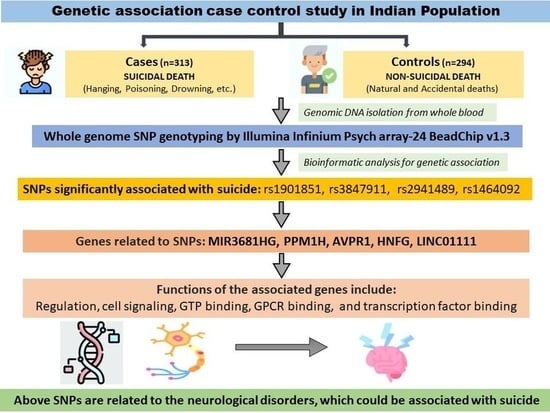
PsychArray-Based Genome Wide Association Study of Suicidal Deaths in India
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.1.1. Suicide Cases (n = 313)
2.1.2. Non-Suicide Controls (n = 294)
2.1.3. SAS Population from 1000 G Project
2.2. Sample Collection and DNA Extraction
2.3. PsychArray Genotyping
2.4. Quality Control (QC)
2.5. Imputation
2.6. Statistical Analysis and Functional Mapping
3. Results
3.1. Psycho-Social Profiles of the Subjects
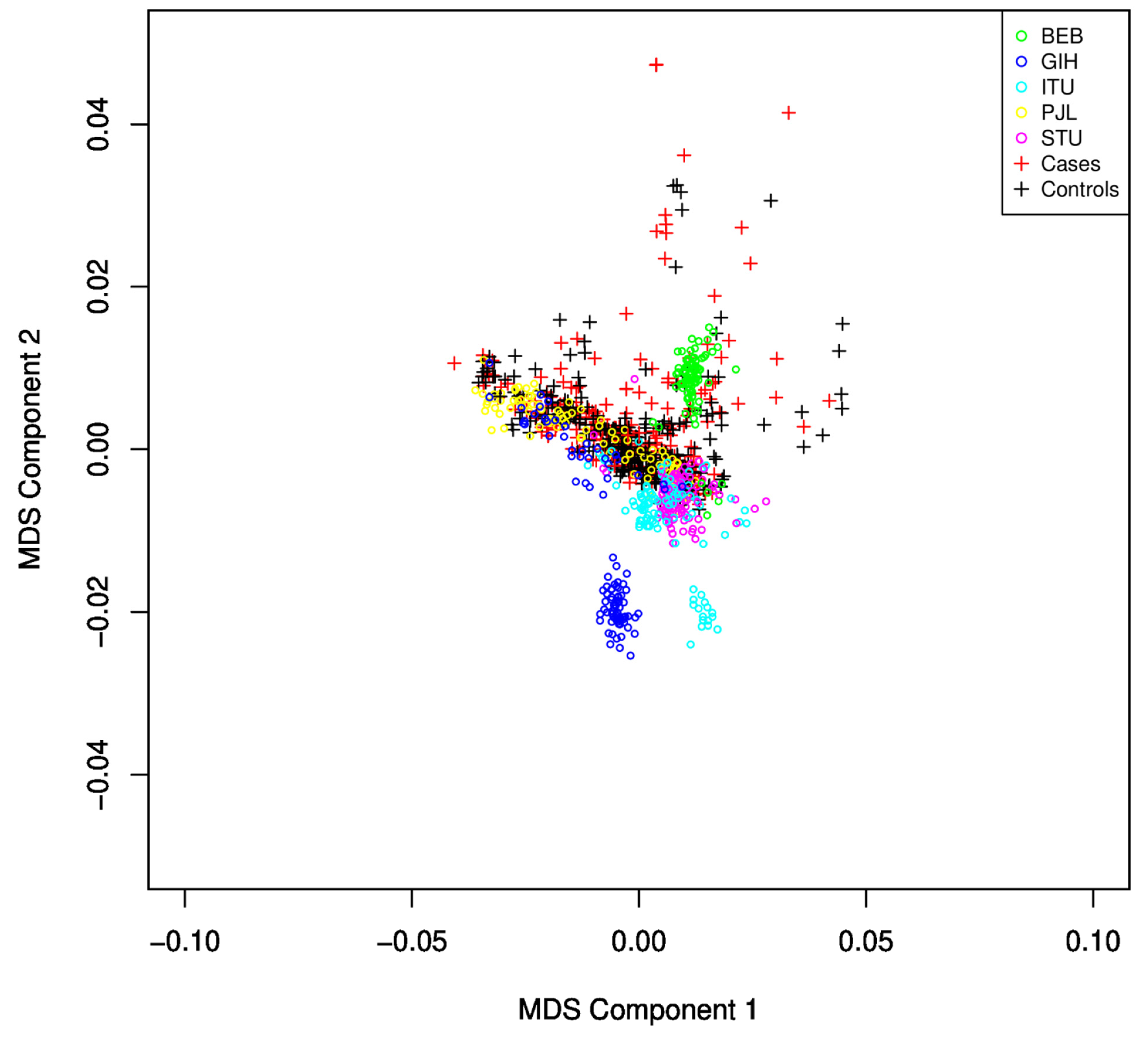
