Abnormal Topological Organization of Structural Covariance Networks in Patients with Temporal Lobe Epilepsy Comorbid Sleep Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. MRI Data Acquisition
2.3. Data Processing
2.3.1. Data Pre-Processing
2.3.2. SCN Construction
2.3.3. Network Parameters
2.3.4. Network Hubs
2.3.5. Degree Distribution
2.3.6. Network Robustness
2.3.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics
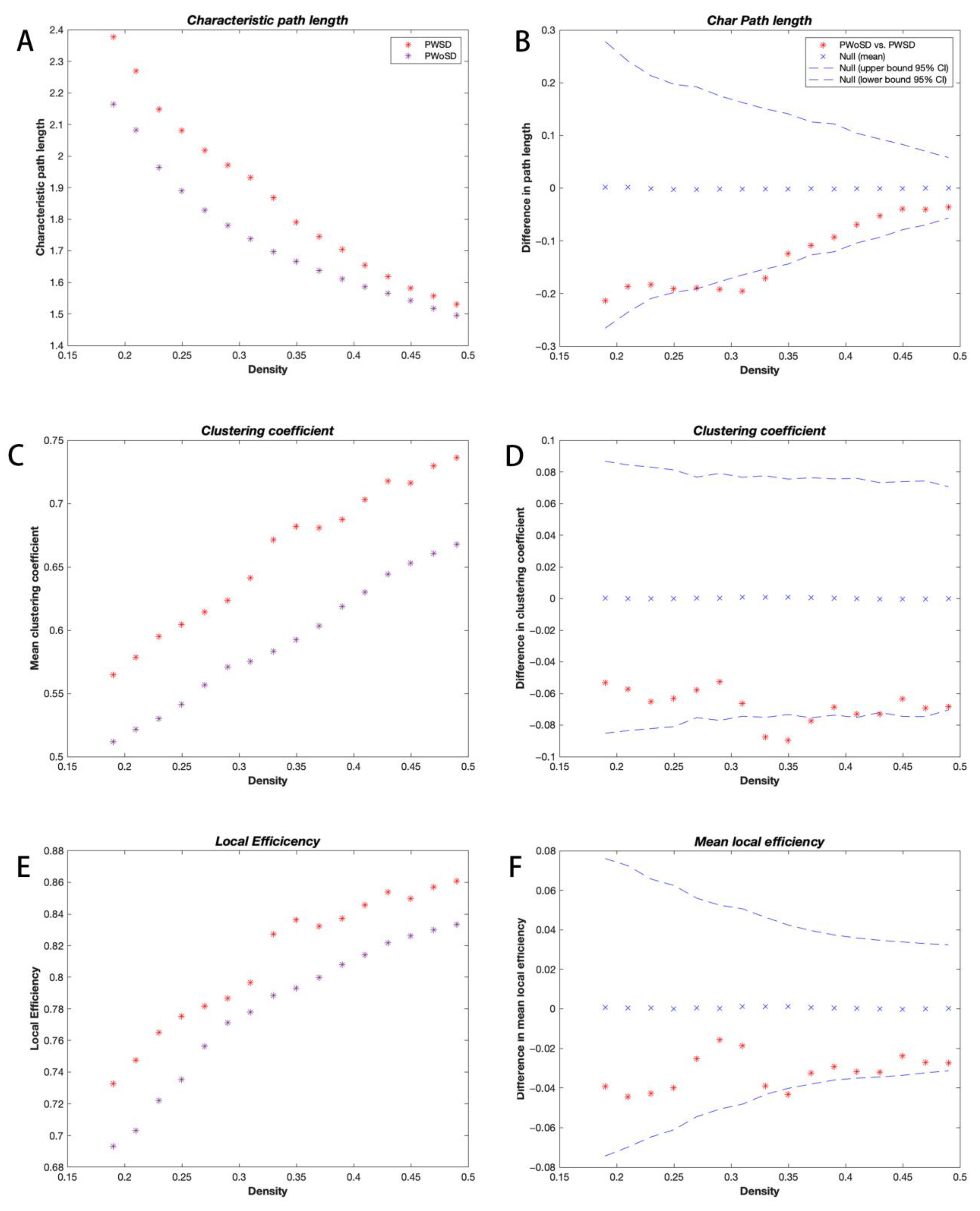
3.2. Global Network Analysis
3.3. Regional Network Analysis
3.4. Network Hub Analysis
3.5. Degree Distribution Analysis
3.6. Network Robustness Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Derry, C.P.; Duncan, S. Sleep and epilepsy. Epilepsy Behav. 2013, 26, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Nobili, L.; de Weerd, A.; Rubboli, G.; Beniczky, S.; Derry, C.; Eriksson, S.; Halasz, P.; Högl, B.; Santamaria, J.; Khatami, R.; et al. Standard procedures for the diagnostic pathway of sleep-related epilepsies and comorbid sleep disorders: An EAN, ESRS and ILAE-Europe consensus review. Eur. J. Neurol. 2021, 28, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Ismayilova, V.; Demir, A.U.; Tezer, F.I. Subjective sleep disturbance in epilepsy patients at an outpatient clinic: A questionnaire-based study on prevalence. Epilepsy Res. 2015, 115, 119–125. [Google Scholar] [CrossRef]
- Macedo, P.; Oliveira, P.S.; Foldvary-Schaefer, N.; Gomes, M.D.M. Insomnia in people with epilepsy: A review of insomnia prevalence, risk factors and associations with epilepsy-related factors. Epilepsy Res. 2017, 135, 158–167. [Google Scholar] [CrossRef]
- Gutter, T.; Callenbach, P.M.C.; Brouwer, O.F.; de Weerd, A.W. Prevalence of sleep disturbances in people with epilepsy and the impact on quality of life: A survey in secondary care. Seizure Eur. J. Epilep. 2019, 69, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Nobili, L.; Beniczky, S.; Eriksson, S.H.; Romigi, A.; Ryvlin, P.; Toledo, M.; Rosenzweig, I. Expert Opinion: Managing sleep disturbances in people with epilepsy. Epilepsy Behav. 2021, 124, 108341. [Google Scholar] [CrossRef]
- Dede, H.O.; Bebek, N.; Akarsu, E.O. Relationship between sleep characteristics and sudden unexplained death risk in epilepsy. Neurologist 2019, 24, 170, Erratum in Neurologist 2020, 25, 17. [Google Scholar] [CrossRef]
- Moeller, F.; Muthuraman, M.; Stephani, U.; Deuschl, G.; Raethjen, J.; Siniatchkin, M. Representation and propagation of epileptic activity in absences and generalized photoparoxysmal responses. Hum. Brain Mapp. 2013, 34, 1896–1909. [Google Scholar] [CrossRef]
- Chiosa, V.; Ciolac, D.; Groppa, S.; Koirala, N.; Pintea, B.; Vataman, A.; Winter, Y.; Gonzalez-Escamilla, G.; Muthuraman, M.; Groppa, S. Large-scale network architecture and associated structural cortico-subcortical abnormalities in patients with sleep/awake-related seizures. Sleep 2019, 42, zsz006. [Google Scholar] [CrossRef]
