Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Expression Analysis
2.2. Tumor Characteristics of TK1 Expression Heterogeneity
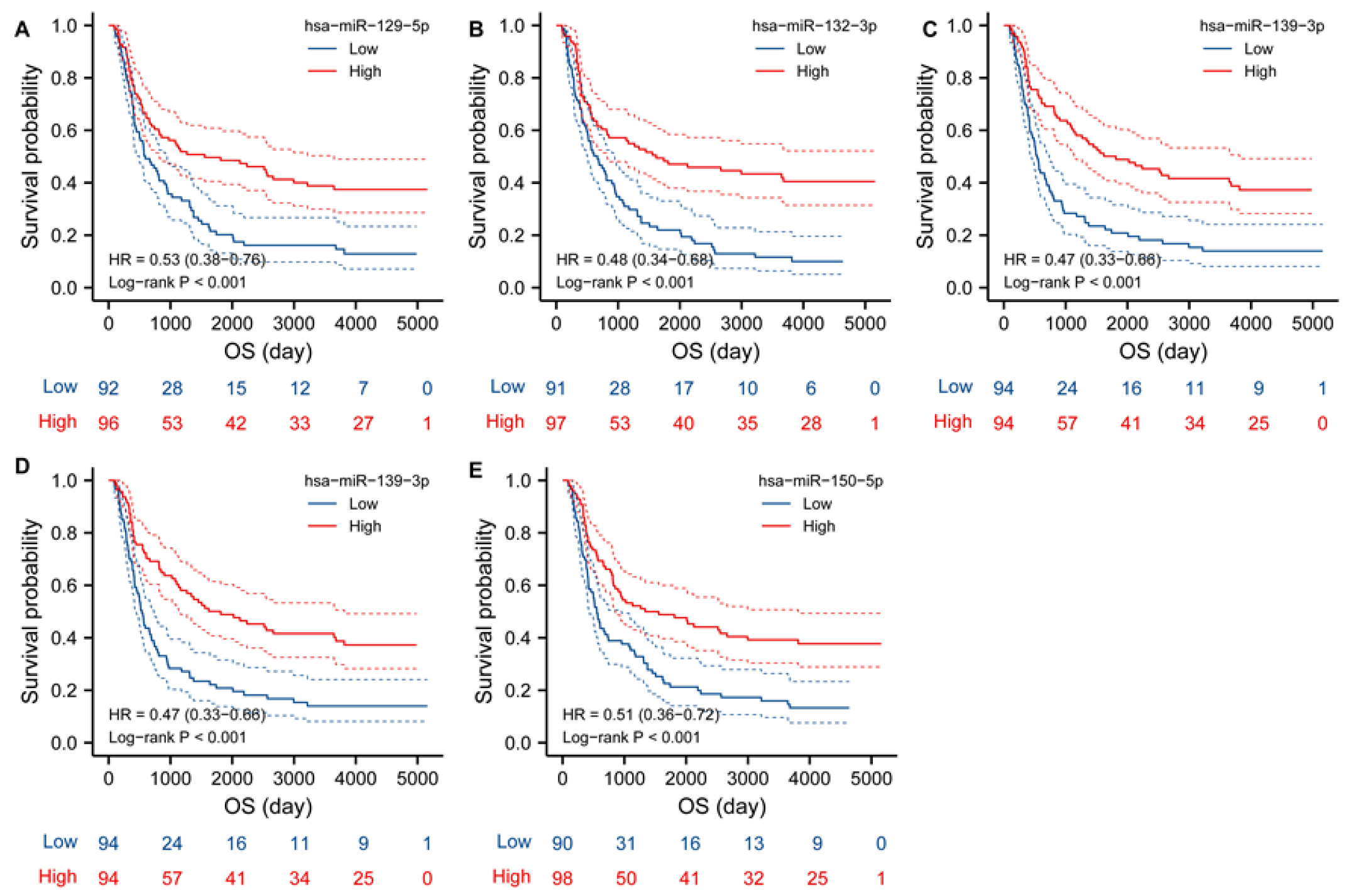
2.3. Prognostic Value of TK1 Expression in Glioma
