Immediate Early Gene c-fos in the Brain: Focus on Glial Cells
Abstract
:1. Introduction
2. c-fos in the Brain
3. Glial Cells
4. c-fos and Glial Cells: Evidence and Perspectives
4.1. c-fos in Astrocytes
4.2. c-fos in Oligodendrocytes
4.3. c-fos in Microglia
5. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Curran, T.; Peters, G.; Van Beveren, C.; Teich, N.M.; Verma, I.M. FBJ murine osteosarcoma virus: Identification and molecular cloning of biologically active proviral DNA. J. Virol. 1982, 44, 674–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, H.M.; Hansen, S.K.; Mikkelsen, J.D.; Hyttel, P.; Stummann, T.C. Alpha7 nicotinic acetylcholine receptors and neural network synaptic transmission in human induced pluripotent stem cell-derived neurons. Stem Cell Res. 2019, 41, 101642. [Google Scholar] [CrossRef] [PubMed]
- Groves, A.; Kihara, Y.; Jonnalagadda, D.; Rivera, R.; Kennedy, G.; Mayford, M.; Chun, J. A Functionally Defined In Vivo Astrocyte Population Identified by c-Fos Activation in a Mouse Model of Multiple Sclerosis Modulated by S1P Signaling: Immediate-Early Astrocytes (ieAstrocytes). eNeuro 2018, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Ortega, E.; Canadas, F.; Carvajal, F.; Cardona, D. In vivo stimulation of locus coeruleus: Effects on amygdala subnuclei. Acta Neurobiol. Exp. 2017, 77, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Bussolino, D.F.; de Arriba Zerpa, G.A.; Grabois, V.R.; Conde, C.B.; Guido, M.E.; Caputto, B.L. Light affects c-fos expression and phospholipid synthesis in both retinal ganglion cells and photoreceptor cells in an opposite way for each cell type. Brain Res. Mol. Brain Res. 1998, 58, 10–15. [Google Scholar] [CrossRef]
- Piechaczyk, M.; Blanchard, J.M. C-fos proto-oncogene regulation and function. Crit. Rev. Oncol. Hematol. 1994, 17, 93–131. [Google Scholar] [CrossRef]
- Distel, R.J.; Spiegelman, B.M. Protooncogene c-fos as a transcription factor. Adv. Cancer Res. 1990, 55, 37–55. [Google Scholar] [CrossRef]
- Wilson, T.; Treisman, R. Fos C-terminal mutations block down-regulation of c-fos transcription following serum stimulation. EMBO J. 1988, 7, 4193–4202. [Google Scholar] [CrossRef] [PubMed]
- Wisdon, R.; Verma, I.M. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 1993, 13, 7429–7438. [Google Scholar] [CrossRef] [Green Version]
- Curran, T.; Morgan, J.I. Fos: An immediate-early transcription factor in neurons. J. Neurobiol. 1995, 26, 403–412. [Google Scholar] [CrossRef]
- Rubio, N. Interferon-gamma induces the expression of immediate early genes c-fos and c-jun in astrocytes. Immunology 1997, 91, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Cochran, B.H.; Reffel, A.C.; Stiles, C.D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell 1983, 33, 939–947. [Google Scholar] [CrossRef]
- Miao, G.G.; Curran, T. Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol. Cell. Biol. 1994, 14, 4295–4310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goelet, P.; Castellucci, V.F.; Schacher, S.; Kandel, E.R. The long and the short of long-term memory—A molecular framework. Nature 1986, 322, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Gonda, T.J.; Metcalf, D. Expression of myb, myc and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 1984, 310, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.E.; Ziff, E.B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 1984, 311, 433–438. [Google Scholar] [CrossRef]
- Morgan, J.I.; Curran, T. Stimulus-transcription coupling in the nervous system: Involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 1991, 14, 421–451. [Google Scholar] [CrossRef]
- Kanda, N.; Watanabe, S. 17Beta-estradiol enhances the production of nerve growth factor in THP-1-derived macrophages or peripheral blood monocyte-derived macrophages. J. Investig. Dermatol. 2003, 121, 771–780. [Google Scholar] [CrossRef] [Green Version]
- Magistretti, P.J.; Cardinaux, J.R.; Martin, J.L. VIP and PACAP in the CNS: Regulators of glial energy metabolism and modulators of glutamatergic signaling. Ann. N. Y. Acad. Sci. 1998, 865, 213–225. [Google Scholar] [CrossRef]
- Peng, L.; Li, B.; Du, T.; Kong, E.K.; Hu, X.; Zhang, S.; Shan, X.; Zhang, M. Astrocytic transactivation by alpha2A-adrenergic and 5-HT2B serotonergic signaling. Neurochem. Int. 2010, 57, 421–431. [Google Scholar] [CrossRef]
- Hudson, A.E. Genetic Reporters of Neuronal Activity: C-Fos and G-CaMP6. Methods Enzymol. 2018, 603, 197–220. [Google Scholar] [CrossRef] [PubMed]
- Campeau, S.; Akil, H.; Watson, S.J. Lesions of the medial geniculate nuclei specifically block corticosterone release and induction of c-fos mRNA in the forebrain associated with audiogenic stress in rats. J. Neurosci. 1997, 17, 5979–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campeau, S.; Dolan, D.; Akil, H.; Watson, S.J. C-fos mRNA induction in acute and chronic audiogenic stress: Possible role of the orbitofrontal cortex in habituation. Stress 2002, 5, 121–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Umezu, T.; Shibata, Y. Brain regions and monoaminergic neurotransmitters that are involved in mouse ambulatory activity promoted by bupropion. Toxicol. Rep. 2016, 3, 552–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fetterly, T.L.; Basu, A.; Nabit, B.P.; Awad, E.; Williford, K.M.; Centanni, S.W.; Matthews, R.T.; Silberman, Y.; Winder, D.G. Alpha2A-Adrenergic Receptor Activation Decreases Parabrachial Nucleus Excitatory Drive onto BNST CRF Neurons and Reduces Their Activity In Vivo. J. Neurosci. 2019, 39, 472–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, S.O.K.; Zhang, L.; Chow, B.K.C. In vivo actions of SCTR/AT1aR heteromer in controlling Vp expression and release via cFos/cAMP/CREB pathway in magnocellular neurons of PVN. FASEB J. 2019, 33, 5389–5398. [Google Scholar] [CrossRef]
