; Wang, C.; Hui Zheng, H.; Deneen, B.; Wu, J. Brain region-specific gene signatures revealed by distinct astrocyte subpopulations unveil links to glioma and neurodegenerative diseases. eNeuro 2019, 6, ENEURO.0288-0218.2019. [Google Scholar] [CrossRef]
- Vasile, E.; Dossi, N. Human astrocytes: Structure and functions in the healthy brain. Brain Struct. Funct. 2017, 222, 2017–2029. [Google Scholar] [CrossRef]
- Chai, H.; Diaz-Castro, B.; Shigetomi, E.; Monte, E.; Octeau, J.; Yu, X.; Cohn, W.; Rajendran, P.; Vondriska, T.; Whitelegge, J.; et al. Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological and functional evidence. Neuron 2018, 95, 531–549.e539. [Google Scholar] [CrossRef]
- Tsai, H.; Li, H.; Fuentealba, L.; Molofsky, A.; Taveira-Marques, R.; Zhuang, H.; Tenney, A.; Murnen, A.; Fancy, S.; Merkle, F.; et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 2012, 337, 358–362. [Google Scholar] [CrossRef]
- Clavreul, S.; Abdeladim, L.; Hernández-Garzón, E.; Niculescu, D.; Durand, J.; Ieng, S.; Barry, R.; Bonvento, G.; Beaurepaire, E.; Livet, J.; et al. Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nat. Commun. 2019, 10, 4884. [Google Scholar] [CrossRef] [PubMed]
- Lanjakornsiripan, D.; Pior, B.; Kawaguchi, D.; Furutachi, S.; Tahara, T.; Katsuyama, Y.; Suzuki, Y.; Fukazawa, Y.; Gotoh, Y. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 2018, 9, 1623. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.; Taha, D.; Tyzack, G.; Patani, R. Regionally encoded functional heterogeneity of astrocytes in health and disease: A perspective. Glia 2020, 69, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Oberheim, N.; Wang, X.; Goldman, S.; Nedergaard, M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 2006, 29, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Chen, M.; Wang, F.; Windrem, M.; Wang, S.; Shanz, S.; Xu, Q.; Oberheim, N.; Bekar, L.; Betstadt, S.; et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem. Cell 2013, 12, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Peteri, U.; Niukkanen, M.; Castrén, M. Astrocytes in neuropathologies affecting the frontal cortex. Front. Cell Neurosci 2019. [Google Scholar] [CrossRef]
- Khakh, B.; Deneen, B. The emerging nature of astrocyte diversity. Annu. Rev. Neurosci. 2019, 42, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Okita, K.; Nakagawa, M.; Yamanaka, S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2007, 2, 3081–3089. [Google Scholar] [CrossRef] [PubMed]
- Krencik, R.; Weick, J.; Liu, Y.; Zhang, Z.; Zhanga, S.-C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011, 29, 528–534. [Google Scholar] [CrossRef]
- Krencik, R.; Ullian, E. A cellular star atlas: Using astrocytes from human pluripotent stem cells for disease studies. Front. Cell Neurosci. 2013, 7, 25. [Google Scholar] [CrossRef]
- Roybon, L.; Lamas, N.; Garcia, A.; Yang, E.; Sattler, R.; Lewis, V.; Kim, Y.; Kachel, C.; Rothstein, J.; Przedborski, S.; et al. Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep. 2013, 4, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Tcw, J.; Wang, M.; Pimenova, A.; Bowles, K.; Hartley, B.; Lacin, E.; Machlovi, S.; Abdelaal, R.; Karch, C.; Phatnani, H.; et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem. Cell Rep. 2017, 9, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.; Avci, H.; Leist, M.; Kobolák, J.; Dinnyés, A. Astrocyte differentiation of human pluripotent stem cells: New tools for neurological disorder research. Front. Cell Neurosci. 2016, 215. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, Q.; Zhao, C.; Da, Y.; Zhang, H.; Chen, Z. Differentiation of glial cells From hiPSCs: Potential applications in neurological diseases and cell replacement therapy. Front. Cell Neurosci. 2018, 12, 239. [Google Scholar] [CrossRef] [PubMed]
- Asikainen, S.; Heikkinen, L.; Juhila, J.; Holm, F.; Weltner, J.; Trokovic, R.; Mikkola, M.; Toivonen, S.; Balboa, D.; Lampela, R.; et al. Selective microRNA-Offset RNA expression in human embryonic stem cells. PLoS ONE 2015, 10, e0116668. [Google Scholar] [CrossRef]