- Vespa, S.; Heyse, J.; Stumpp, L.; Liberati, G.; Ferrao Santos, S.; Rooijakkers, H.; Nonclercq, A.; Mouraux, A.; van Mierlo, P.; El Tahry, R. Vagus Nerve Stimulation Elicits Sleep EEG Desynchronization and Network Changes in Responder Patients in Epilepsy. Neurotherapeutics 2021, 18, 2623–2638. [Google Scholar] [CrossRef]
- Zhang, Z.; Liao, W.; Xu, Q.; Wei, W.; Zhou, H.J.; Sun, K.; Yang, F.; Mantini, D.; Ji, X.; Lu, G. Hippocampus-associated causal network of structural covariance measuring structural damage progression in temporal lobe epilepsy. Hum. Brain Mapp. 2017, 38, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Alexander-Bloch, A.; Giedd, J.N.; Bullmore, E.T. Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci. 2013, 14, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Buysse, D.J.; Reynolds, C.F., 3rd; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The Pittsburgh Sleep Quality Index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, A.C.; Hickie, I.B.; Scott, J.; Duffy, S.L.; Norrie, L.; Terpening, Z.; Grunstein, R.R.; Lagopoulos, J.; Batchelor, J.; Lewis, S.J.; et al. Current sleep disturbance in older people with a lifetime history of depression is associated with increased connectivity in the Default Mode Network. J. Affect. Disord. 2018, 229, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.G.; Wang, X.D.; Zuo, X.N.; Zang, Y.F. DPABI: Data Processing & Analysis for (Resting-State) Brain Imaging. Neuroinformatics 2016, 14, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.M.; Hoeft, F.; Kesler, S.R. GAT: A graph-theoretical analysis toolbox for analyzing between-group differences in large-scale structural and functional brain networks. PLoS ONE 2012, 7, e40709. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiu, T.; Yuan, X.; Zhang, J.; Wang, Y.; Zhang, N.; Zhou, C.; Luo, C.; Zhang, J. Abnormal topological organization of structural covariance networks in amyotrophic lateral sclerosis. Neuroimage Clin. 2019, 21, 101619. [Google Scholar] [CrossRef]
- Wang, E.; Jia, Y.; Ya, Y.; Xu, J.; Mao, C.; Luo, W.; Fan, G.; Jiang, Z. Abnormal Topological Organization of Sulcal Depth-Based Structural Covariance Networks in Parkinson’s Disease. Front. Aging Neurosci. 2020, 12, 575672. [Google Scholar] [CrossRef]
- Xia, M.; Wang, J.; He, Y. BrainNet Viewer: A network visualization tool for human brain connectomics. PLoS ONE 2013, 8, e68910. [Google Scholar] [CrossRef]
- He, Y.; Chen, Z.J.; Evans, A.C. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex 2007, 17, 2407–2419. [Google Scholar] [CrossRef]
- Hosseini, S.H.; Mazaika, P.; Mauras, N.; Buckingham, B.; Weinzimer, S.A.; Tsalikian, E.; White, N.H.; Reiss, A.L.; Diabetes Research in Children Network (DirecNet). Altered Integration of Structural Covariance Networks in Young Children with Type 1 Diabetes. Hum. Brain Mapp. 2016, 37, 4034–4046. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Han, X.; Song, Y.; Han, Y.; Zhou, C.; Zhou, D.; Zhang, F.; Xue, Q.; Liu, J.; Zhao, L.; et al. Toward neuroimaging-based network biomarkers for transient ischemic attack. Hum. Brain Mapp. 2019, 40, 3347–3361. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Vasilakos, A.V.; He, Y. Small-world human brain networks: Perspectives and challenges. Neurosci. Biobehav. Rev. 2017, 77, 286–300. [Google Scholar] [CrossRef]