3.2. Population Stratification/Multidimensional Scaling Analysis
3.3. Inflation Rate of the Data/QQ Plot
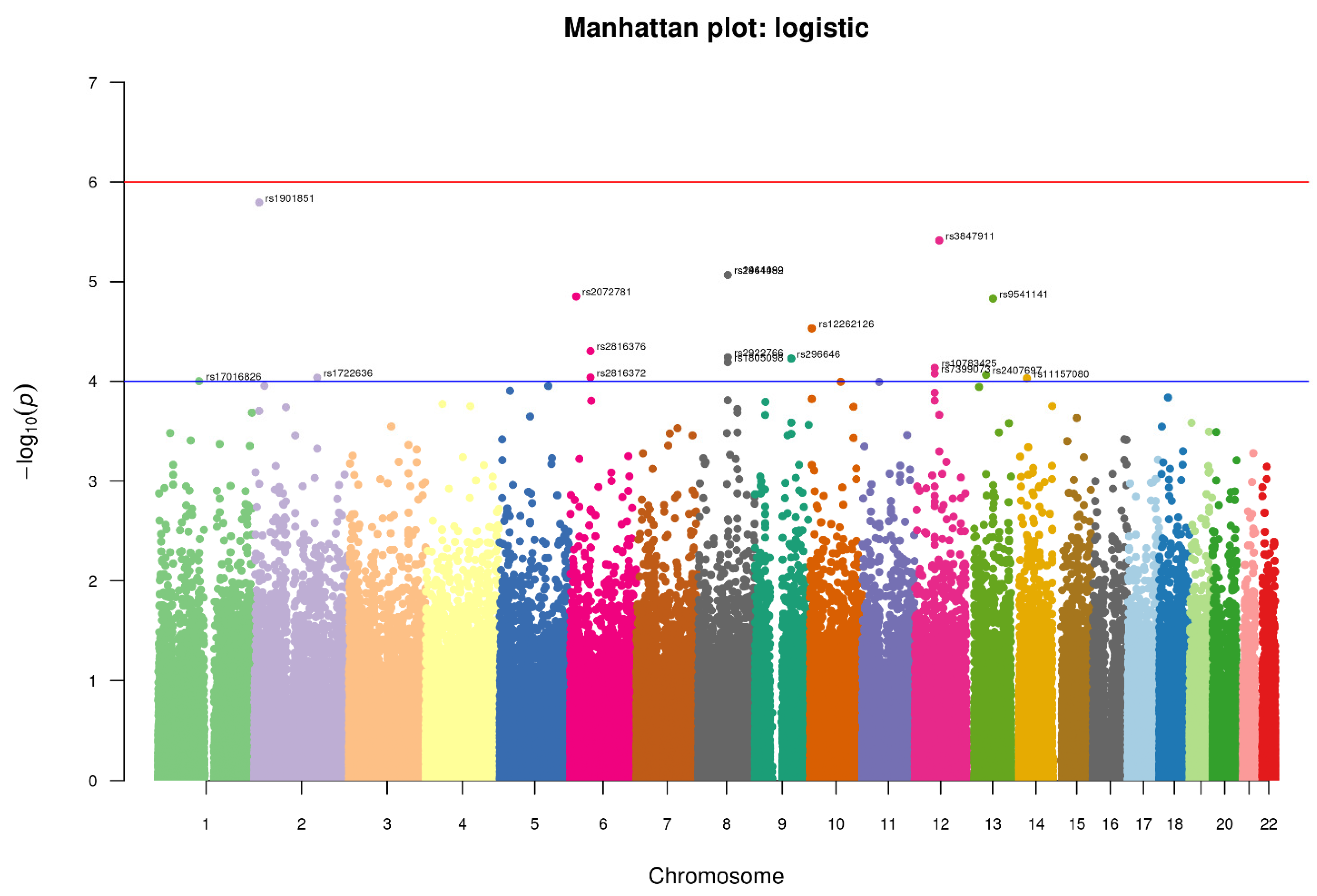
3.4. Association of SNPs with the Suicidal Deaths
4. Discussion
5. Conclusions
6. Limitation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Suicide Worldwide in 2019: Global Health Estimates; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- NCRB 2021. Available online: https://ncrb.gov.in/sites/default/files/ADSI-2021/adsi2021_Chapter-2-Suicides.pdf (accessed on 1 September 2022).