2.4. Immune Associated Analysis
2.5. Multi-Omics Analysis
2.6. Gene Co-Expression and Gene Set Enrichment Analysis
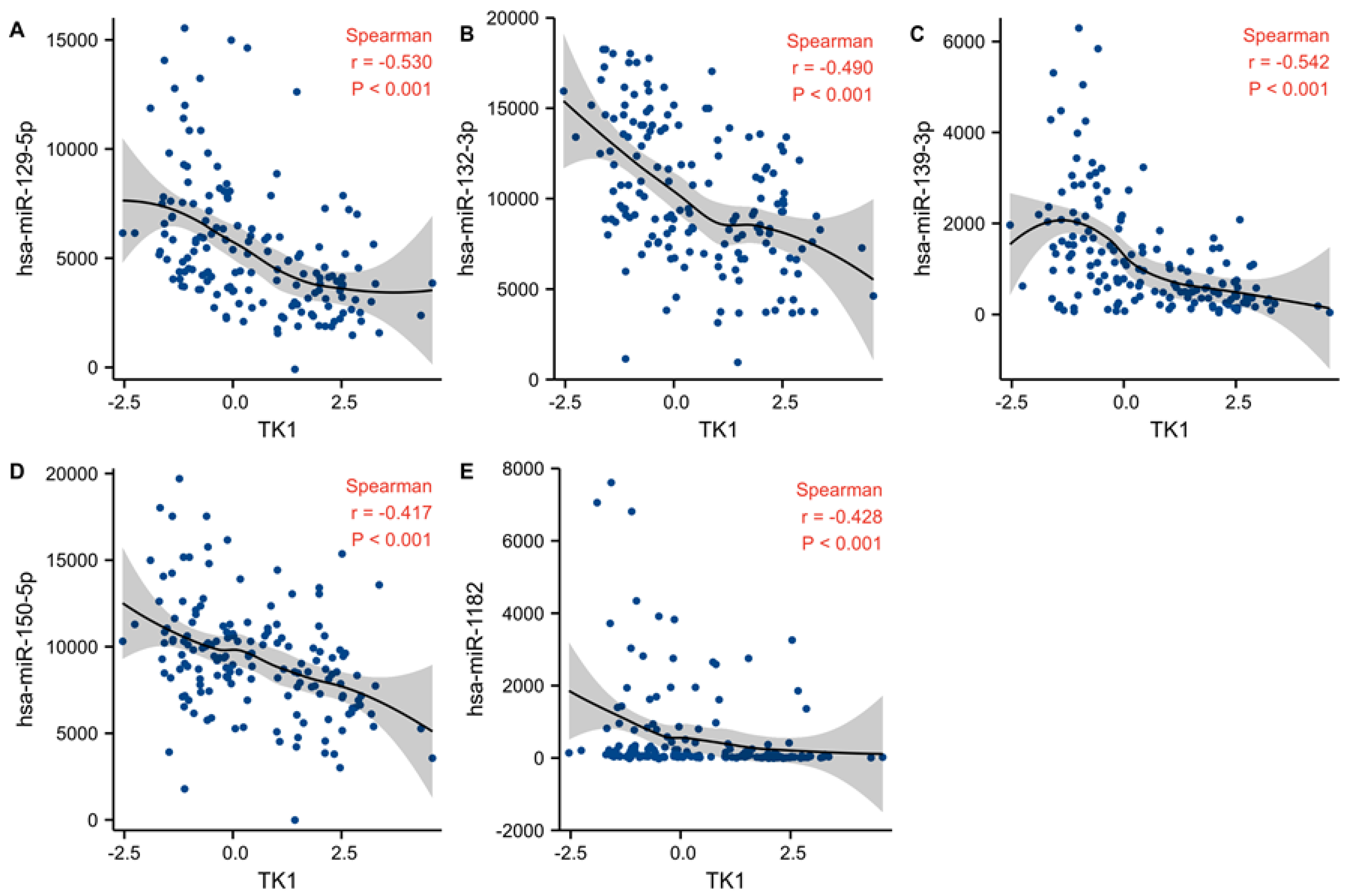
2.7. TK1 Associated miRNAs