- Trokovic, R.; Weltner, J.; Otonkoski, T. Generation of iPSC line HEL24.3 from human neonatal foreskin fibroblasts. Stem. Cell Res. 2015, 15, 266–268. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, S.; Lehtonen, Š.; Chumarina, M.; Puttonen, K.; Azevedo, C.; Lebedeva, O.; Ruponen, M.; Oksanen, M.; Djelloul, M.; Collin, A.; et al. Creation of a library of induced pluripotent stem cells from Parkinsonian patients. NPJ Parkinsons. Dis. 2016, 2, 16009. [Google Scholar] [CrossRef]
- Utami, K.; Skotte, N.; Colaço, A.; Yusof, N.; Bernice Sim, B.; Yeo, X.; Bae, H.-G.; Garcia-Miralles, M.; Radulescu, C.; Chen, Q.; et al. Integrative analysis identifies key molecular signatures underlying neurodevelopmental deficits in fragile X syndrome. Biol. Psychiatry 2020, 88, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.; Fasano, C.; Papapetrou, E.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Glinka, A.; Wu, W.; Delius, H.; Monaghan, A.; Blumenstock, C.; Niehrs, C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 1998, 391, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Bonni, A.; Sun, Y.; Nadal-Vicens, M.; Bhatt, A.; Frank, D.; Rozovsky, I.; Stahl, N.; Yancopoulos, G.; Greenberg, M. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 1997, 278, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Magistri, M.; Khoury, N.; Mazza, E.; Velmeshev, D.; Lee, J.; Bicciato, S.; Tsoulfas, P.; Faghihi, M. A comparative transcriptomic analysis of astrocytes differentiation from human neural progenitor cells. Eur. J. Neurosci. 2016, 44, 2858–2870. [Google Scholar] [CrossRef]
- Zhang, S.; Wernig, M.; Duncan, I.; Brüstle, O.; Thomson, J. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.; Tanaseichuk, O.; Benner, C.; Chanda, S. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Yoshida, M.; Suda, Y.; Matsuo, I.; Miyamoto, N.; Takeda, N.; Kuratani, S.; Aizawa, S. Emx1 and Emx2 functions in development of dorsal telencephalon. Development 1997, 124, 101–111. [Google Scholar]
- Godbole, G.; Shetty, A.; Roy, A.; D’Souza, L.; Chen, B.; Miyoshi, G.; Fishell, G.; Tole, S. Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development 2018, 145, dev154583. [Google Scholar] [CrossRef] [PubMed]
- Deneen, B.; Ho, R.; Lukaszewicz, A.; Hochstim, C.; Gronostajski, R.; Anderson, D. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 2006, 52, 953–968. [Google Scholar] [CrossRef]
- Kleiderman, S.; Sá, J.; Teixeira, A.; Brito, C.; Gutbier, S.; Evje, L.; Hadera, M.; Glaab, E.; Henry, M.; Sachinidis, A.; et al. Functional and phenotypic differences of pure populations of stem cell-derived astrocytes and neuronal precursor cells. Glia 2016, 64, 695–715. [Google Scholar] [CrossRef] [PubMed]
- Benediktsson, A.; Marrs, G.; Tu, J.; Worley, P.; Rothstein, J.; Bergles, D.; Dailey, M. Neuronal activity regulates glutamate transporter dynamics in developing astrocytes. Glia 2012, 60, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Swanson, R.; Liu, J.; Miller, J.; Rothstein, J.; Farrell, K.; Stein, B.; Longuemare, M. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 1997, 17, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Welser-Alves, J.; Crocker, S.; Milner, R. A dual role for microglia in promoting tissue inhibitor of metalloproteinase (TIMP) expression in glial cells in response to neuroinflammatory stimuli. J. Neuroinflamm. 2011, 61. [Google Scholar] [CrossRef]
- Choi, S.; Lee, H.; Lim, I.; Satoh, J.; Kim, S. Human astrocytes: Secretome profiles of cytokines and chemokines. PLoS ONE 2014, 9, e92325. [Google Scholar] [CrossRef]