- Mann, J.J.; Waternaux, C.; Haas, G.L.; Malone, K.M. Toward a Clinical Model of Suicidal Behavior in Psychiatric Patients. Am. J. Psychiatry 1999, 156, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Andrade, C. Suicide: An Indian Perspective. Indian J. Psychiatry 2012, 54, 304. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Segal, N.L.; Centerwall, B.S.; Robinette, C.D. Suicide in Twins. Arch. Gen. Psychiatry 1991, 48, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Brent, D.A.; Bridge, J.; Johnson, B.A.; Connolly, J. Suicidal Behavior Runs in Families: A Controlled Family Study of Adolescent Suicide Victims. Arch. Gen. Psychiatry 1996, 53, 1145–1152. [Google Scholar] [CrossRef]
- Brent, D.A.; Mann, J.J. Family Genetic Studies, Suicide, and Suicidal Behavior. Am. J. Med. Genet. Semin. Med. Genet. 2005, 133, 13–24. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, P.; Perroud, N.; Uher, R.; Butler, A.; Aitchison, K.J.; Craig, I.; Lewis, C.; Farmer, A. The Genetics of Affective Disorder and Suicide. Eur. Psychiatry 2010, 25, 275–277. [Google Scholar] [CrossRef]
- Mann, J.J.; Currier, D. Effects of Genes and Stress on the Neurobiology of Depression. Int. Rev. Neurobiol. 2006, 73, 153–189. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.J.; Bortinger, J.; Oquendo, M.A.; Currier, D.; Li, S.; Brent, D.A. Family History of Suicidal Behavior and Mood Disorders in Probands with Mood Disorders. Am. J. Psychiatry 2005, 162, 1672–1679. [Google Scholar] [CrossRef]
- McGuffin, P.; Riley, B.; Plomin, R. Toward Behavioral Genomics. Science 2001, 291, 1232–1249. [Google Scholar] [CrossRef]
- Mann, J.J. Neurobiology of Suicidal Behaviour. Nat. Rev. Neurosci. 2003, 4, 819–828. [Google Scholar] [CrossRef]
- Rujescu, D.; Giegling, I. The Genetics of Neurosystems in Mental Ill-Health and Suicidality: Beyond Serotonin. Eur. Psychiatry 2010, 25, 272–274. [Google Scholar] [CrossRef] [PubMed]
- Rujescu, D.; Thalmeier, A.; Möller, H.J.; Bronisch, T.; Giegling, I. Molecular Genetic Findings in Suicidal Behavior: What Is beyond the Serotonergic System? Arch. Suicide Res. 2007, 11, 17–40. [Google Scholar] [CrossRef] [PubMed]
- Kasper, S.; Hamon, M. Beyond the Monoaminergic Hypothesis: Agomelatine, a New Antidepressant with an Innovative Mechanism of Action. World J. Biol. Psychiatry 2009, 10, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yoshikawa, A.; Meltzer, H.Y. Replication of Rs300774, a Genetic Biomarker near ACP1, Associated with Suicide Attempts in Patients with Schizophrenia: Relation to Brain Cholesterol Biosynthesis. J. Psychiatr. Res. 2017, 94, 54–61. [Google Scholar] [CrossRef]
- Stein, M.B.; Ware, E.B.; Mitchell, C.; Chen, C.Y.; Borja, S.; Cai, T.; Dempsey, C.L.; Fullerton, C.S.; Gelernter, J.; Heeringa, S.G.; et al. Genomewide Association Studies of Suicide Attempts in US Soldiers. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Kimbrel, N.A.; Garrett, M.E.; Dennis, M.F.; Hauser, M.A.; Ashley-Koch, A.E.; Beckham, J.C. A Genome-Wide Association Study of Suicide Attempts and Suicidal Ideation in U.S. Military Veterans. Psychiatry Res. 2018, 269, 64. [Google Scholar] [CrossRef] [PubMed]
