Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Behavioral Testing
2.2.1. Experimental Design for Gene Expression Studies
2.2.2. Acute AMPH Treatment Studies
2.3. Gene and Protein Expression Studies
2.3.1. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time-PCR (qPCR)
2.3.2. Primer Design
2.3.3. Western Blot
2.4. Statistical Analyses
3. Results
3.1. Body Weight and Locomotor Activity
3.1.1. Body Weight Change during Experiment
3.1.2. Locomotor Activity in RMT Batch
3.1.3. Amphetamine-Induced Locomotor Stimulation
3.2. Gene Expression Data
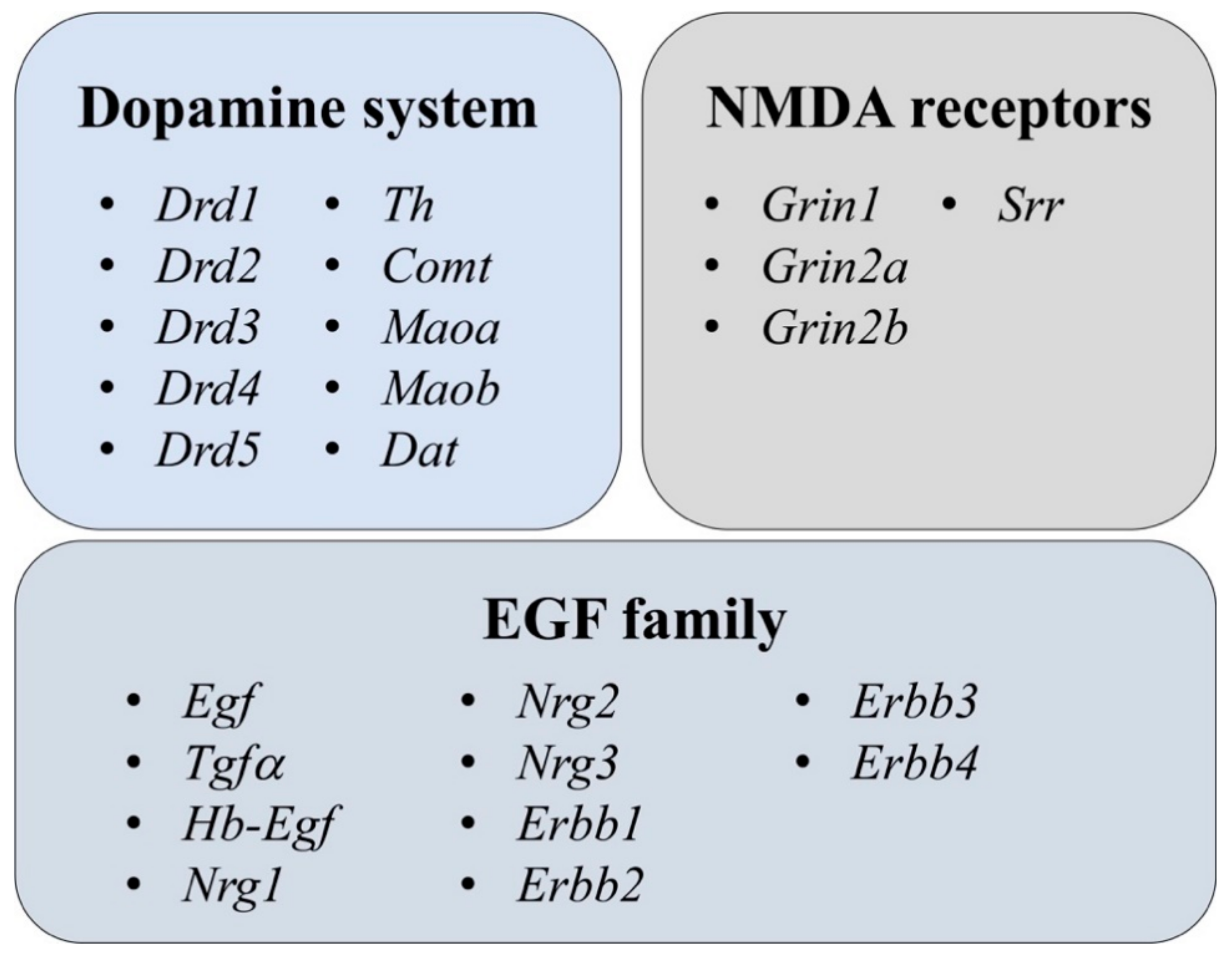
3.2.1. NMDA and DA Systems
Frontal Cortex (Figure 6, Table S1)
Hippocampus (Figure 7, Table S1)
Ventral striatum (Figure 8, Table S1)
Dorsal Striatum (Figure 9, Table S1)
3.2.2. EGF Family
Frontal Cortex (Figure 10, Table S1)
Hippocampus (Figure 11, Table S1)
Ventral Striatum (Figure 12, Table S1)
Dorsal Striatum (Figure 13, Table S1)
3.2.3. Gene Expression Alterations in the Midbrain (Table S1)
3.3. Measurement of EGF Family and NMDA Protein Levels in the Frontal Cortex and Hippocampus Using Western Blot Analysis (Figure 14 and Figure 15)
4. Discussion
4.1. Differences between 129Sv and Bl6 Strains in Weight Gain, Locomotor Activity and Amphetamine-Induced Hyperlocomotion
4.2. Gene Expression Differences between Bl6 and 129Sv in HCC and RMT Groups
4.2.1. Dopamine System
4.2.2. NMDA System
4.2.3. EGF Family
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abramov, U.; Puussaar, T.; Raud, S.; Kurrikoff, K.; Vasar, E. Behavioural Differences between C57BL/6 and 129S6/SvEv Strains Are Reinforced by Environmental Enrichment. Neurosci. Lett. 2008, 443, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Contet, C.; Rawlins, J.N.P.; Bannerman, D.M. Faster Is Not Surer—A Comparison of C57BL/6J and 129S2/Sv Mouse Strains in the Watermaze. Behav. Brain Res. 2001, 125, 261–267. [Google Scholar] [CrossRef]
- Heinla, I.; Leidmaa, E.; Visnapuu, T.; Philips, M.-A.; Vasar, E. Enrichment and Individual Housing Reinforce the Differences in Aggressiveness and Amphetamine Response in 129S6/SvEv and C57BL/6 Strains. Behav. Brain Res. 2014, 267, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Võikar, V.; Kõks, S.; Vasar, E.; Rauvala, H. Strain and Gender Differences in the Behavior of Mouse Lines Commonly Used in Transgenic Studies. Physiol. Behav. 2001, 72, 271–281. [Google Scholar] [CrossRef]
- Belknap, J.K.; Crabbe, J.C.; Young, E.R. Voluntary Consumption of Ethanol in 15 Inbred Mouse Strains. Psychopharmacology 1993, 112, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, M.; Park, S.; Gnegy, M.E. C57BL/6J Mice Show Greater Amphetamine-Induced Locomotor Activation and Dopamine Efflux in the Striatum than 129S2/SvHsd Mice. Pharmacol. Biochem. Behav. 2007, 87, 158–163. [Google Scholar] [CrossRef] [Green Version]