2.8. Single-Cell Function Analysis
2.9. Cell Culture
2.10. Protein Expression
2.11. Statistical Analysis
3. Results
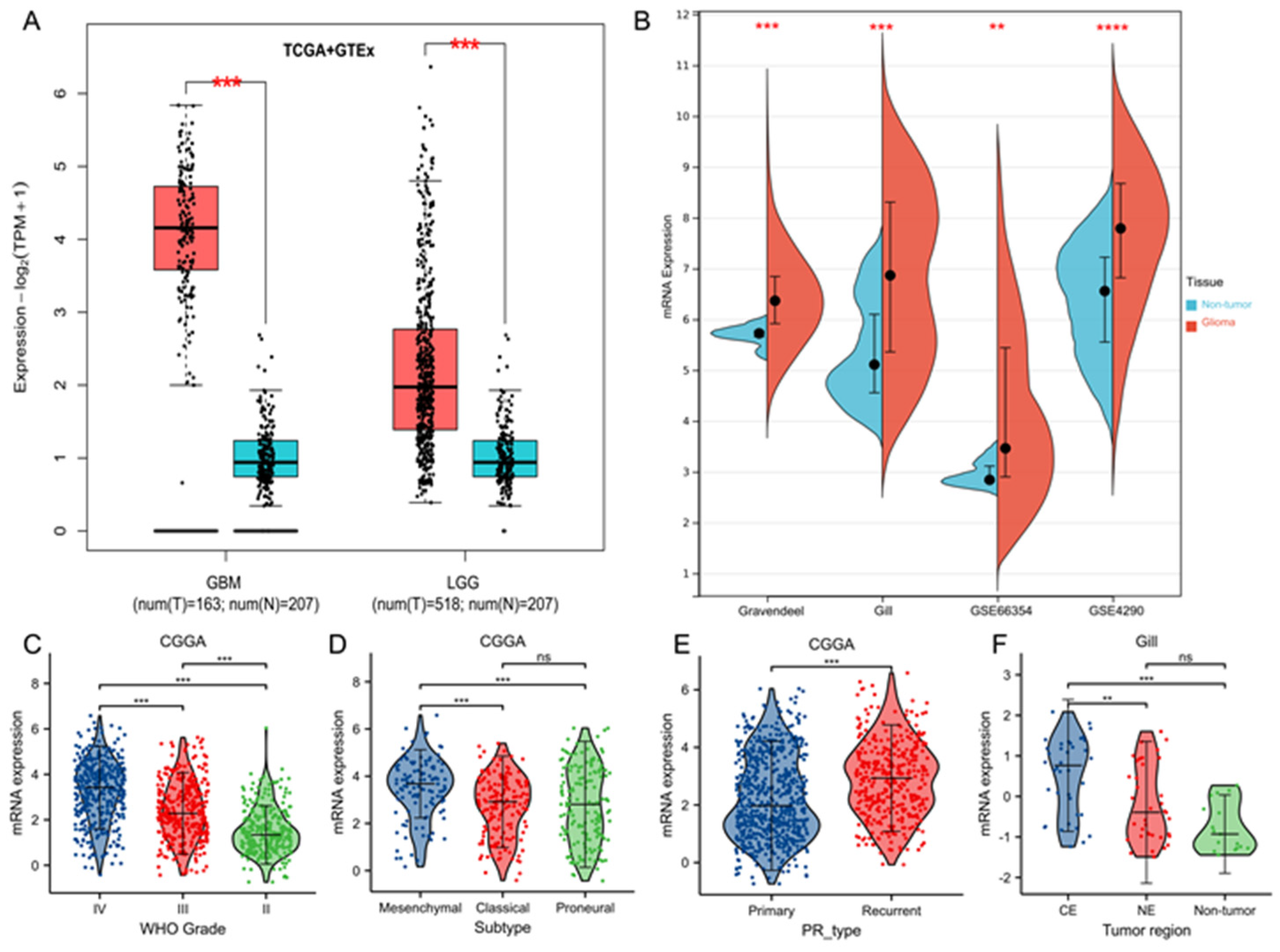
3.1. TK1 Overexpression in Glioma Tissues and Other