- Schmidt, K.T.; Makhijani, V.H.; Boyt, K.M.; Cogan, E.S.; Pati, D.; Pina, M.M.; Bravo, I.M.; Locke, J.L.; Jones, S.R.; Besheer, J.; et al. Stress-Induced Alterations of Norepinephrine Release in the Bed Nucleus of the Stria Terminalis of Mice. ACS Chem. Neurosci. 2019, 10, 1908–1914. [Google Scholar] [CrossRef]
- Hashimoto, K.; Parker, A.; Malone, P.; Gabelt, B.T.; Rasmussen, C.; Kaufman, P.S.; Hernandez, M.R. Long-term activation of c-Fos and c-Jun in optic nerve head astrocytes in experimental ocular hypertension in monkeys and after exposure to elevated pressure in vitro. Brain Res. 2005, 1054, 103–115. [Google Scholar] [CrossRef]
- Prosser, R.A.; Macdonald, E.S.; Heller, H.C. C-fos mRNA in the suprachiasmatic nuclei in vitro shows a circadian rhythm and responds to a serotonergic agonist. Brain Res. Mol. Brain Res. 1994, 25, 151–156. [Google Scholar] [CrossRef]
- Koya, E.; Margetts-Smith, G.; Hope, B.T. Daun02 Inactivation of Behaviorally Activated Fos-Expressing Neuronal Ensembles. Curr. Protoc. Neurosci. 2016, 76, 8–36. [Google Scholar] [CrossRef]
- Warren, B.L.; Mendoza, M.P.; Cruz, F.C.; Leao, R.M.; Caprioli, D.; Rubio, F.J.; Whitaker, L.R.; McPherson, K.B.; Bossert, J.M.; Shaham, Y.; et al. Distinct Fos-Expressing Neuronal Ensembles in the Ventromedial Prefrontal Cortex Mediate Food Reward and Extinction Memories. J. Neurosci. 2016, 36, 6691–6703. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, L.R.; Warren, B.L.; Venniro, M.; Harte, T.C.; McPherson, K.B.; Beidel, J.; Bossert, J.M.; Shaham, Y.; Bonci, A.; Hope, B.T. Bidirectional Modulation of Intrinsic Excitability in Rat Prelimbic Cortex Neuronal Ensembles and Non-Ensembles after Operant Learning. J. Neurosci. 2017, 37, 8845–8856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, F.; Bauer, A.; Perl, S.; Schulz, A.; Richter, A. Optogenetic augmentation of the hypercholinergic endophenotype in DYT1 knock-in mice induced erratic hyperactive movements but not dystonia. EBioMedicine 2019, 41, 649–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moolenaar, W.H.; Kruijer, W.; Tilly, B.C.; Verlaan, I.; Bierman, A.J.; de Laat, S.W. Growth factor-like action of phosphatidic acid. Nature 1986, 323, 171–173. [Google Scholar] [CrossRef]
- Greenberg, M.E.; Greene, L.A.; Ziff, E.B. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J. Biol. Chem. 1985, 260, 14101–14110. [Google Scholar] [CrossRef]
- Gauthier-Rouviere, C.; Fernandez, A.; Lamb, N.J. Ras-induced c-fos expression and proliferation in living rat fibroblasts involves C-kinase activation and the serum response element pathway. EMBO J. 1990, 9, 171–180. [Google Scholar] [CrossRef]
- Janknecht, R.; Cahill, M.A.; Nordheim, A. Signal integration at the c-fos promoter. Carcinogenesis 1995, 16, 443–450. [Google Scholar] [CrossRef] [Green Version]
- Andre, V.; Pineau, N.; Motte, J.E.; Marescaux, C.; Nehlig, A. Mapping of neuronal networks underlying generalized seizures induced by increasing doses of pentylenetetrazol in the immature and adult rat: A c-Fos immunohistochemical study. Eur. J. Neurosci. 1998, 10, 2094–2106. [Google Scholar] [CrossRef]
- Yang, H.; Shan, W.; Zhu, F.; Yu, T.; Fan, J.; Guo, A.; Li, F.; Yang, X.; Wang, Q. C-Fos mapping and EEG characteristics of multiple mice brain regions in pentylenetetrazol-induced seizure mice model. Neurol. Res. 2019, 41, 749–761. [Google Scholar] [CrossRef]
- Krisztin-Peva, B.; Mihaly, A.; Toth, Z. Differential expression of the c-fos protein and synaptophysin in zebrin II positive and zebrin II negative cerebellar cortical areas in 4-aminopyridine seizures. Acta Neurobiol. Exp. 2019, 79, 238–250. [Google Scholar] [CrossRef] [Green Version]
- Wallengren, C.; Li, S.; Morris, M.J.; Jupp, B.; O’Brien, T.J. Aggravation of absence seizures by carbamazepine in a genetic rat model does not induce neuronal c-Fos activation. Clin. Neuropharmacol. 2005, 28, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, A.; Maletta, T.; Carr, J.; Lehmann, H.; Fournier, N.M. Spatial exploration induced expression of immediate early genes Fos and Zif268 in adult-born neurons Is reduced after pentylenetetrazole kindling. Brain Res. Bull. 2019, 152, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Burnsed, J.; Skwarzynska, D.; Wagley, P.K.; Isbell, L.; Kapur, J. Neuronal Circuit Activity during Neonatal Hypoxic-Ischemic Seizures in Mice. Ann. Neurol. 2019, 86, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Thomas, K.L.; Everitt, B.J. Fear memory retrieval induces CREB phosphorylation and Fos expression within the amygdala. Eur. J. Neurosci. 2001, 13, 1453–1458. [Google Scholar] [CrossRef]
- Vann, S.D.; Brown, M.W.; Erichsen, J.T.; Aggleton, J.P. Fos imaging reveals differential patterns of hippocampal and parahippocampal subfield activation in rats in response to different spatial memory tests. J. Neurosci. 2000, 20, 2711–2718. [Google Scholar] [CrossRef] [Green Version]
- Corbett, B.F.; You, J.C.; Zhang, X.; Pyfer, M.S.; Tosi, U.; Iascone, D.M.; Petrof, I.; Hazra, A.; Fu, C.H.; Stephens, G.S.; et al. DeltaFosB Regulates Gene Expression and Cognitive Dysfunction in a Mouse Model of Alzheimer’s Disease. Cell Rep. 2017, 20, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Calais, J.B.; Valvassori, S.S.; Resende, W.R.; Feier, G.; Athie, M.C.; Ribeiro, S.; Gattaz, W.F.; Quevedo, J.; Ojopi, E.B. Long-term decrease in immediate early gene expression after electroconvulsive seizures. J. Neural Transm. 2013, 120, 259–266. [Google Scholar] [CrossRef]