- Vanaveski, T.; Narvik, J.; Innos, J.; Philips, M.-A.; Ottas, A.; Plaas, M.; Haring, L.; Zilmer, M.; Vasar, E. Repeated Administration of D-Amphetamine Induces Distinct Alterations in Behavior and Metabolite Levels in 129Sv and Bl6 Mouse Strains. Front Neurosci. 2018, 12, 399. [Google Scholar] [CrossRef]
- Mekada, K.; Abe, K.; Murakami, A.; Nakamura, S.; Nakata, H.; Moriwaki, K.; Obata, Y.; Yoshiki, A. Genetic Differences among C57BL/6 Substrains. Exp. Anim. 2009, 58, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Mukaida, N.; Ishikawa, Y.; Lkeda, N.; Fujioka, N.; Watanabe, S.; Kuno, K.; Matsushima, K. Novel Insight into Molecular Mechanism of Endotoxin Shock: Biochemical Analysis of LPS Receptor Signaling in a Cell-Free System Targeting NF-ΚB and Regulation of Cytokine Production/Action through β2 Integrin In Vivo. J. Leukoc. Biol. 1996, 59, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Koike, H.; Arguello, P.A.; Kvajo, M.; Karayiorgou, M.; Gogos, J.A. Disc1 Is Mutated in the 129S6/SvEv Strain and Modulates Working Memory in Mice. Proc. Natl. Acad. Sci. USA 2006, 103, 3693–3697. [Google Scholar] [CrossRef] [Green Version]
- Trossbach, S.V.; Bader, V.; Hecher, L.; Pum, M.E.; Masoud, S.T.; Prikulis, I.; Schäble, S.; de Souza Silva, M.A.; Su, P.; Boulat, B.; et al. Misassembly of Full-Length Disrupted-in-Schizophrenia 1 Protein Is Linked to Altered Dopamine Homeostasis and Behavioral Deficits. Mol. Psychiatry 2016, 21, 1561–1572. [Google Scholar] [CrossRef] [Green Version]
- Chubb, J.E.; Bradshaw, N.J.; Soares, D.C.; Porteous, D.J.; Millar, J.K. The DISC Locus in Psychiatric Illness. Mol. Psychiatry 2008, 13, 36–64. [Google Scholar] [CrossRef]
- Onishi, T.; Sakamoto, H.; Namiki, S.; Hirose, K. The Altered Supramolecular Structure of Dopamine D2 Receptors in Disc1-Deficient Mice. Sci. Rep. 2018, 8, 1692. [Google Scholar] [CrossRef]
- Ortiz, J. Biochemical Adaptations in the Mesolimbic Dopamine System in Response to Repeated Stress. Neuropsychopharmacology 1996, 14, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Haber, S.N. Corticostriatal Circuitry. Dialogues Clin. Neurosci. 2016, 18, 7–21. [Google Scholar]
- Gonon, F.G. Nonlinear Relationship between Impulse Flow and Dopamine Released by Rat Midbrain Dopaminergic Neurons as Studied by in Vivo Electrochemistry. Neuroscience 1988, 24, 19–28. [Google Scholar] [CrossRef]
- Zweifel, L.S.; Argilli, E.; Bonci, A.; Palmiter, R.D. Role of NMDA Receptors in Dopamine Neurons for Plasticity and Addictive Behaviors. Neuron 2008, 59, 486–496. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.P.; Li, F.; Wang, D.; Xie, K.; Wang, D.; Shen, X.; Tsien, J.Z. NMDA Receptors in Dopaminergic Neurons Are Crucial for Habit Learning. Neuron 2011, 72, 1055–1066. [Google Scholar] [CrossRef] [Green Version]