- Ruderfer, D.M.; Walsh, C.G.; Aguirre, M.W.; Tanigawa, Y.; Ribeiro, J.D.; Franklin, J.C.; Rivas, M.A. Significant Shared Heritability Underlies Suicide Attempt and Clinically Predicted Probability of Attempting Suicide. Mol. Psychiatry 2020, 25, 2422–2430. [Google Scholar] [CrossRef]
- Mirkovic, B.; Laurent, C.; Podlipski, M.A.; Frebourg, T.; Cohen, D.; Gerardin, P. Genetic Association Studies of Suicidal Behavior: A Review of the Past 10 Years, Progress, Limitations, and Future Directions. Front. Psychiatry 2016, 7, 158. [Google Scholar] [CrossRef]
- Bunney, W.E.; Bunney, B.G.; Vawter, M.P.; Tomita, H.; Li, J.; Evans, S.J.; Choudary, P.V.; Myers, R.M.; Jones, E.G.; Watson, S.J.; et al. Microarray Technology: A Review of New Strategies to Discover Candidate Vulnerability Genes in Psychiatric Disorders. Am. J. Psychiatry 2003, 160, 657–666. [Google Scholar] [CrossRef]
- Lipshutz, R.J.; Fodor, S.P.A.; Gingeras, T.R.; Lockhart, D.J. High Density Synthetic Oligonucleotide Arrays. Nat. Genet. 1999, 21, 20–24. [Google Scholar] [CrossRef]
- Matsuzaki, H.; Dong, S.; Loi, H.; Di, X.; Liu, G.; Hubbell, E.; Law, J.; Berntsen, T.; Chadha, M.; Hui, H.; et al. Genotyping over 100,000 SNPs on a Pair of Oligonucleotide Arrays. Nat. Methods 2004, 1, 109–111. [Google Scholar] [CrossRef]
- Ebert, B.W. Guide to Conducting a Psychological Autopsy. Prof. Psychol. Res. Pract. 1987, 18, 52–56. [Google Scholar] [CrossRef]
- Shneidman, E.S. The Psychological Autopsy. Am. Psychol. 1994, 49, 75–76. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar]
- Zhao, S.; Jing, W.; Samuels, D.C.; Sheng, Q.; Shyr, Y.; Guo, Y. Strategies for Processing and Quality Control of Illumina Genotyping Arrays. Brief. Bioinform. 2018, 19, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; He, J.; Zhao, S.; Wu, H.; Zhong, X.; Sheng, Q.; Samuels, D.C.; Shyr, Y.; Long, J. Illumina Human Exome Genotyping Array Clustering and Quality Control. Nat. Protoc. 2014, 9, 2643–2662. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 2643–2662. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-Generation Genotype Imputation Service and Methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Howie, B.; Fuchsberger, C.; Stephens, M.; Marchini, J.; Abecasis, G.R. Fast and Accurate Genotype Imputation in Genome-Wide Association Studies through Pre-Phasing. Nat. Genet. 2012, 44, 955–959. [Google Scholar] [CrossRef]
- Loh, P.R.; Palamara, P.F.; Price, A.L. Fast and Accurate Long-Range Phasing in a UK Biobank Cohort. Nat. Genet. 2016, 48, 811–816. [Google Scholar] [CrossRef]
- Santy GitHub—Santy-8128/DosageConvertor: DosageConvertor Is a C++ Tool to Convert Dosage Files (in VCF Format) from Minimac3/4 to Other Formats Such as MaCH or PLINK. Available online: https://github.com/Santy-8128/DosageConvertor (accessed on 23 August 2022).