- Yang, S.; Wu, Y.; Sun, L.; You, X.; Wu, Y. Reorganization of brain networks in patients with temporal lobe epilepsy and comorbid headache. Epilepsy Behav. 2023, 140, 109101. [Google Scholar] [CrossRef]
- Cai, L.; Dong, Q.; Niu, H. The development of functional network organization in early childhood and early adolescence: A resting-state fNIRS study. Dev. Cogn. Neurosci. 2018, 30, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Achard, S.; Bullmore, E.T. Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 2007, 3, 174–183. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, J.; Zhang, J.; Wang, Y.; Zhang, Y.; Wang, J.; Li, G.; Hu, Q.; Zhang, Y. Abnormalities in Structural Covariance of Cortical Gyrification in Parkinson’s Disease. Front. Neuroanat. 2017, 11, 12. [Google Scholar] [CrossRef]
- Latora, V.; Marchiori, M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001, 87, 198701. [Google Scholar] [CrossRef]
- Fallani, F.D.V.; Astolfi, L.; Cincotti, F.; Mattia, D.; Marciani, M.G.; Salinari, S.; Kurths, J.; Gao, S.; Cichocki, A.; Colosimo, A.; et al. Cortical functional connectivity networks in normal and spinal cord injured patients: Evaluation by graph analysis. Hum. Brain Mapp. 2007, 28, 1334–1346. [Google Scholar] [CrossRef]
- Liu, H.; Li, H.; Wang, Y.L.; Lei, X. Enhanced brain small-worldness after sleep deprivation: A compensatory effect. J. Sleep Res. 2014, 23, 554–563. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhang, P.; Wang, C.H.; Yang, L.; Zhang, X.Z. Cortical Thinning and Abnormal Structural Covariance Network After Three Hours Sleep Restriction. Front. Psychiatry 2021, 12, 664811. [Google Scholar] [CrossRef] [PubMed]
- Ponten, S.C.; Bartolomei, F.; Stam, C.J. Small-world networks and epilepsy: Graph theoretical analysis of intracerebrally recorded mesial temporal lobe seizures. Clin. Neurophysiol. 2007, 118, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Schindler, K.A.; Bialonski, S.; Horstmann, M.T.; Elger, C.E.; Lehnertz, K. Evolving functional network properties and synchronizability during human epileptic seizures. Chaos 2008, 18, 033119. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.A.; Eden, U.T.; Kolaczyk, E.D.; Zepeda, R.; Eskandar, E.N.; Cash, S.S. Coalescence and fragmentation of cortical networks during focal seizures. J. Neurosci. 2010, 30, 10076–10085. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, R.; Guan, M.; Wang, E.; Qian, T.; Zhao, C.; Zou, Z.; Beck, T.; Shi, D.; Wang, M.; et al. Disrupted brain network topology in chronic insomnia disorder: A resting-state fMRI study. Neuroimage Clin. 2018, 18, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.G.; Wang, D.; Liu, K.; Weng, J.; Guan, Y.; Chan, K.C.; Chu, W.C.; Shi, L. Brain Structure Network Analysis in Patients with Obstructive Sleep Apnea. PLoS ONE 2015, 10, e0139055. [Google Scholar] [CrossRef]