- Jastrzębska, K.; Walczak, M.; Cieślak, P.E.; Szumiec, Ł.; Turbasa, M.; Engblom, D.; Błasiak, T.; Parkitna, J.R. Loss of NMDA Receptors in Dopamine Neurons Leads to the Development of Affective Disorder-like Symptoms in Mice. Sci. Rep. 2016, 6, 37171. [Google Scholar] [CrossRef]
- Willetts, J.; Balster, R.L.; Leander, J.D. The Behavioral Pharmacology of NMDA Receptor Antagonists. Trends Pharmacol. Sci. 1990, 11, 423–428. [Google Scholar] [CrossRef]
- Kato, T.; Abe, Y.; Sotoyama, H.; Kakita, A.; Kominami, R.; Hirokawa, S.; Ozaki, M.; Takahashi, H.; Nawa, H. Transient Exposure of Neonatal Mice to Neuregulin-1 Results in Hyperdopaminergic States in Adulthood: Implication in Neurodevelopmental Hypothesis for Schizophrenia. Mol. Psychiatry 2011, 16, 307–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawa, H.; Sotoyama, H.; Iwakura, Y.; Takei, N.; Namba, H. Neuropathologic Implication of Peripheral Neuregulin-1 and EGF Signals in Dopaminergic Dysfunction and Behavioral Deficits Relevant to Schizophrenia: Their Target Cells and Time Window. Biomed. Res. Int. 2014, 2014, 697935. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, M.; Sotoyama, H.; Namba, H.; Shibuya, M.; Eda, T.; Wang, R.; Okubo, T.; Nagata, K.; Iwakura, Y.; Nawa, H. ErbB Inhibitors Ameliorate Behavioral Impairments of an Animal Model for Schizophrenia: Implication of Their Dopamine-Modulatory Actions. Transl. Psychiatry 2013, 3, e252. [Google Scholar] [CrossRef] [PubMed]
- Skirzewski, M.; Karavanova, I.; Shamir, A.; Erben, L.; Garcia-Olivares, J.; Shin, J.H.; Vullhorst, D.; Alvarez, V.A.; Amara, S.G.; Buonanno, A. ErbB4 Signaling in Dopaminergic Axonal Projections Increases Extracellular Dopamine Levels and Regulates Spatial/Working Memory Behaviors. Mol. Psychiatry 2018, 23, 2227–2237. [Google Scholar] [CrossRef] [Green Version]
- Skirzewski, M.; Cronin, M.E.; Murphy, R.; Fobbs, W.; Kravitz, A.V.; Buonanno, A. ErbB4 Null Mice Display Altered Mesocorticolimbic and Nigrostriatal Dopamine Levels as Well as Deficits in Cognitive and Motivational Behaviors. eNeuro 2020, 7. [Google Scholar] [CrossRef]
- Ting, A.K.; Chen, Y.; Wen, L.; Yin, D.-M.; Shen, C.; Tao, Y.; Liu, X.; Xiong, W.-C.; Mei, L. Neuregulin 1 Promotes Excitatory Synapse Development and Function in GABAergic Interneurons. J. Neurosci. 2011, 31, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-M.; Zhang, J.; Chen, X.-J.; Geng, H.-Y.; Ye, M.; Spitzer, N.C.; Luo, J.-H.; Duan, S.-M.; Li, X.-M. Development of GABA Circuitry of Fast-Spiking Basket Interneurons in the Medial Prefrontal Cortex of Erbb4-Mutant Mice. J. Neurosci. 2013, 33, 19724–19733. [Google Scholar] [CrossRef] [Green Version]