- Calais, J.B.; Ojopi, E.B.; Morya, E.; Sameshima, K.; Ribeiro, S. Experience-dependent upregulation of multiple plasticity factors in the hippocampus during early REM sleep. Neurobiol. Learn. Mem. 2015, 122, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Nikolaev, E.; Kaczmarek, L.; Zhu, S.W.; Winblad, B.; Mohammed, A.H. Environmental manipulation differentially alters c-Fos expression in amygdaloid nuclei following aversive conditioning. Brain Res. 2002, 957, 91–98. [Google Scholar] [CrossRef]
- Su, Y.; Shin, J.; Zhong, C.; Wang, S.; Roychowdhury, P.; Lim, J.; Kim, D.; Ming, G.L.; Song, H. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 2017, 20, 476–483. [Google Scholar] [CrossRef] [Green Version]
- West, A.E.; Greenberg, M.E. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 2011, 3, a005744. [Google Scholar] [CrossRef] [PubMed]
- Cattane, N.; Minelli, A.; Milanesi, E.; Maj, C.; Bignotti, S.; Bortolomasi, M.; Chiavetto, L.B.; Gennarelli, M. Altered gene expression in schizophrenia: Findings from transcriptional signatures in fibroblasts and blood. PLoS ONE 2015, 10, e0116686. [Google Scholar] [CrossRef] [PubMed]
- Covington, H.E., 3rd; Lobo, M.K.; Maze, I.; Vialou, V.; Hyman, J.M.; Zaman, S.; LaPlant, Q.; Mouzon, E.; Ghose, S.; Tamminga, C.A.; et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J. Neurosci. 2010, 30, 16082–16090. [Google Scholar] [CrossRef] [PubMed]
- MacGibbon, G.A.; Lawlor, P.A.; Bravo, R.; Dragunow, M. Clozapine and haloperidol produce a differential pattern of immediate early gene expression in rat caudate-putamen, nucleus accumbens, lateral septum and islands of Calleja. Brain Res. Mol. Brain Res. 1994, 23, 21–32. [Google Scholar] [CrossRef]
- Verma, V.; Rasmussen, K.; Dawe, G.S. Effects of short-term and chronic olanzapine treatment on immediate early gene protein and tyrosine hydroxylase immunoreactivity in the rat locus coeruleus and medial prefrontal cortex. Neuroscience 2006, 143, 573–585. [Google Scholar] [CrossRef]
- Quezada, D.F. Cambios Cognitivos, Expresión de C-Fos y Arborización Dendrítica Después de la Exposición a Ruido Ambiental Crónico en Ratas Macho Adultas; Universidad de Guadalajara: Guadalajara, México, 2020. [Google Scholar]
- Fernández-Quezada, D.; Luquín, S.; Ruvalcaba-Delgadillo, Y.; García-Estrada, J.; Jauregui-Huerta, F. Sex Differences in the Expression of c-fos in a Rat Brain after Exposure to Environmental Noise. Sustainability 2022, 14, 2798. [Google Scholar] [CrossRef]
- Ons, S.; Rotllant, D.; Marin-Blasco, I.J.; Armario, A. Immediate-early gene response to repeated immobilization: Fos protein and arc mRNA levels appear to be less sensitive than c-fos mRNA to adaptation. Eur. J. Neurosci. 2010, 31, 2043–2052. [Google Scholar] [CrossRef]
- Vialou, V.; Robison, A.J.; Laplant, Q.C.; Covington, H.E., 3rd; Dietz, D.M.; Ohnishi, Y.N.; Mouzon, E.; Rush, A.J., 3rd; Watts, E.L.; Wallace, D.L.; et al. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat. Neurosci. 2010, 13, 745–752. [Google Scholar] [CrossRef] [Green Version]
- Vialou, V.; Bagot, R.C.; Cahill, M.E.; Ferguson, D.; Robison, A.J.; Dietz, D.M.; Fallon, B.; Mazei-Robison, M.; Ku, S.M.; Harrigan, E.; et al. Prefrontal cortical circuit for depression- and anxiety-related behaviors mediated by cholecystokinin: Role of DeltaFosB. J. Neurosci. 2014, 34, 3878–3887. [Google Scholar] [CrossRef] [Green Version]
- Perrotti, L.I.; Hadeishi, Y.; Ulery, P.G.; Barrot, M.; Monteggia, L.; Duman, R.S.; Nestler, E.J. Induction of deltaFosB in reward-related brain structures after chronic stress. J. Neurosci. 2004, 24, 10594–10602. [Google Scholar] [CrossRef] [Green Version]
- Machida, M.; Lonart, G.; Sanford, L.D. Effects of stressor controllability on transcriptional levels of c-fos, Arc, and brain-derived neurotrophic factor in mouse amygdala and medial prefrontal cortex. Neuroreport 2018, 29, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Abbud, R.; Lee, W.S.; Hoffman, G.; Smith, M.S. Lactation inhibits hippocampal and cortical activation of cFos expression by nivida but not kainate receptor agonists. Mol. Cell. Neurosci. 1992, 3, 244–250. [Google Scholar] [CrossRef]
- Wosiski-Kuhn, M.; Bota, M.; Snider, C.A.; Wilson, S.P.; Venkataraju, K.U.; Osten, P.; Stranahan, A.M. Hippocampal brain-derived neurotrophic factor determines recruitment of anatomically connected networks after stress in diabetic mice. Hippocampus 2018, 28, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Houben, T.; Coomans, C.P.; Meijer, J.H. Regulation of circadian and acute activity levels by the murine suprachiasmatic nuclei. PLoS ONE 2014, 9, e110172. [Google Scholar] [CrossRef]
- Caldelas, I.; Poirel, V.J.; Sicard, B.; Pevet, P.; Challet, E. Circadian profile and photic regulation of clock genes in the suprachiasmatic nucleus of a diurnal mammal Arvicanthis ansorgei. Neuroscience 2003, 116, 583–591. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Nunez, A.A.; Smale, L. Rhythmic cFos expression in the ventral subparaventricular zone influences general activity rhythms in the Nile grass rat, Arvicanthis niloticus. Chronobiol. Int. 2009, 26, 1290–1306. [Google Scholar] [CrossRef]