- Koenis, M.M.; Romeijn, N.; Piantoni, G.; Verweij, I.; Van der Werf, Y.D.; Van Someren, E.J.; Stam, C.J. Does sleep restore the topology of functional brain networks? Hum. Brain Mapp. 2013, 34, 487–500. [Google Scholar] [CrossRef]
- Ferri, R.; Rundo, F.; Bruni, O.; Terzano, M.G.; Stam, C.J. The functional connectivity of different EEG bands moves towards small-world network organization during sleep. Clin. Neurophysiol. 2008, 119, 2026–2036. [Google Scholar] [CrossRef]
- Chen, L.L.; Qi, X.L.; Zheng, J.Y. Altered Regional Cortical Brain Activity in Healthy Subjects After Sleep Deprivation: A Functional Magnetic Resonance Imaging Study. Front. Neurol. 2018, 9, 588. [Google Scholar] [CrossRef]
- Chee, M.W.; Tan, J.C.; Zheng, H.; Parimal, S.; Weissman, D.H.; Zagorodnov, V.; Dinges, D.F. Lapsing during sleep deprivation is associated with distributed changes in brain activation. J. Neurosci. 2008, 28, 5519–5528. [Google Scholar] [CrossRef]
- Vecchio, F.; Miraglia, F.; Gorgoni, M.; Ferrara, M.; Iberite, F.; Bramanti, P.; De Gennaro, L.; Rossini, P.M. Cortical connectivity modulation during sleep onset: A study via graph theory on EEG data. Hum. Brain Mapp. 2017, 38, 5456–5464. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Rolls, E.T.; Ruan, H.T.; Feng, J.F. Functional Connectivities in the Brain That Mediate the Association Between Depressive Problems and Sleep Quality. JAMA Psychiatry 2018, 75, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Zikopoulos, B.; Barbas, H. Prefrontal projections to the thalamic reticular nucleus form a unique circuit for attentional mechanisms. J. Neurosci. 2006, 26, 7348–7361. [Google Scholar] [CrossRef]
- Datta, S.; MacLean, R.R. Neurobiological mechanisms for the regulation of mammalian sleep-wake behavior: Reinterpretation of historical evidence and inclusion of contemporary cellular and molecular evidence. Neurosci. Biobehav. R. 2007, 31, 775–824. [Google Scholar] [CrossRef]
- Palaniyappan, L.; Hodgson, O.; Balain, V.; Iwabuchi, S.; Gowland, P.; Liddle, P. Structural covariance and cortical reorganisation in schizophrenia: A MRI-based morphometric study. Psychol. Med. 2019, 49, 412–420. [Google Scholar] [CrossRef]
- Crossley, N.A.; Mechelli, A.; Scott, J.; Carletti, F.; Fox, P.T.; McGuire, P.; Bullmore, E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 2014, 137, 2382–2395. [Google Scholar] [CrossRef]
- Bonilha, L.; Tabesh, A.; Dabbs, K.; Hsu, D.A.; Stafstrom, C.E.; Hermann, B.P.; Lin, J.J. Neurodevelopmental alterations of large-scale structural networks in children with new-onset epilepsy. Hum. Brain Mapp. 2014, 35, 3661–3672. [Google Scholar] [CrossRef]
- Tijms, B.M.; Möller, C.; Vrenken, H.; Wink, A.M.; de Haan, W.; van der Flier, W.M.; Stam, C.J.; Scheltens, P.; Barkhof, F. Single-subject grey matter graphs in Alzheimer’s disease. PLoS ONE 2013, 8, e58921. [Google Scholar] [CrossRef]
- Drenthen, G.S.; Backes, W.H.; Rouhl, R.P.W.; Vlooswijk, M.C.G.; Majoie, M.H.J.M.; Hofman, P.A.M.; Aldenkamp, A.P.; Jansen, J.F.A. Structural covariance networks relate to the severity of epilepsy with focal-onset seizures. Neuroimage Clin. 2018, 20, 861–867. [Google Scholar] [CrossRef]