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef]
- Nishimura, D. GeneCards. Biotech Softw. Internet Rep. 2004, 2, 47–49. [Google Scholar] [CrossRef]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a Knowledgebase of Human Genes and Genetic Disorders. Nucleic. Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Stein, T.I.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An Amalgamated Human Disease Compendium with Diverse Clinical and Genetic Annotation and Structured Search. Nucleic. Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Neale, B.M.; Medland, S.; Ripke, S.; Anney, R.J.L.; Asherson, P.; Buitelaar, J.; Franke, B.; Gill, M.; Kent, L.; Holmans, P.; et al. Case-Control Genome-Wide Association Study of Attention-Deficit/Hyperactivity Disorder. J. Am. Acad. Child Adolesc. Psychiatry 2010, 49, 906–920. [Google Scholar] [CrossRef]
- Roach, K.L.; Hershberger, P.E.; Rutherford, J.N.; Molokie, R.E.; Wang, Z.J.; Wilkie, D.J. The AVPR1A Gene and Its Single Nucleotide Polymorphism Rs10877969: A Literature Review of Associations with Health Conditions and Pain. Pain Manag. Nurs. 2018, 19, 430–444. [Google Scholar] [CrossRef]
- Sasaki, S.; Urabe, M.; Maeda, T.; Suzuki, J.; Irie, R.; Suzuki, M.; Tomaru, Y.; Sakaguchi, M.; Gonzalez, F.J.; Inoue, Y. Induction of Hepatic Metabolic Functions by a Novel Variant of Hepatocyte Nuclear Factor 4γ. Mol. Cell Biol. 2018, 38, e00213-18. [Google Scholar] [CrossRef]
- Yan, T.T.; Yin, R.X.; Li, Q.; Huang, P.; Zeng, X.N.; Huang, K.K.; Wu, D.F.; Aung, L.H.H. Association of MYLIP Rs3757354 SNP and Several Environmental Factors with Serum Lipid Levels in the Guangxi Bai Ku Yao and Han Populations. Lipids Health Dis. 2012, 11, 141. [Google Scholar] [CrossRef]
- Papiha, S.S. Genetic Variation in India. Hum. Biol. 1996, 68, 607–628. [Google Scholar]


| Characteristics | Cases Suicide Deaths (n = 313) | Controls Non-Suicidal Deaths (n = 294) | p-Value |
|---|---|---|---|
| Gender (Male) | 220 (70.29%) | 254 (86.39) | <1 × 10−5 a |
| Age (years (SD) | 30.26 (11.58) | 40.5 (14.48) | <1 × 10−5 b |
| Married Unmarried Separated | 187 (59.74%) 125 (39.93%) 1 (0.31%) | 226 (76.87%) 65 (22.10%) 3 (0.10%) | <1 × 10−5 a <1 × 10−5 a 0.346 |
| Means of death Hanging Poisoning Burn Fall from height Natural Accidental Homicide | 291 (92.97%) 17 (5.43%) 1 (0.31%) 4 (1.27%) | none none none none 216 (73.46%) 76 (25.85%) 2 (0.6%) | NA |
| Psychiatric illness | 18 (5.75%) | none | NA |
| Family history of suicide | 19 (6.07%) | none | NA |
| Previous suicide attempts | 38 (12.14%) | none | NA |
| Suicide note | 13 (4.15%) | NA | NA |
| Alcohol/Drug abuse | 153 (48.88%) | 134 (45.57%) | 0.463 a |
| S.No. | CHR | SNP | A1 (Minor) | A2 | Cases Genotype | Controls Genotype | MAF Case | MAF Control | OR (95% CI) | p-Value | MAF 1 KG | MAF SAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | rs1901851 | A | C | 110/128/62 | 50/127/87 | 0.58 | 0.42 | 1.799 | 1.606 × 10−6 | A = 0.5178 C = 0.4822 | A = 0.518 C = 0.482 |
| 2 | 12 | rs3847911 | G | T | 68/136/96 | 24/124/116 | 0.453 | 0.327 | 1.814 | 3.86 × 10−6 | G = 0.2670 T = 0.7330 | G = 0.403 T = 0.597 |
| 3 | 8 | rs2941489 | C | T | 79/139/82 | 33/123/108 | 0.495 | 0.3579 | 1.754 | 8.572 × 10−6 | C = 0.4127 T = 0.5873 | C = 0.452 T = 0.548 |
| 4 | 8 | rs1464092 | C | T | 79/139/82 | 33/123/108 | 0.495 | 0.3579 | 1.754 | 8.572 × 10−6 | C = 0.4012 T = 0.5988 | C = 0.453 T = 0.547 |
| 5 | 6 | rs2072781 | C | T | 5/72/223 | 1/31/232 | 0.1367 | 0.0625 | 2.681 | 1.405 × 10−5 | T = 0.8297 C = 0.1703 | T = 0.876 C = 0.124 |
| 6 | 13 | rs9541141 | T | C | 40/129/131 | 19/87/158 | 0.3483 | 0.2367 | 1.8 | 1.478 × 10−5 | C = 0.7544 T = 0.2456 | C = 0.706 T = 0.294 |