- Langel, J.; Yan, L.; Nunez, A.A.; Smale, L. Behavioral Masking and cFos Responses to Light in Day- and Night-Active Grass Rats. J. Biol. Rhythm. 2014, 29, 192–202. [Google Scholar] [CrossRef]
- Shuboni-Mulligan, D.D.; Cavanaugh, B.L.; Tonson, A.; Shapiro, E.M.; Gall, A.J. Functional and anatomical variations in retinorecipient brain areas in Arvicanthis niloticus and Rattus norvegicus: Implications for the circadian and masking systems. Chronobiol. Int. 2019, 36, 1464–1481. [Google Scholar] [CrossRef]
- Schottner, K.; Vuillez, P.; Challet, E.; Pevet, P.; Weinert, D. Light-induced c-Fos expression in the SCN and behavioural phase shifts of Djungarian hamsters with a delayed activity onset. Chronobiol. Int. 2015, 32, 596–607. [Google Scholar] [CrossRef]
- Zheng, W.; Niu, L.; Zhang, C.; Zhu, C.; Xie, F.; Cao, C.; Li, G. Brain edema and protein expression of c-Fos and c-Jun in the brain after diffused brain injury. Int. J. Clin. Exp. Pathol. 2014, 7, 2809–2817. [Google Scholar]
- Telford, S.; Wang, S.; Redgrave, P. Analysis of nociceptive neurones in the rat superior colliculus using c-fos immunohistochemistry. J. Comp. Neurol. 1996, 375, 601–617. [Google Scholar] [CrossRef]
- Pinto, M.; Lima, D.; Tavares, I. Correlation of noxious evoked c-fos expression in areas of the somatosensory system during chronic pain: Involvement of spino-medullary and intra-medullary connections. Neurosci. Lett. 2006, 409, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Sandor, K.; Kormos, V.; Botz, B.; Imreh, A.; Bolcskei, K.; Gaszner, B.; Markovics, A.; Szolcsanyi, J.; Shintani, N.; Hashimoto, H.; et al. Impaired nocifensive behaviours and mechanical hyperalgesia, but enhanced thermal allodynia in pituitary adenylate cyclase-activating polypeptide deficient mice. Neuropeptides 2010, 44, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Araujo, P.; Coelho, C.A.; Oliveira, M.G.; Tufik, S.; Andersen, M.L. Neonatal Sleep Restriction Increases Nociceptive Sensitivity in Adolescent Mice. Pain Physician 2018, 21, E137–E148. [Google Scholar]
- Flores, E.N.; Duggan, A.; Madathany, T.; Hogan, A.K.; Márquez, F.G.; Kumar, G.; Seal, R.P.; Edwards, R.H.; Liberman, M.C.; García-Añoveros, J. A Non-Canonical Pathway from Cochlea to Brain Signals Tissue-Damaging Noise. Curr. Biol. 2015, 25, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.; Hummel, V.; Weilbach, F.X.; Kieseier, B.C.; Gold, R. Phagocytosis of apoptotic inflammatory cells downregulates microglial chemoattractive function and migration of encephalitogenic T cells. J. Neurosci. Res. 2006, 84, 1217–1224. [Google Scholar] [CrossRef]
- Jauregui-Huerta, F.; Ruvalcaba-Delgadillo, Y.; Gonzalez-Castañeda, R.; Garcia-Estrada, J.; Gonzalez-Perez, O.; Luquin, S. Responses of glial cells to stress and glucocorticoids. Curr. Immunol. Rev. 2010, 6, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Sominsky, L.; De Luca, S.; Spencer, S.J. Microglia: Key players in neurodevelopment and neuronal plasticity. Int. J. Biochem. Cell Biol. 2018, 94, 56–60. [Google Scholar] [CrossRef]
- Palmada, M.; Centelles, J.J. Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front. Biosci. 1998, 3, d701–d718. [Google Scholar] [CrossRef] [Green Version]
- Daneman, R.; Prat, A. The blood-brain barrier. Cold Spring Harb. Perspect. Biol. 2015, 7, a020412. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.A.; Alvarez-Buylla, A. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb. Perspect. Biol. 2016, 8, a018820. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.Y.; Rosenberg, S.S.; Fancy, S.P.; Zhao, C.; Shen, Y.A.; Hahn, A.T.; McGee, A.W.; Xu, X.; Zheng, B.; Zhang, L.I.; et al. Neurite outgrowth inhibitor Nogo-A establishes spatial segregation and extent of oligodendrocyte myelination. Proc. Natl. Acad. Sci. USA 2012, 109, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jessen, K.R. Glial cells. Int. J. Biochem. Cell Biol. 2004, 36, 1861–1867. [Google Scholar] [CrossRef] [PubMed]
- Hassinger, T.D.; Atkinson, P.B.; Strecker, G.J.; Whalen, L.R.; Dudek, F.E.; Kossel, A.H.; Kater, S.B. Evidence for glutamate-mediated activation of hippocampal neurons by glial calcium waves. J. Neurobiol. 1995, 28, 159–170. [Google Scholar] [CrossRef]
- Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G. Tripartite synapses: Glia, the unacknowledged partner. Trends Neurosci. 1999, 22, 208–215. [Google Scholar] [CrossRef]
- Cui, W.; Allen, N.D.; Skynner, M.; Gusterson, B.; Clark, A.J. Inducible ablation of astrocytes shows that these cells are required for neuronal survival in the adult brain. Glia 2001, 34, 272–282. [Google Scholar] [CrossRef]
- McAllister, M.S.; Krizanac-Bengez, L.; Macchia, F.; Naftalin, R.J.; Pedley, K.C.; Mayberg, M.R.; Marroni, M.; Leaman, S.; Stanness, K.A.; Janigro, D. Mechanisms of glucose transport at the blood-brain barrier: An in vitro study. Brain Res. 2001, 904, 20–30. [Google Scholar] [CrossRef]
- Gourine, A.V.; Kasparov, S. Astrocytes as brain interoceptors. Exp. Physiol. 2011, 96, 411–416. [Google Scholar] [CrossRef]
- Lois, C.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Chain migration of neuronal precursors. Science 1996, 271, 978–981. [Google Scholar] [CrossRef]
- Kaneko, N.; Sawada, M.; Sawamoto, K. Mechanisms of neuronal migration in the adult brain. J. Neurochem. 2017, 141, 835–847. [Google Scholar] [CrossRef] [Green Version]
- Walz, W. Role of astrocytes in the clearance of excess extracellular potassium. Neurochem. Int. 2000, 36, 291–300. [Google Scholar] [CrossRef]