| Healthy Controls | PWoSD | PWSD | p-Value | |
|---|---|---|---|---|
| Number of participants | 17 | 33 | 34 | - |
| Age (year/mean ± SD) | 25.18 ± 3.01 | 22.30 ± 7.30 | 26.26 ± 9.27 | 0.10 a |
| Gender (male/female) | 9/8 | 21/12 | 15/19 | 0.28 b |
| Handedness | 22R | 32R | 25R | - |
| Seizure duration (year/mean ± SD) | - | 7.15 ± 5.44 | 7.38 ± 6.19 | 0.87 c |
| Seizure frequency (N/%) | ||||
| Seizure-free > 1 year | - | 5(15.15) | 3(8.82) | 0.42 b |
| 1–2 per year | - | 7(21.21) | 2(5.88) | 0.07 b |
| 1–5 in last 6 months | - | 3(9.09) | 2(5.88) | 0.62 b |
| 1–5 per month | - | 13(39.39) | 17(50.00) | 0.38 b |
| >5 per month | - | 5(15.15) | 10(29.41) | 0.16 b |
| Period of seizure (N/%) | ||||
| Sleep | - | 7(21.21) | 15(44.12) | 0.046 b |
| Awake | - | 17(51.51) | 6(17.65) | 0.004 b |
| Both sleep and awake | - | 9(27.27) | 13(38.24) | 0.34 b |
| PSQI (mean ± SD) | 3.47 ± 1.18 | 3.24 ± 1.66 | 9.06 ± 2.16 | <0.0001 a |
| ASMs (number/mean ± SD) | - | 1.49 ± 0.80 | 1.59 ± 0.70 | 0.57 c |
| Hub Regions | Abbreviation |
|---|---|
| Healthy controls | |
| L precental gyrus | PreCG.L |
| R inferior frontal gyrus, opercular part | IFGoperc.R |
| L inferior frontal gyrus, triangular part | IFGtriang.L |
| L insula | ORBsupmed.R |
| R insula | INS.L |
| L calcarine fissure and surrounding cortex | INS.R |
| PWoSD | |
| L middle frontal gyrus | MFG.L |
| R middle frontal gyrus | MFG.R |
| R insula | INS.R |
| R inferior parietal, but supramarginal and angular gyri | IPL.R |
| R angular gyrus | ANG.R |
| PWSD | |
| L precental gyrus | PreCG.L |
| R superior frontal gyrus, dorsolateral | SFGdor.R |
| L middle frontal gyrus | MFG.L |
| R middle frontal gyrus | MFG.R |
| R superior occipital gyrus | SOG.R |
| R middle occipital gyrus | MOG.R |
| L superior parietal gyrus | SPG.L |
| R angular gyrus | ANG.R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Wu, Y.; Sun, L.; Lu, Y.; Qian, K.; Kuang, H.; Meng, J.; Wu, Y. Abnormal Topological Organization of Structural Covariance Networks in Patients with Temporal Lobe Epilepsy Comorbid Sleep Disorder. Brain Sci. 2023, 13, 1493. https://doi.org/10.3390/brainsci13101493
Yang S, Wu Y, Sun L, Lu Y, Qian K, Kuang H, Meng J, Wu Y. Abnormal Topological Organization of Structural Covariance Networks in Patients with Temporal Lobe Epilepsy Comorbid Sleep Disorder. Brain Sciences. 2023; 13(10):1493. https://doi.org/10.3390/brainsci13101493
Chicago/Turabian StyleYang, Shengyu, Ying Wu, Lanfeng Sun, Yuling Lu, Kai Qian, Huimin Kuang, Jie Meng, and Yuan Wu. 2023. "Abnormal Topological Organization of Structural Covariance Networks in Patients with Temporal Lobe Epilepsy Comorbid Sleep Disorder" Brain Sciences 13, no. 10: 1493. https://doi.org/10.3390/brainsci13101493
APA StyleYang, S., Wu, Y., Sun, L., Lu, Y., Qian, K., Kuang, H., Meng, J., & Wu, Y. (2023). Abnormal Topological Organization of Structural Covariance Networks in Patients with Temporal Lobe Epilepsy Comorbid Sleep Disorder. Brain Sciences, 13(10), 1493. https://doi.org/10.3390/brainsci13101493