| 7 | 10 | rs12262126 | A | G | 86/147/67 | 40/135/89 | 0.5317 | 0.4071 | 1.696 | 2.944 × 10−5 | G = 0.4343 A = 0.5657 | G = 0.472 A = 0.528 |
| 8 | 6 | rs2816376 | C | T | 43/142/115 | 22/94/148 | 0.380 | 0.2613 | 1.71 | 4.979 × 10−5 | T = 0.6296 C = 0.3704 | T = 0.623 C = 0.377 |
| 9 | 8 | rs2922766 | T | C | 72/156/72 | 36/136/92 | 0.5 | 0.3939 | 1.701 | 5.732 × 10−5 | T = 0.3770 C = 0.6230 | T = 0.473 C = 0.527 |
| 10 | 9 | rs296646 | C | T | 58/134/108 | 22/113/129 | 0.4167 | 0.2973 | 1.683 | 5.915 × 10−5 | T = 0.6100 C = 0.3900 | T = 0.598 C = 0.402 |
| 11 | 8 | rs1805098 | G | A | 89/143/68 | 89/115/60 | 0.535 | 0.6079 | 1.663 | 6.447 × 10−5 | G = 0.3852 A = 0.6148 | G = 0.496 A = 0.504 |
| 12 | 12 | rs10783425 | C | T | 65/147/88 | 29/126/109 | 0.4617 | 0.3484 | 1.661 | 7.325 × 10−5 | C = 0.4998 T = 0.5002 | C = 0.394 T = 0.606 |
| 13 | 12 | rs7399073 | C | A | 43/144/113 | 17/111/136 | 0.3833 | 0.2746 | 1.709 | 8.376 × 10−5 | C = 0.3844 A = 0.6156 | C = 0.307 A = 0.693 |
| 14 | 13 | rs2407697 | G | T | 17/133/150 | 10/73/181 | 0.2783 | 0.1761 | 1.838 | 8.627 × 10−5 | G = 0.1683 T = 0.8317 | G = 0.225 T = 0.775 |
| 15 | 6 | rs2816372 | G | A | 60/145/95 | 32/106/126 | 0.4417 | 0.3219 | 1.634 | 9.14 × 10−5 | A = 0.6148 G = 0.3852 | A = 0.578 G = 0.422 |
| 16 | 2 | rs1722636 | T | C | 40/134/126 | 14/104/146 | 0.3567 | 0.25 | 1.703 | 9.19 × 10−5 | C = 0.7113 T = 0.2887 | C = 0.691 T = 0.309 |
| 17 | 14 | rs11157080 | T | C | 67/168/65 | 42/125/97 | 0.5033 | 0.3958 | 1.655 | 9.338 × 10−5 | C = 0.6410 T = 0.3590 | C = 0.540 T = 0.460 |
| 18 | 1 | rs17016826 | C | A | 6/78/216 | 2/39/223 | 0.15 | 0.0814 | 2.242 | 9.984 × 10−5 | A = 0.8986 C = 0.1014 | A = 0.860 C = 0.140 |
| S.No. | CHR | SNP | Function of Reference Gene | Nearest Gene (s) | Function | Diseases Associated with Gene (s) |
|---|---|---|---|---|---|---|
| 1 | 2 | rs1901851 | Intron Variant | MIR3681HG | RNA Gene, and is affiliated with the lncRNA class. | |
| 2 | 12 | rs3847911 | Intergenic | PPM1H;AVPR1A | PPM1H (Protein Phosphatase, Mg2+/Mn2+ Dependent 1H) is a Protein Coding gene. The protein encoded by AVPR1A gene acts as receptor for arginine vasopressin | PPM1H: Diseases associated with PPM1H include Multiple Endocrine Neoplasia, Type Iv and Attention Deficit-Hyperactivity Disorder. Gene Ontology (GO) annotations related to this gene include phosphoprotein phosphatase activity. An important paralog of this gene is PPM1J; AVPR1A: Diseases associated with AVPR1A include Acth-Independent Macronodular Adrenal Hyperplasia and Diabetes Insipidus. Among its related pathways are RET signaling and Signaling by GPCR. |
| 3 | 8 | rs2941489 | intergenic | HNF4G;LINC01111 | HNF4G (Hepatocyte Nuclear Factor 4 Gamma) is a Protein Coding gene; LINC01111 (Long Intergenic Non-Protein Coding RNA 1111) is an RNA Gene, and is affiliated with the lncRNA class | Diseases associated with HNF4G include Maturity-Onset Diabetes Of The Young and Hyperuricemia. Among its related pathways are regulation of beta-cell development and Gene Expression. Gene Ontology (GO) annotations related to this gene include DNA-binding transcription factor activity and steroid hormone receptor activity. An important paralog of this gene is HNF4A. Diseases associated with LINC01111 include Chromosome 8Q21.11 Deletion Syndrome and Sclerocornea. |
| 4 | 8 | rs1464092 | intergenic | HNF4G;LINC01111 | ||
| 5 | 8 | rs2922766 | intergenic | HNF4G;LINC01111 | ||
| 6 | 6 | rs2072781 | UTR3 | MYLIP | MYLIP protein interacts with myosin regulatory light chain and inhibits neurite outgrowth. | Diseases associated with MYLIP include Deafness, Autosomal Dominant 31, and Deafness, Autosomal Dominant 21. Among its related pathways are Lipoprotein metabolism and Innate Immune System. |