- Wilson, C.S.; Mongin, A.A. The signaling role for chloride in the bidirectional communication between neurons and astrocytes. Neurosci. Lett. 2019, 689, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Dehouck, M.P.; Meresse, S.; Delorme, P.; Fruchart, J.C.; Cecchelli, R. An easier, reproducible, and mass-production method to study the blood-brain barrier in vitro. J. Neurochem. 1990, 54, 1798–1801. [Google Scholar] [CrossRef] [PubMed]
- Doetsch, F.; Caille, I.; Lim, D.A.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 1999, 97, 703–716. [Google Scholar] [CrossRef] [Green Version]
- Falk, S.; Götz, M. Glial control of neurogenesis. Curr. Opin. Neurobiol. 2017, 47, 188–195. [Google Scholar] [CrossRef]
- Czopka, T.; Ffrench-Constant, C.; Lyons, D.A. Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths in vivo. Dev. Cell 2013, 25, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Larson, V.A.; Mironova, Y.; Vanderpool, K.G.; Waisman, A.; Rash, J.E.; Agarwal, A.; Bergles, D.E. Oligodendrocytes control potassium accumulation in white matter and seizure susceptibility. eLife 2018, 7, e34829. [Google Scholar] [CrossRef]
- Bloch, B.; Popovici, T.; Levin, M.J.; Tuil, D.; Kahn, A. Transferrin gene expression visualized in oligodendrocytes of the rat brain by using in situ hybridization and immunohistochemistry. Proc. Natl. Acad. Sci. USA 1985, 82, 6706–6710. [Google Scholar] [CrossRef] [Green Version]
- De los Monteros, A.E.; Peña, L.A.; de Vellis, J. Does transferrin have a special role in the nervous system? J. Neurosci. Res. 1989, 24, 125–136. [Google Scholar] [CrossRef]
- Baumann, N.; Pham-Dinh, D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001, 81, 871–927. [Google Scholar] [CrossRef]
- Philips, T.; Rothstein, J.D. Oligodendroglia: Metabolic supporters of neurons. J. Clin. Investig. 2017, 127, 3271–3280. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Nave, K.A. Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb. Perspect. Biol. 2015, 8, a020479. [Google Scholar] [CrossRef] [PubMed]
- Boda, E.; Di Maria, S.; Rosa, P.; Taylor, V.; Abbracchio, M.P.; Buffo, A. Early phenotypic asymmetry of sister oligodendrocyte progenitor cells after mitosis and its modulation by aging and extrinsic factors. Glia 2015, 63, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Hesp, Z.C.; Yoseph, R.Y.; Suzuki, R.; Jukkola, P.; Wilson, C.; Nishiyama, A.; McTigue, D.M. Proliferating NG2-Cell-Dependent Angiogenesis and Scar Formation Alter Axon Growth and Functional Recovery after Spinal Cord Injury in Mice. J. Neurosci. 2018, 38, 1366–1382. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Su, Q.; Jie, X.; Liu, A.; Wang, H.; He, B.; Jiang, H. NG2 expression in microglial cells affects the expression of neurotrophic and proinflammatory factors by regulating FAK phosphorylation. Sci. Rep. 2016, 6, 27983. [Google Scholar] [CrossRef]
- Liu, Y.; Aguzzi, A. NG2 glia are required for maintaining microglia homeostatic state. Glia 2020, 68, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Raff, M.C.; Miller, R.H.; Noble, M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature 1983, 303, 390–396. [Google Scholar] [CrossRef]
- Robins, S.C.; Trudel, E.; Rotondi, O.; Liu, X.; Djogo, T.; Kryzskaya, D.; Bourque, C.W.; Kokoeva, M.V. Evidence for NG2-glia derived, adult-born functional neurons in the hypothalamus. PLoS ONE 2013, 8, e78236. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Zhang, Z.; Zhou, H.; Zhou, J. Differential Modulators of NG2-Glia Differentiation into Neurons and Glia and Their Crosstalk. Cell. Mol. Neurobiol. 2020, 41, 1–15. [Google Scholar] [CrossRef]
- Morest, D.K.; Silver, J. Precursors of neurons, neuroglia, and ependymal cells in the CNS: What are they? Where are they from? How do they get where they are going? Glia 2003, 43, 6–18. [Google Scholar] [CrossRef]
- Sawamoto, K.; Wichterle, H.; Gonzalez-Perez, O.; Cholfin, J.A.; Yamada, M.; Spassky, N.; Murcia, N.S.; Garcia-Verdugo, J.M.; Marin, O.; Rubenstein, J.L.; et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 2006, 311, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Spassky, N.; Merkle, F.T.; Flames, N.; Tramontin, A.D.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J. Neurosci. 2005, 25, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Saijo, K.; Glass, C.K. Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 2011, 11, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Choi, G.E. Microglia: Physiological Functions Revealed through Morphological Profiles. Folia Biol. 2017, 63, 85–90. [Google Scholar]
- Zhang, L.; Zhang, J.; You, Z. Switching of the Microglial Activation Phenotype is a Possible Treatment for Depression Disorder. Front. Cell. Neurosci. 2018, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Pósfai, B.; Cserép, C.; Orsolits, B.; Dénes, Á. New Insights into Microglia-Neuron Interactions: A Neuron’s Perspective. Neuroscience 2019, 405, 103–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karperien, A.; Ahammer, H.; Jelinek, H.F. Quantitating the subtleties of microglial morphology with fractal analysis. Front. Cell. Neurosci. 2013, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Dragunow, M.; Currie, R.W.; Robertson, H.A.; Faull, R.L. Heat shock induces c-fos protein-like immunoreactivity in glial cells in adult rat brain. Exp. Neurol. 1989, 106, 105–109. [Google Scholar] [CrossRef]
- Dragunow, M.; Robertson, H.A. Brain injury induces c-fos protein(s) in nerve and glial-like cells in adult mammalian brain. Brain Res. 1988, 455, 295–299. [Google Scholar] [CrossRef]