- Holmqvist, S.; Brouwer, M.; Djelloul, M.; Diaz, A.; Devine, M.; Hammarberg, A.; Fog, K.; Kunath, T.; Roybon, L. Generation of human pluripotent stem cell reporter lines for the isolation of and reporting on astrocytes generated from ventral midbrain and ventral spinal cord neural progenitors. Stem. Cell Res. 2015, 15, 203–220. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Takahashi, N.; Forrest, A.; Shin, J.; Kinoshita, Y.; Suzuki, H.; Hayashizaki, Y. CC chemokine ligand 2 and leukemia inhibitory factor cooperatively promote pluripotency in mouse induced pluripotent cells. Stem. Cells 2011, 29, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Bradley, R.; Shireman, J.; McFalls, C.; Choi, J.; Canfield, S.; Dong, Y.; Liu, K.; Lisota, B.; Jones, J.; Petersen, A.; et al. Regionally specified human pluripotent stem cell-derived astrocytes exhibit different molecular signatures and functional properties. Development 2019, 146, dev170910. [Google Scholar] [CrossRef] [PubMed]
- Bushong, E.; Martone, M.; Jones, Y.; Ellisman, M. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002, 22, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Hattori, T.; Fukuda, M.; Kitamura, T.; Fujita, S. Quantitative studies on proliferative changes of reactive astrocytes in mouse cerebral cortex. Brain Res. 1988, 451, 133–138. [Google Scholar] [CrossRef]
- Colombo, J.; Sherwood, C.; Hof, P. Interlaminar astroglial processes in the cerebral cortex of great apes. Anat. Embryol. 2004, 208, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Hochstim, C.; Deneen, B.; Lukaszewicz, A.; Zhou, Q.; Anderson, D. The spinal cord sontains positionally distinct astrocyte subtypes whose identities are specified by a homeodomain trnscriptional code. Cell 2008, 133, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Morel, L.; Chiang, M.; Higashimori, H.; Shoneye, T.; Iyer, L.; Yelick, J.; Tai, A.; Yang, Y. Molecular and functional properties of regional astrocytes in the adult brain. J. Neurosci. 2017, 37, 8706–8717. [Google Scholar] [CrossRef] [PubMed]
- Poitry-Yamate, C.; Vutskits, L.; Rauen, T. Neuronal-induced and glutamate-dependent acivation of glial glutamate transporter function. J. Neuroschem. 2002, 82, 987–997. [Google Scholar] [CrossRef] [PubMed]
- Schlag, B.; Vondrasek, J.; Munir, M.; Kalandadze, A.; Zelenaia, O.; Rothstein, J.; Robinson, M. Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol. Pharmacol. 1998, 53, 355–369. [Google Scholar] [CrossRef]
- Mahad, D.; Ransohoff, R. The role of MCP-1 (CCL2) and CCR2 in multiple sclerosis and experimental autoimmune encephalomyelitis (EAE). Sem. Immunol. 2003, 15, 23–32. [Google Scholar] [CrossRef]
- Simhal, A.; Zuo, Y.; Perez, M.; Madison, D.; Sapiro, G.; Micheva, K. Multifaceted changes in synaptic composition and astrocytic involvement in a mouse model of fragile X syndrome. Sci. Rep. 2019, 9, 13855. [Google Scholar] [CrossRef] [PubMed]
- Brigida, A.; Siniscalco, D. Induced pluripotent stem cells as a cellular model for studying Down Syndrome. J. Stem. Cells Regen. Med. 2016, 12, 54–60. [Google Scholar] [CrossRef]
- Russo, F.; Freitas, B.; Pignatari, G.; Fernandes, I.; Sebat, J.; Muotri, A.; Beltrão-Braga, P. Modeling the interplay between neurons and astrocytes in autism using human induced pluripotent stem cells. Biol. Psychiatry 2018, 83, 569–578. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peteri, U.-K.; Pitkonen, J.; Utami, K.H.; Paavola, J.; Roybon, L.; Pouladi, M.A.; Castrén, M.L. Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity. Brain Sci. 2021, 11, 209. https://doi.org/10.3390/brainsci11020209
Peteri U-K, Pitkonen J, Utami KH, Paavola J, Roybon L, Pouladi MA, Castrén ML. Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity. Brain Sciences. 2021; 11(2):209. https://doi.org/10.3390/brainsci11020209
Chicago/Turabian StylePeteri, Ulla-Kaisa, Juho Pitkonen, Kagistia Hana Utami, Jere Paavola, Laurent Roybon, Mahmoud A. Pouladi, and Maija L. Castrén. 2021. "Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity" Brain Sciences 11, no. 2: 209. https://doi.org/10.3390/brainsci11020209
APA StylePeteri, U.-K., Pitkonen, J., Utami, K. H., Paavola, J., Roybon, L., Pouladi, M. A., & Castrén, M. L. (2021). Generation of the Human Pluripotent Stem-Cell-Derived Astrocyte Model with Forebrain Identity. Brain Sciences, 11(2), 209. https://doi.org/10.3390/brainsci11020209