| 7 | 13 | rs9541141 | intergenic | LINC00364 | LINC00364 (Long Intergenic Non-Protein Coding RNA 364) is an RNA Gene and is affiliated with the lncRNA class. | - |
| 8 | 10 | rs12262126 | intergenic | CALML3;LINC02657 | CALML3 may function as a specific light chain of unconventional myosin-10 (MYO10), also enhances MYO10 translation, possibly by acting as a chaperone for the emerging MYO10 heavy chain protein. LINC02657 (LASTR) is an RNA Gene, and is affiliated with the lncRNA class. | Diseases associated with CALML3 include Alzheimer’s disease. Among its related pathways are tuberculosis and Inositol phosphate metabolism (KEGG). Gene Ontology (GO) annotations related to this gene include calcium ion binding. |
| 9 | 6 | rs2816376 | Intergenic | GCM1;ELOVL5 | GCM1 encodes a DNA-binding protein with a gcm-motif (glial cell missing motif) ELOVL5. It is highly expressed in the adrenal gland and testis and encodes a multi-pass membrane protein that is localized in the endoplasmic reticulum. This protein is involved in the elongation of long-chain polyunsaturated fatty acids. | Diseases associated with GCM1 include Cardiomyopathy, Familial Restrictive, 2 and Pre-Eclampsia. Among its related pathways are Human Early Embryo Development and Parathyroid hormone synthesis, secretion, and action. Diseases associated with ELOVL5 include Spinocerebellar Ataxia 38 and Intermittent Squint. Among its related pathways are alpha-linolenic (omega3) and linoleic (omega6) acid metabolism and Metabolism. |
| 10 | 6 | rs2816372 | Intergenic | GCM1;ELOVL5 | ||
| 11 | 9 | rs296646 | intergenic | SYK;LOC100129316 | SYK gene encodes a member of the family of non-receptor type Tyr protein kinases. This protein is widely expressed in hematopoietic cells and is involved in coupling activated immunoreceptors to downstream signaling events that mediate diverse cellular responses, including proliferation, differentiation, and phagocytosis. LOC100129316 is a disease associated with CALML3, including Alzheimer’s Disease. Among its related pathways are Tuberculosis and Inositol phosphate metabolism (KEGG). Gene Ontology (GO) annotations related to this gene include calcium ion binding RNA Gene and is affiliated with the lncRNA class. | Diseases associated with SYK include Peripheral T-Cell Lymphoma and Hantavirus Pulmonary Syndrome. Among its related pathways are B cell receptor signaling pathway (KEGG) and signaling by GPCR. |
| 12 | 8 | rs1805098 | Exonic (mis sense) | HNF4G | HNF4G (Hepatocyte Nuclear Factor 4 Gamma) is a Protein Coding gene | Diseases associated with HNF4G include Maturity-Onset Diabetes Of The Young and Hyperuricemia. Among its related pathways are regulation of beta-cell development and Gene Expression. Gene Ontology (GO) annotations related to this gene include DNA-binding transcription factor activity and steroid hormone receptor activity. |
| 13 | 12 | rs10783425 | intergenic | POU6F1;DAZAP2 | POU6F1: DNA-binding transcription factor activity. An important paralog of this gene is POU6F2. DAZAP2. This gene encodes a proline-rich protein which interacts with the deleted in azoospermia (DAZ) and the deleted in azoospermia-like gene through the DAZ-like repeats. This protein also interacts with the transforming growth factor-beta signaling molecule SARA (Smad anchor for receptor activation), eukaryotic initiation factor 4G, and an E3 ubiquitinase that regulates its stability in splicing factor containing nuclear speckles. The encoded protein may function in various biological and pathological processes, including spermatogenesis, cell signaling and transcription regulation, formation of stress granules