- Arenander, A.T.; Lim, R.W.; Varnum, B.C.; Cole, R.; de Vellis, J.; Herschman, H.R. TIS gene expression in cultured rat astrocytes: Multiple pathways of induction by mitogens. J. Neurosci. Res. 1989, 23, 257–265. [Google Scholar] [CrossRef]
- Arenander, A.T.; de Vellis, J.; Herschman, H.R. Induction of c-fos and TIS genes in cultured rat astrocytes by neurotransmitters. J. Neurosci. Res. 1989, 24, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Hisanaga, K.; Sagar, S.M.; Hicks, K.J.; Swanson, R.A.; Sharp, F.R. c-fos proto-oncogene expression in astrocytes associated with differentiation or proliferation but not depolarization. Brain Res. Mol. Brain Res. 1990, 8, 69–75. [Google Scholar] [CrossRef]
- Ladenheim, R.G.; Lacroix, I.; Foignant-Chaverot, N.; Strosberg, A.D.; Couraud, P.O. Endothelins stimulate c-fos and nerve growth factor expression in astrocytes and astrocytoma. J. Neurochem. 1993, 60, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Haas, C.A.; Reddington, M.; Kreutzberg, G.W. Calcitonin Gene-related Peptide Stimulates the Induction of c-fos Gene Expression in Rat Astrocyte Cultures. Eur. J. Neurosci. 1991, 3, 708–712. [Google Scholar] [CrossRef]
- Fraczek, L.A.; Martin, C.B.; Martin, B.K. C-Jun and c-Fos regulate the complement factor H promoter in murine astrocytes. Mol. Immunol. 2011, 49, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Suh, H.W.; Choi, S.S.; Lee, J.K.; Lee, H.K.; Han, E.J.; Lee, J. Regulation of c-fos and c-jun gene expression by lipopolysaccharide and cytokines in primary cultured astrocytes: Effect of PKA and PKC pathways. Arch. Pharm. Res. 2004, 27, 396–401. [Google Scholar] [CrossRef]
- Simi, A.; Edling, Y.; Ingelman-Sundberg, M.; Tindberg, N. Activation of c-fos by lipopolysaccharide in glial cells via p38 mitogen-activated protein kinase-dependent activation of serum or cyclic AMP/calcium response element. J. Neurochem. 2005, 92, 915–924. [Google Scholar] [CrossRef]
- Rubio, N.; Martin-Clemente, B. Binding of adenovirus to its receptors in mouse astrocytes induces c-fos proto-oncogene and apoptosis. Virology 2002, 297, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.Y.; Yu, A.C. Quercetin inhibits c-fos, heat shock protein, and glial fibrillary acidic protein expression in injured astrocytes. J. Neurosci. Res. 2000, 62, 730–736. [Google Scholar] [CrossRef]
- Yu, A.C.; Lee, Y.L.; Fu, W.Y.; Eng, L.F. Gene expression in astrocytes during and after ischemia. Prog. Brain Res. 1995, 105, 245–253. [Google Scholar] [CrossRef]
- Hung, A.C.; Huang, H.M.; Tsay, H.J.; Lin, T.N.; Kuo, J.S.; Sun, S.H. ATP-stimulated c-fos and zif268 mRNA expression is inhibited by chemical hypoxia in a rat brain-derived type 2 astrocyte cell line, RBA-2. J. Cell. Biochem. 2000, 77, 323–332. [Google Scholar] [CrossRef]
- Pechan, P.A.; Chowdhury, K.; Gerdes, W.; Seifert, W. Glutamate induces the growth factors NGF, bFGF, the receptor FGF-R1 and c-fos mRNA expression in rat astrocyte culture. Neurosci. Lett. 1993, 153, 111–114. [Google Scholar] [CrossRef]
- Edling, Y.; Ingelman-Sundberg, M.; Simi, A. Glutamate activates c-fos in glial cells via a novel mechanism involving the glutamate receptor subtype mGlu5 and the transcriptional repressor DREAM. Glia 2007, 55, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Delaney, J.; Chiarello, R.; Villar, D.; Kandalam, U.; Castejon, A.M.; Clark, M.A. Regulation of c-fos, c-jun and c-myc gene expression by angiotensin II in primary cultured rat astrocytes: Role of ERK1/2 MAP kinases. Neurochem. Res. 2008, 33, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Jia, S.; Yue, T.; Yang, L.; Huang, C.; Verkhratsky, A.; Peng, L. Biphasic Regulation of Caveolin-1 Gene Expression by Fluoxetine in Astrocytes: Opposite Effects of PI3K/AKT and MAPK/ERK Signaling Pathways on c-fos. Front. Cell. Neurosci. 2017, 11, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamsky, A.; Kol, A.; Kreisel, T.; Doron, A.; Ozeri-Engelhard, N.; Melcer, T.; Refaeli, R.; Horn, H.; Regev, L.; Groysman, M.; et al. Astrocytic Activation Generates De Novo Neuronal Potentiation and Memory Enhancement. Cell 2018, 174, 59–71.e14. [Google Scholar] [CrossRef] [Green Version]
- Fan, F.; Li, L.; Liu, W.; Yang, M.; Ma, X.; Sun, H. Astrocytes and neurons in locus coeruleus mediate restraint water immersion stress-induced gastric mucosal damage through the ERK1/2 signaling pathway. Neurosci. Lett. 2018, 675, 95–102. [Google Scholar] [CrossRef]
- Eng, L.F.; Lee, Y.L.; Murphy, G.M.; Yu, A.C. A RT-PCR study of gene expression in a mechanical injury model. Prog. Brain Res. 1995, 105, 219–229. [Google Scholar] [CrossRef]
- Matsunaga, W.; Osawa, S.; Miyata, S.; Kiyohara, T. Astrocytic Fos expression in the rat posterior pituitary following LPS administration. Brain Res. 2001, 898, 215–223. [Google Scholar] [CrossRef]
- Hsieh, H.L.; Wang, H.H.; Wu, C.Y.; Yang, C.M. Reactive Oxygen Species-Dependent c-Fos/Activator Protein 1 Induction Upregulates Heme Oxygenase-1 Expression by Bradykinin in Brain Astrocytes. Antioxid. Redox Signal. 2010, 13, 1829–1844. [Google Scholar] [CrossRef]