- Mei, L.; Nave, K.-A. Neuregulin-ERBB Signaling in the Nervous System and Neuropsychiatric Diseases. Neuron 2014, 83, 27–49. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Ye, M.; Du, Y.; Qiu, X.; Lv, X.; Yang, W.; Luo, J. EGFR Signaling Upregulates Surface Expression of the GluN2B-Containing NMDA Receptor and Contributes to Long-Term Potentiation in the Hippocampus. Neuroscience 2015, 304, 109–121. [Google Scholar] [CrossRef]
- Neddens, J.; Vullhorst, D.; Buonanno, A. Neuregulin Links Dopaminergic and Glutamatergic Neurotransmission to Control Hippocampal Synaptic Plasticity. Commun. Integr. Biol. 2009, 2, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Pan, W.X.; Mao, T.; Dudman, J.T. Inputs to the Dorsal Striatum of the Mouse Reflect the Parallel Circuit Architecture of the Forebrain. Front. Neuroanat. 2010, 4, 147. [Google Scholar] [CrossRef] [Green Version]
- Sotres-Bayon, F.; Quirk, G.J. Prefrontal Control of Fear: More than Just Extinction. Curr. Opin Neurobiol. 2010, 20, 231–235. [Google Scholar] [CrossRef] [Green Version]
- Zemla, R.; Basu, J. Hippocampal Function in Rodents. Curr. Opin. Neurobiol. 2017, 43, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Franklin, K.B.J.; Paxinos, G. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates, 4th ed.; Academic Press, an Imprint of Elsevier: Amsterdam, The Netherlands, 2013; ISBN 978-0-12-391057-8. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Raud, S.; Sütt, S.; Luuk, H.; Plaas, M.; Innos, J.; Kõks, S.; Vasar, E. Relation between Increased Anxiety and Reduced Expression of Alpha1 and Alpha2 Subunits of GABAA Receptors in Wfs1-Deficient Mice. Neurosci. Lett. 2009, 460, 138–142. [Google Scholar] [CrossRef]
- Clapcote, S.J.; Roder, J.C. Deletion Polymorphism of Disc1 Is Common to All 129 Mouse Substrains: Implications for Gene-Targeting Studies of Brain Function. Genetics 2006, 173, 2407–2410. [Google Scholar] [CrossRef] [Green Version]
- Dahoun, T.; Trossbach, S.V.; Brandon, N.J.; Korth, C.; Howes, O.D. The Impact of Disrupted-in-Schizophrenia 1 (DISC1) on the Dopaminergic System: A Systematic Review. Transl. Psychiatry 2017, 7, e1015. [Google Scholar] [CrossRef]
- Thomson, P.A.; Duff, B.; Blackwood, D.H.R.; Romaniuk, L.; Watson, A.; Whalley, H.C.; Li, X.; Dauvermann, M.R.; Moorhead, T.W.J.; Bois, C.; et al. Balanced Translocation Linked to Psychiatric Disorder, Glutamate, and Cortical Structure/Function. NPJ Schizophr. 2016, 2, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wied, C.C.G.; Jansen, L.M.C. The Stress-Vulnerability Hypothesis in Psychotic Disorders: Focus on the Stress Response Systems. Curr. Psychiatry Rep. 2002, 4, 166–170. [Google Scholar] [CrossRef]