during translation arrest, RNA splicing, and pathogenesis of multiple myeloma. Multiple transcript variants encoding different isoforms have been found for this gene. | POU6F1: Diseases associated with POU6F1 include Clear Cell Adenocarcinoma Of The Ovary and Clear Cell Adenocarcinoma. DAZAP2: Diseases associated with DAZAP2 include Thyroid Hormone Resistance, Selective Pituitary, and Azoospermia. Among its related pathways are Diurnally Regulated Genes with Circadian Orthologs. Gene Ontology (GO) annotations related to this gene include WW domain binding |
| 14 | 12 | rs7399073 | Intergenic (2KB Upstream Variant) | POU6F1;DAZAP2 | ||
| 15 | 13 | rs2407697 | intronic | RCBTB1 | In rats, over-expression of this gene in vascular smooth muscle cells induced cellular hypertrophy. In rats, the C-terminus of RCBTB1 interacts with the angiotensin II receptor-1A | Diseases associated with RCBTB1 include Retinal Dystrophy With Or Without Extraocular Anomalies and Reticular Dystrophy Of Retinal Pigment Epithelium. An important paralog of this gene is RCBTB2. |
| 16 | 2 | rs1722636 | intergenic | RBMS1;TANK | RBMS1 (RNA Binding Motif Single Stranded Interacting Protein 1) is a Protein Coding gene encodes a member of a small family of proteins which bind single stranded DNA/RNA. The TRAF (tumor necrosis factor receptor-associated factor) family of proteins associate with and transduce signals from members of the tumor necrosis factor receptor superfamily. The protein encoded by this gene is found in the cytoplasm and can bind to TRAF1, TRAF2, or TRAF3, thereby inhibiting TRAF function by sequestering the TRAFs in a latent state in the cytoplasm. | Diseases associated with RBMS1 include Blue Toe Syndrome and Diffuse Glomerulonephritis. Gene Ontology (GO) annotations related to this gene include nucleic acid binding and RNA binding. An important paralog of this gene is RBMS3. Diseases associated with TANK include Nipah Virus Encephalitis. Among its related pathways are Activated TLR4 signaling and TRAF Pathway. Gene Ontology (GO) annotations related to this gene include ubiquitin protein ligase binding. |
| 17 | 14 | rs11157080 | intergenic | FBXO33;LINC02315 | FBXO33 may be associated with placental RNAse inhibitor, and locus may be associated with copy number variation of UGT2B17 (GeneID 7367), which has been associated with susceptibility to osteoporosis (bone disease). LINC02315: Long Intergenic Non-Protein Coding | Diseases associated with FBXO33 include Protoplasmic Astrocytoma and Attention Deficit-Hyperactivity Disorder |
| 18 | 1 | rs17016826 | Intergenic | LINC01677;LINC01661 | LncRNA | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behera, C.; Kaushik, R.; Bharti, D.R.; Nayak, B.; Bhardwaj, D.N.; Pradhan, D.; Singh, H. PsychArray-Based Genome Wide Association Study of Suicidal Deaths in India. Brain Sci. 2023, 13, 136. https://doi.org/10.3390/brainsci13010136
Behera C, Kaushik R, Bharti DR, Nayak B, Bhardwaj DN, Pradhan D, Singh H. PsychArray-Based Genome Wide Association Study of Suicidal Deaths in India. Brain Sciences. 2023; 13(1):136. https://doi.org/10.3390/brainsci13010136
Chicago/Turabian StyleBehera, Chittaranjan, Ruchika Kaushik, Deepak Ramkumar Bharti, Baibaswata Nayak, Daya Nand Bhardwaj, Dibyabhaba Pradhan, and Harpreet Singh. 2023. "PsychArray-Based Genome Wide Association Study of Suicidal Deaths in India" Brain Sciences 13, no. 1: 136. https://doi.org/10.3390/brainsci13010136
APA StyleBehera, C., Kaushik, R., Bharti, D. R., Nayak, B., Bhardwaj, D. N., Pradhan, D., & Singh, H. (2023). PsychArray-Based Genome Wide Association Study of Suicidal Deaths in India. Brain Sciences, 13(1), 136. https://doi.org/10.3390/brainsci13010136