- Morioka, N.; Suekama, K.; Zhang, F.F.; Kajitani, N.; Hisaoka-Nakashima, K.; Takebayashi, M.; Nakata, Y. Amitriptyline up-regulates connexin43-gap junction in rat cultured cortical astrocytes via activation of the p38 and c-Fos/AP-1 signalling pathway. Br. J. Pharmacol. 2014, 171, 2854–2867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yester, J.W.; Bryan, L.; Waters, M.R.; Mierzenski, B.; Biswas, D.D.; Gupta, A.S.; Bhardwaj, R.; Surace, M.J.; Eltit, J.M.; Milstien, S.; et al. Sphingosine-1-phosphate inhibits IL-1-induced expression of C-C motif ligand 5 via c-Fos-dependent suppression of IFN-beta amplification loop. FASEB J. 2015, 29, 4853–4865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhat, N.R.; Hauser, K.F.; Kindy, M.S. Cell proliferation and protooncogene induction in oligodendroglial progenitors. J. Neurosci. Res. 1992, 32, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, M.; Almazan, G. Protein kinases mediate basic fibroblast growth factor’s stimulation of proliferation and c-fos induction in oligodendrocyte progenitors. Brain Res. Mol. Brain Res. 1994, 24, 118–128. [Google Scholar] [CrossRef]
- Prasad, A.; Teh, D.B.L.; Blasiak, A.; Chai, C.; Wu, Y.; Gharibani, P.M.; Yang, I.H.; Phan, T.T.; Lim, K.L.; Yang, H.; et al. Static Magnetic Field Stimulation Enhances Oligodendrocyte Differentiation and Secretion of Neurotrophic Factors. Sci. Rep. 2017, 7, 6743. [Google Scholar] [CrossRef]
- Liu, H.N.; Almazan, G. Glutamate induces c-fos proto-oncogene expression and inhibits proliferation in oligodendrocyte progenitors: Receptor characterization. Eur. J. Neurosci. 1995, 7, 2355–2363. [Google Scholar] [CrossRef]
- Cohen, R.I.; Molina-Holgado, E.; Almazan, G. Carbachol stimulates c-fos expression and proliferation in oligodendrocyte progenitors. Brain Res. Mol. Brain Res. 1996, 43, 193–201. [Google Scholar] [CrossRef]
- Khorchid, A.; Larocca, J.N.; Almazan, G. Characterization of the signal transduction pathways mediating noradrenaline-stimulated MAPK activation and c-fos expression in oligodendrocyte progenitors. J. Neurosci. Res. 1999, 58, 765–778. [Google Scholar] [CrossRef]
- Goldenberg-Cohen, N.; Guo, Y.; Margolis, F.; Cohen, Y.; Miller, N.R.; Bernstein, S.L. Oligodendrocyte dysfunction after induction of experimental anterior optic nerve ischemia. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2716–2725. [Google Scholar] [CrossRef] [Green Version]
- Reissig, C.J.; Rabin, R.A.; Winter, J.C.; Dlugos, C.A. D-LSD-induced c-Fos expression occurs in a population of oligodendrocytes in rat prefrontal cortex. Eur. J. Pharmacol. 2008, 583, 40–47. [Google Scholar] [CrossRef]
- Bichenkov, E.; Ellingson, J.S. Ethanol alters the expressions of c-Fos and myelin basic protein in differentiating oligodendrocytes. Alcohol 2009, 43, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Eun, S.Y.; Hong, Y.H.; Kim, E.H.; Jeon, H.; Suh, Y.H.; Lee, J.E.; Jo, C.; Jo, S.A.; Kim, J. Glutamate receptor-mediated regulation of c-fos expression in cultured microglia. Biochem. Biophys. Res. Commun. 2004, 325, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Du, M.; Lu, Y.; Zeng, C.Q. Effects of triptolide on the expression of MHC II in microglia in kainic acidinduced epilepsy. Mol. Med. Rep. 2018, 17, 8357–8362. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, X.; Chen, M.; Chen, J.; Gao, T.; Yao, S. Dexmedetomidine inhibits inflammation in microglia cells under stimulation of LPS and ATP by c-Fos/NLRP3/caspase-1 cascades. EXCLI J. 2018, 17, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Rigillo, G.; Vilella, A.; Benatti, C.; Schaeffer, L.; Brunello, N.; Blom, J.M.C.; Zoli, M.; Tascedda, F. LPS-induced histone H3 phospho(Ser10)-acetylation(Lys14) regulates neuronal and microglial neuroinflammatory response. Brain Behav. Immun. 2018, 74, 277–290. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, J.; Xu, Y.; Zhang, X. Paraquat-induced inflammatory response of microglia through HSP60/TLR4 signaling. Hum. Exp. Toxicol. 2018, 37, 1161–1168. [Google Scholar] [CrossRef]
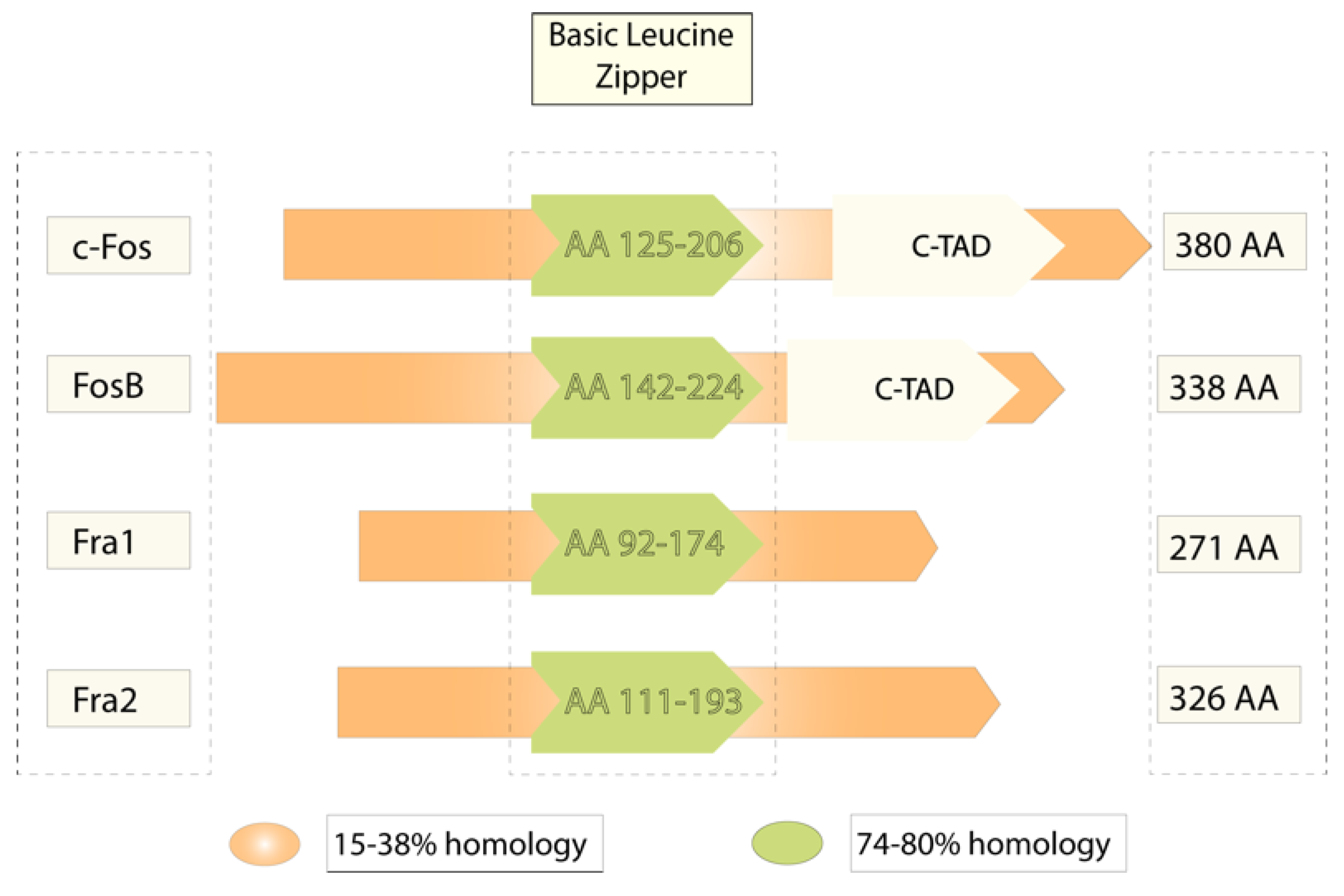
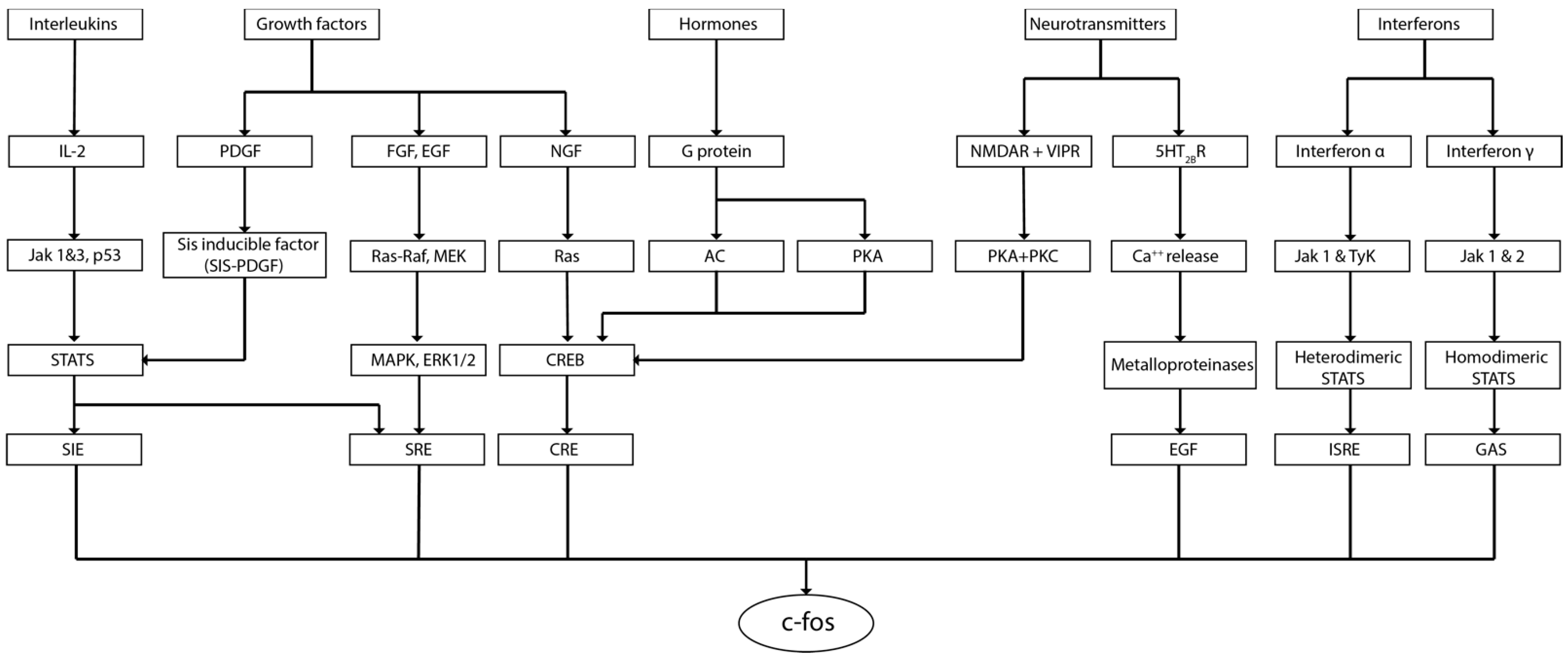
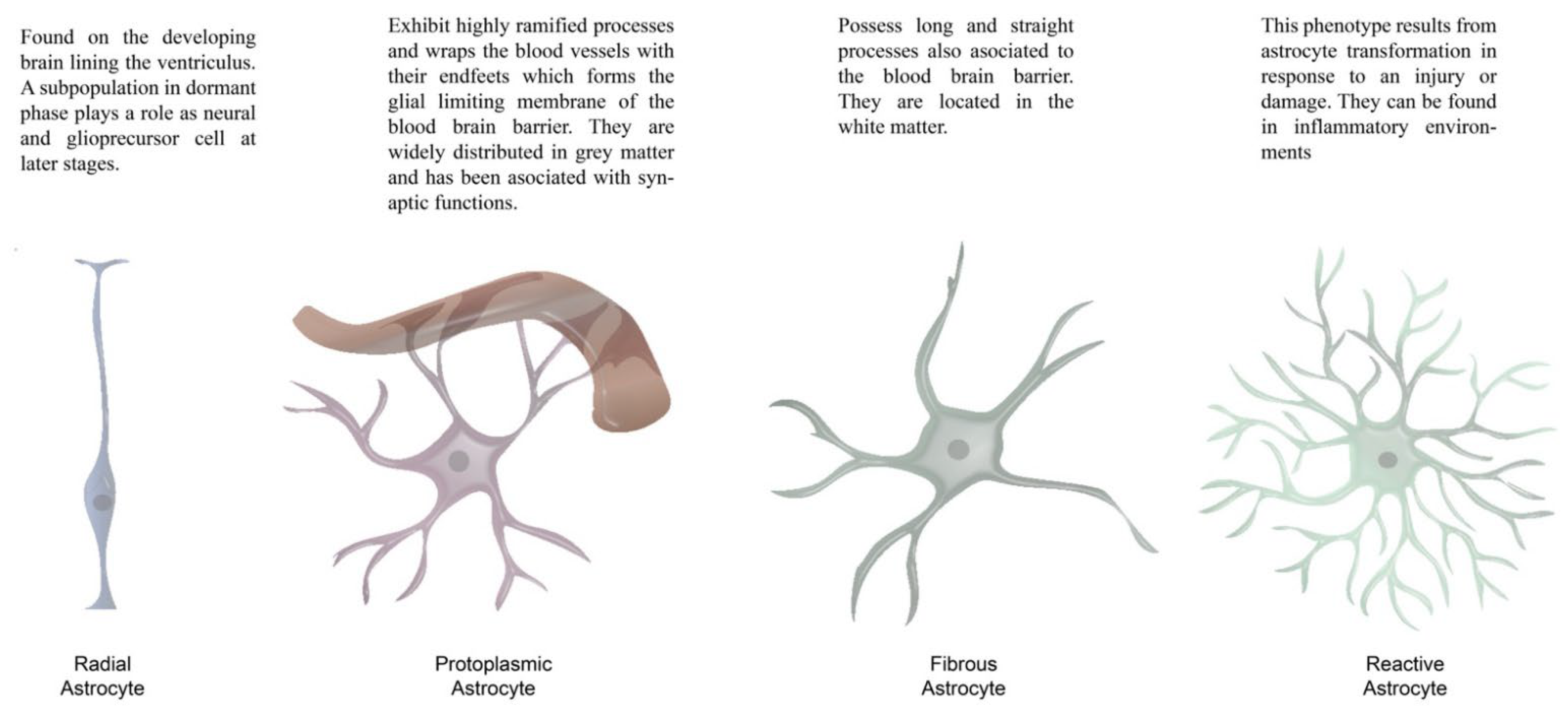

| Paper | Effect | Model | Approach | Methodology | Studied Area | Species |
|---|---|---|---|---|---|---|
| [119] | ↑ After 1 h | Heat shock insult | In vivo | Immunohistochemical | Thalamus, hippocampus, corpus callosum, internal capsule, and fornix/fimbria | Rat |
| [121] | ↑ After 30 min | Mitogens and growth factor exposure | In vitro | Northern blot | Primary cultures of cortical astrocytes (In secondary cultures) | Rat |
| [122] | ↑ After 30–60 min | Muscarinic and adrenergic agonist exposure | In vitro | Northern blot | Primary cultures of cortical astrocytes (In secondary cultures) | Rat |
| [123] | -↑ After 30 min mRNA and 2 h protein -No change | -Mitogen exposure -Depolarizing conditions | In vitro | Northern blot and immunohistochemical | Primary cultures of neocortical astrocytes | Rat |
| [125] | ↑After 30 min | Damage-associated molecular pattern (DAMPs) | In vitro | Northern blot | Primary cultures of cortical astrocytes | Rat |
| [124] | ↑After 30 min | Endothelin exposure | In vitro | Northern blot | Rat astrocytoma C6 cells (C6-S and C6-V subclones) and primary cultures of cortical and striatal astrocytes | Rat/Mouse |
| [139] | ↑After 30–60 min | Scratch wound of culture astrocytes | In vitro | Quantitative reverse transcriptase polymerase chain reaction (RT-PCR | Primary cultures of cortical astrocytes | Rat |
| [131] | ↑0.5–2 h, peak at 1 h | Ischemic model (mineral oil) | In vitro | RT-PCR | Primary cultures of cortical astrocytes | Rat |
| [11] | ↑After 30 min | Proinflammatory factor exposure | In vitro | Northern blot and flow cytometry | Primary cultures of cortical astrocytes | Mouse |
| [132] | ↑After 15–60 min | Chemical hypoxia (0.5 mM cyanide for 1 h) | In vitro | Northern blot | RBA-2 type 2 astrocytes cell line | Rat |