- Lampis, V.; Maziade, M.; Battaglia, M. Animal Models of Human Anxiety Disorders: Reappraisal from a Developmental Psychopathology Vantage Point. Pediatric. Res. 2011, 69, 77R–84R. [Google Scholar] [CrossRef]
- Pani, L.; Porcella, A.; Gessa, G.L. The Role of Stress in the Pathophysiology of the Dopaminergic System. Mol. Psychiatry 2000, 5, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Wieduwilt, M.J.; Moasser, M.M. The Epidermal Growth Factor Receptor Family: Biology Driving Targeted Therapeutics. Cell. Mol. Life Sci. 2008, 65, 1566–1584. [Google Scholar] [CrossRef] [Green Version]
- Iwakura, Y.; Nawa, H. ErbB1-4-Dependent EGF/Neuregulin Signals and Their Cross Talk in the Central Nervous System: Pathological Implications in Schizophrenia and Parkinson’s Disease. Front. Cell. Neurosci. 2013, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, K.; Hayashi, Y.; Yoshida, T.; Kashiwagi, M.; Nakagawa, N.; Michikawa, T.; Tanaka, M.; Ando, R.; Huang, A.; Hosoya, T.; et al. Schizophrenia-like Phenotypes in Mice with NMDA Receptor Ablation in Intralaminar Thalamic Nucleus Cells and Gene Therapy-Based Reversal in Adults. Transl. Psychiatry 2017, 7, e1047. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Iwakura, Y.; Sotoyama, H.; Kitayama, E.; Takei, N.; Someya, T.; Nawa, H. Clozapine-Dependent Inhibition of EGF/Neuregulin Receptor (ErbB) Kinases. Transl. Psychiatry 2019, 9, 181. [Google Scholar] [CrossRef] [Green Version]
- Li, C.-T.; Yang, K.-C.; Lin, W.-C. Glutamatergic Dysfunction and Glutamatergic Compounds for Major Psychiatric Disorders: Evidence from Clinical Neuroimaging Studies. Front. Psychiatry 2019, 9, 767. [Google Scholar] [CrossRef] [Green Version]
- Kahlig, K.M.; Binda, F.; Khoshbouei, H.; Blakely, R.D.; McMahon, D.G.; Javitch, J.A.; Galli, A. Amphetamine Induces Dopamine Efflux through a Dopamine Transporter Channel. Proc. Natl. Acad. Sci. USA 2005, 102, 3495–3500. [Google Scholar] [CrossRef] [Green Version]
- Shih, J.C.; Chen, K.; Ridd, M.J. Role of MAO A and B in Neurotransmitter Metabolism and Behavior. Pol. J. Pharm. 1999, 51, 25–29. [Google Scholar]
- Fornai, F.; Chen, K.; Giorgi, F.S.; Gesi, M.; Alessandri, M.G.; Shih, J.C. Striatal Dopamine Metabolism in Monoamine Oxidase B-Deficient Mice: A Brain Dialysis Study. J. Neurochem. 2002, 73, 2434–2440. [Google Scholar] [CrossRef] [Green Version]
- Inoue, H.; Castagnoli, K.; Van Der Schyf, C.; Mabic, S.; Igarashi, K.; Castagnoli, N. Species-Dependent Differences in Monoamine Oxidase A and B-Catalyzed Oxidation of Various C4 Substituted 1-Methyl-4-Phenyl-1,2,3, 6-Tetrahydropyridinyl Derivatives. J. Pharmacol. Exp. Ther. 1999, 291, 856–864. [Google Scholar]
- Sedelis, M.; Hofele, K.; Auburger, G.W.; Morgan, S.; Huston, J.P.; Schwarting, R.K. Evidence for Resistance to MPTP in C57BL/6 x BALB/c F1 Hybrids as Compared with Their Progenitor Strains. Neuroreport 2000, 11, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Przedborski, S.; Jackson-Lewis, V.; Djaldetti, R.; Liberatore, G.; Vila, M.; Vukosavic, S.; Almer, G. The Parkinsonian Toxin MPTP: Action and Mechanism. Restor. Neurol. Neurosci. 2000, 16, 135–142. [Google Scholar] [PubMed]
- Missale, C.; Nash, S.R.; Robinson, S.W.; Jaber, M.; Caron, M.G. Dopamine Receptors: From Structure to Function. Physiol. Rev. 1998, 78, 189–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flanigan, M.; LeClair, K. Shared Motivational Functions of Ventral Striatum D1 and D2 Medium Spiny Neurons. J. Neurosci. 2017, 37, 6177–6179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langen, M.; Kas, M.J.H.; Staal, W.G.; van Engeland, H.; Durston, S. The Neurobiology of Repetitive Behavior: Of Mice…. Neurosci. Biobehav. Rev. 2011, 35, 345–355. [Google Scholar] [CrossRef]
- Calabresi, P.; Pisani, A.; Mercuri, N.B.; Bernardi, G. The Corticostriatal Projection: From Synaptic Plasticity to Dysfunctions of the Basal Ganglia. Trends Neurosci. 1996, 19, 19–24. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-Induced Changes in MiRNA Biogenesis and Functioning. Cell. Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wu, X.; Meng, J.; Yao, J.; Wang, B. Functional Analysis of the 3′ Untranslated Region of the Human GRIN1 Gene in Regulating Gene Expression In Vitro. NDT 2020, 16, 2361–2370. [Google Scholar] [CrossRef]
- Narvik, J.; Vanaveski, T.; Innos, J.; Philips, M.-A.; Ottas, A.; Haring, L.; Zilmer, M.; Vasar, E. Metabolic Profile Associated with Distinct Behavioral Coping Strategies of 129Sv and Bl6 Mice in Repeated Motility Test. Sci. Rep. 2018, 8, 3405. [Google Scholar] [CrossRef] [Green Version]
- Piirsalu, M.; Taalberg, E.; Lilleväli, K.; Tian, L.; Zilmer, M.; Vasar, E. Treatment with Lipopolysaccharide Induces Distinct Changes in Metabolite Profile and Body Weight in 129Sv and Bl6 Mouse Strains. Front. Pharmacol. 2020, 11, 371. [Google Scholar] [CrossRef]
- Tuboly, G.; Tar, L.; Bohar, Z.; Safrany-Fark, A.; Petrovszki, Z.; Kekesi, G.; Vecsei, L.; Pardutz, A.; Horvath, G. The Inimitable Kynurenic Acid: The Roles of Different Ionotropic Receptors in the Action of Kynurenic Acid at a Spinal Level. Brain Res. Bull. 2015, 112, 52–60. [Google Scholar] [CrossRef]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.-Q. Kynurenines in the Mammalian Brain: When Physiology Meets Pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef]