| [130] | ↑ NA | Heat shock insult | In vitro | Western blot | Primary cultures of astrocytes | Mouse |
| [140] | ↑1 h, peak at 2 h | LPS administration | In vivo | Immunohistochemical | Hypothalamic supraoptic nucleus, posterior and anterior pituitary | Rat |
| [129] | ↑0.5–1 h, peak at 30 min | Adenovirus (Ad.βGal) exposure | In vitro | Northern blot | Primary cultures of cortical astrocytes | Mouse |
| [127] | ↑1 h | Proinflammatory factor exposure | In vitro | Northern blot | Primary cultures of astrocytes | Rat |
| [134] | ↑After 15–30 min, peak at 30 min | Glutamate stimulation in excitotoxic levels | In vitro | Northern blot and immunohistochemical | Primary cortical glial cell cultures | Rat |
| [141] | ↑After 1 h | Bradykinin exposure | In vitro | Western blot and RT-PCR | RBA-1 cell line | Rat |
| [126] | c-fos binding to mCFH promoter | NA | In vitro | Electrophoretic mobility shift assay (EMSA) and supershift assay | Astrocyte 2.1 (Ast 2.1) cell line and primary astrocytes, microglia and oligodendrocytes cultures | Mouse |
| [142] | Nuclear translocation | Amitriptyline exposure | In vitro | Western blot and real-time PCR | Primary cultures of astrocytes | Rat |
| [143] | ↑After 1 h | Forskolin and IL-1 exposure | In vitro/ in vivo | Western blot and qPCR | Human cortical astrocytes/KO mouse | Human/ Mouse |
| [136] | ↓ After 1 h at low doses (0.5–1 μM) ↑After 1 h at high doses (5–10 μM) | Fluoxetine exposure | In vitro | Western blot and RT-PCR | Primary cultures of astrocytes | Mouse |
| [137] | ↑ After 90 min | Viral vector injection and CNO administration | In vivo | Immunohistochemical | Hippocampus | Mouse |
| [3] | ↑ NA | Experimental autoimmune encephalomyelitis (EAE) | In vivo | Immunolabeling-enabled three-dimensional imaging of solvent-cleared organs (iDISCO) and flow cytometry | TetTag-cFos reporter mice | Mouse |
| Paper | Effect | Model | Approach | Methodology | Studied Area | Species |
|---|---|---|---|---|---|---|
| [144] | ↑After 30 min | Mitogenic and growth factors exposure | In vitro | Northern blot and immunohistochemical | OPCs isolated from mixed glial cell cultures | Rat |
| [145] | ↑After 0.25–8 h, peak at 1 h | Basic fibroblast growth factor (bFGF) exposure | In vitro | Northern blot and immunohistochemical | OPCs isolated from mixed glial cell cultures | Rat |
| [147] | ↑After 0.25–6 h, peak at 1 h | Glutamate exposure | In vitro | Northern blot | OPCs isolated from mixed glial cell cultures | Rat |
| [148] | ↑After 30–60 min | Carbachol exposure | In vitro | Northern blot and immunohistochemical | OPCs isolated from mixed glial cell cultures | Rat |
| [149] | ↑After 30–60 min | NE exposure | In vitro | Western and Northern blot | OPCs isolated from mixed glial cell cultures | Rat |
| [150] | ↑ 1, 6, and 9 days after induction with progressive increases | Experimental anterior optic nerve ischemia | In vivo | Quantitative real-time PCR (qRT-PCR) and immunohistochemical | Optic nerve | Mouse |
| [151] | ↑After 90 min | d-LSD exposure | In vivo | Immunohistochemical | Prefrontal cortex | Rat |
| [152] | Delayed downregulation of c-fos levels during differentiation | Ethanol administration | In vitro | Western blot | CG-4 glial cell line | Rat |
| [146] | ↑After 2 hours/day of SMF stimulation (0.3 T) for a period of 14 days | Static magnetic field (SMF) stimulation | In vitro | qRT-PCR | Human OPCs derived from induced pluripotent stem cells | Human |
| Paper | Effect | Model | Approach | Methodology | Studied Area | Species |
|---|---|---|---|---|---|---|
| [153] | ↑After 30 min (mRNA) ↑After 2 h postexposure (protein) | 10-50-fold higher doses of glutamate than physiological exposure | In vitro | RT-PCR, Western blot, and immunohistochemical | Primary cortical microglial cells | Rat |
| [154] | ↑p-c-fos after 2 h (KA) | Kainic acid (KA) exposure | In vitro | Western blot | BV-2 microglia cell line | Mouse |
| [155] | ↓ Dose-dependent (Dex) | LPS/ATP and Dexmedetomi-dine (Dex) treatment | In vitro | PCR, Western blot, and immunohistochemical | Human microglia clone 3 cell line (HMC3) | Human |
| [156] | ↑2 h after treatment (BV-2 cells) ↑2–6 h after treatment (hypothalamus) | LPS treatment | In vivo/in vitro | PCR, Western blot, and immunohistochemical | BV-2 microglia cell line/hippocampus and hypothalamus | Mouse/rat |
| [157] | ↑6, 12, and 24 h of treatment | Paraquat and LPS exposure | In vitro | qRT-PCR | BV-2 microglia cell line | Mouse |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Mendoza, F.; Jauregui-Huerta, F.; Aguilar-Delgadillo, A.; García-Estrada, J.; Luquin, S. Immediate Early Gene c-fos in the Brain: Focus on Glial Cells. Brain Sci. 2022, 12, 687. https://doi.org/10.3390/brainsci12060687
Cruz-Mendoza F, Jauregui-Huerta F, Aguilar-Delgadillo A, García-Estrada J, Luquin S. Immediate Early Gene c-fos in the Brain: Focus on Glial Cells. Brain Sciences. 2022; 12(6):687. https://doi.org/10.3390/brainsci12060687
Chicago/Turabian StyleCruz-Mendoza, Fernando, Fernando Jauregui-Huerta, Adriana Aguilar-Delgadillo, Joaquín García-Estrada, and Sonia Luquin. 2022. "Immediate Early Gene c-fos in the Brain: Focus on Glial Cells" Brain Sciences 12, no. 6: 687. https://doi.org/10.3390/brainsci12060687
APA StyleCruz-Mendoza, F., Jauregui-Huerta, F., Aguilar-Delgadillo, A., García-Estrada, J., & Luquin, S. (2022). Immediate Early Gene c-fos in the Brain: Focus on Glial Cells. Brain Sciences, 12(6), 687. https://doi.org/10.3390/brainsci12060687