- Moa, Q.C.; Chen, J.C. Reduced Neuregulin 1 Expression in the Medial Prefrontal Cortex of a Rat Ketamine Model for Schizophrenia. Neuropsychiatry 2017, 7, 370–377. [Google Scholar] [CrossRef]
- Stefansson, H.; Petursson, H.; Sigurdsson, E.; Steinthorsdottir, V.; Bjornsdottir, S.; Sigmundsson, T.; Ghosh, S.; Brynjolfsson, J.; Gunnarsdottir, S.; Ivarsson, O.; et al. Neuregulin 1 and Susceptibility to Schizophrenia. Am. J. Hum. Genet. 2002, 71, 877–892. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Shamir, A.; Skirzewski, M.; Leiva-Salcedo, E.; Kwon, O.B.; Karavanova, I.; Paredes, D.; Malkesman, O.; Bailey, K.R.; Vullhorst, D.; et al. Neuregulin-2 Ablation Results in Dopamine Dysregulation and Severe Behavioral Phenotypes Relevant to Psychiatric Disorders. Mol. Psychiatry 2018, 23, 1233–1243. [Google Scholar] [CrossRef] [Green Version]
- Hayes, L.N.; Shevelkin, A.; Zeledon, M.; Steel, G.; Chen, P.-L.; Obie, C.; Pulver, A.; Avramopoulos, D.; Valle, D.; Sawa, A.; et al. Neuregulin 3 Knockout Mice Exhibit Behaviors Consistent with Psychotic Disorders. Mol. Neuropsychiatry 2016, 2, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Ledonne, A.; Mercuri, N.B. MGluR1-Dependent Long Term Depression in Rodent Midbrain Dopamine Neurons Is Regulated by Neuregulin 1/ErbB Signaling. Front. Mol. Neurosci. 2018, 11, 346. [Google Scholar] [CrossRef]
- Ledonne, A.; Mercuri, N.B. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. IJMS 2019, 21, 275. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Marcotte, E.M. Insights into the Regulation of Protein Abundance from Proteomic and Transcriptomic Analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Ball, C.B.; Rodriguez, K.F.; Stumpo, D.J.; Ribeiro-Neto, F.; Korach, K.S.; Blackshear, P.J.; Birnbaumer, L.; Ramos, S.B.V. The RNA-Binding Protein, ZFP36L2, Influences Ovulation and Oocyte Maturation. PLoS ONE 2014, 9, e97324. [Google Scholar] [CrossRef] [Green Version]














Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varul, J.; Eskla, K.-L.; Piirsalu, M.; Innos, J.; Philips, M.-A.; Visnapuu, T.; Plaas, M.; Vasar, E. Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation. Brain Sci. 2021, 11, 725. https://doi.org/10.3390/brainsci11060725
Varul J, Eskla K-L, Piirsalu M, Innos J, Philips M-A, Visnapuu T, Plaas M, Vasar E. Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation. Brain Sciences. 2021; 11(6):725. https://doi.org/10.3390/brainsci11060725
Chicago/Turabian StyleVarul, Jane, Kattri-Liis Eskla, Maria Piirsalu, Jürgen Innos, Mari-Anne Philips, Tanel Visnapuu, Mario Plaas, and Eero Vasar. 2021. "Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation" Brain Sciences 11, no. 6: 725. https://doi.org/10.3390/brainsci11060725