Cancers Compared with Normal Tissues
3.2. Tumor Characteristics of TK1 Heterogeneity in Gliomas
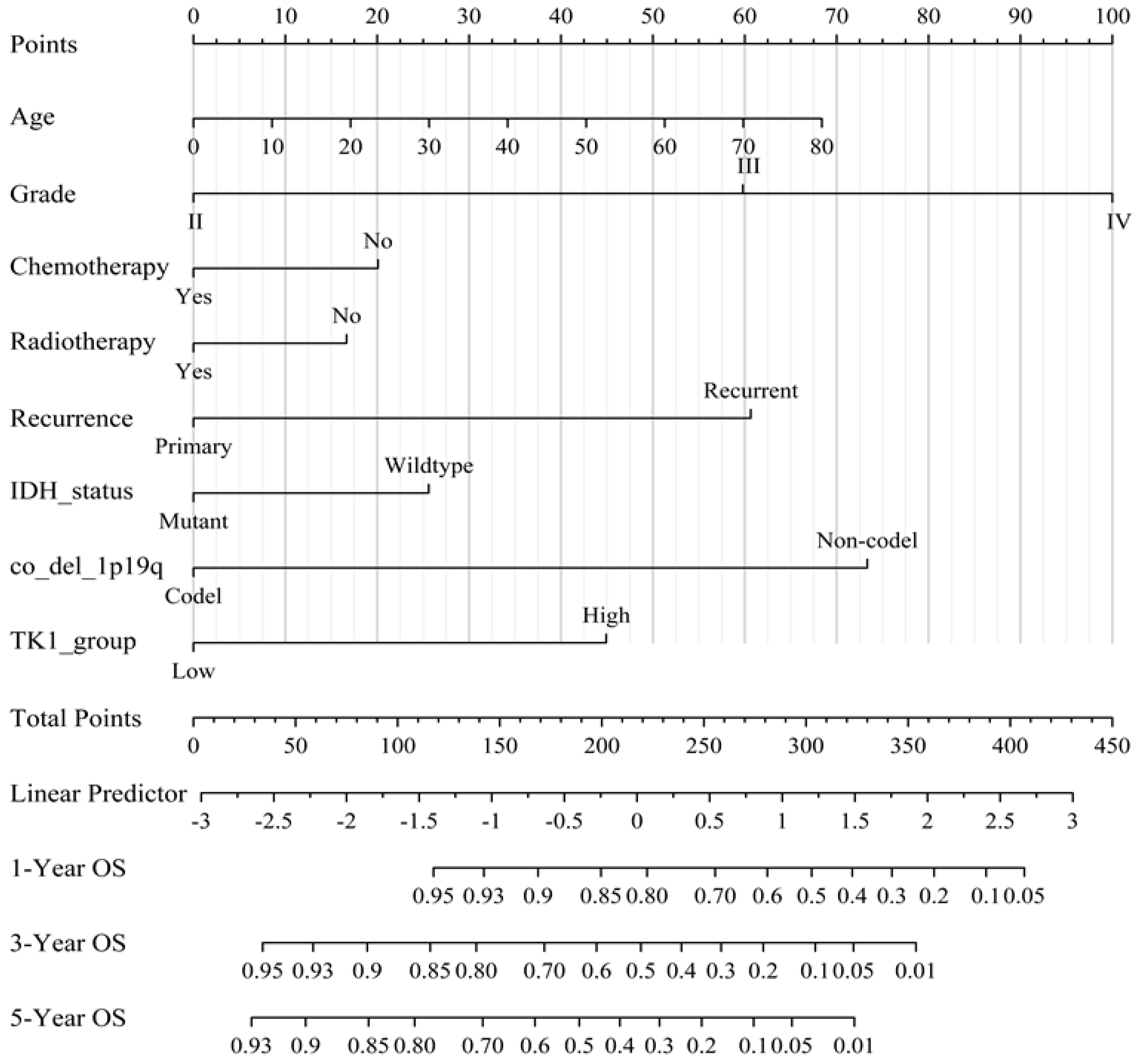
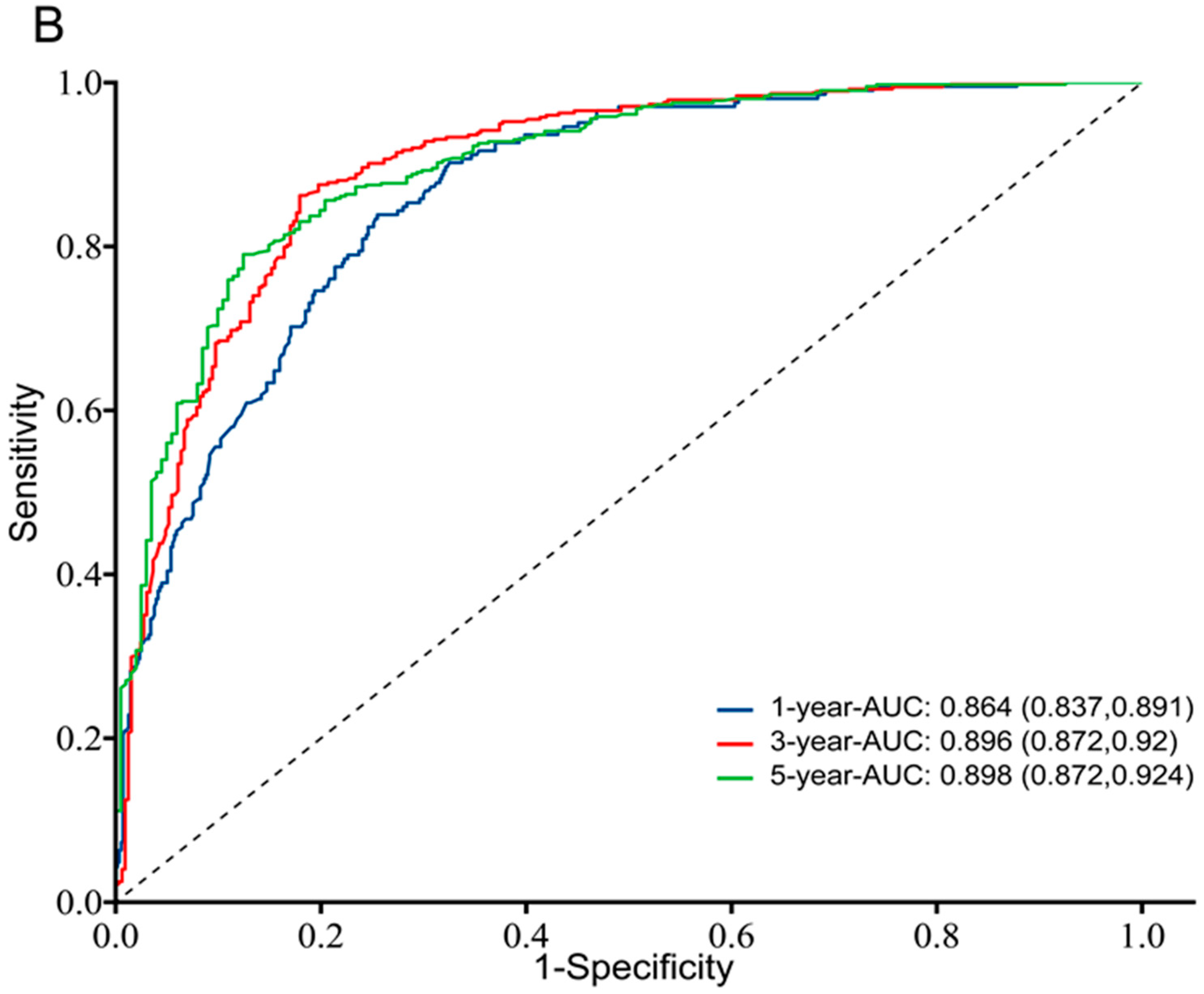
3.3. Prognostic Value of TK1 Expression in Glioma
3.4. Ascertainment of Protein Expression in Glioma Tissue, Glioma Cell Lines, and Other Cancers Compared with Normal Tissues
3.5. Immune Associated Analysis
3.6. Multi-Omics Analysis
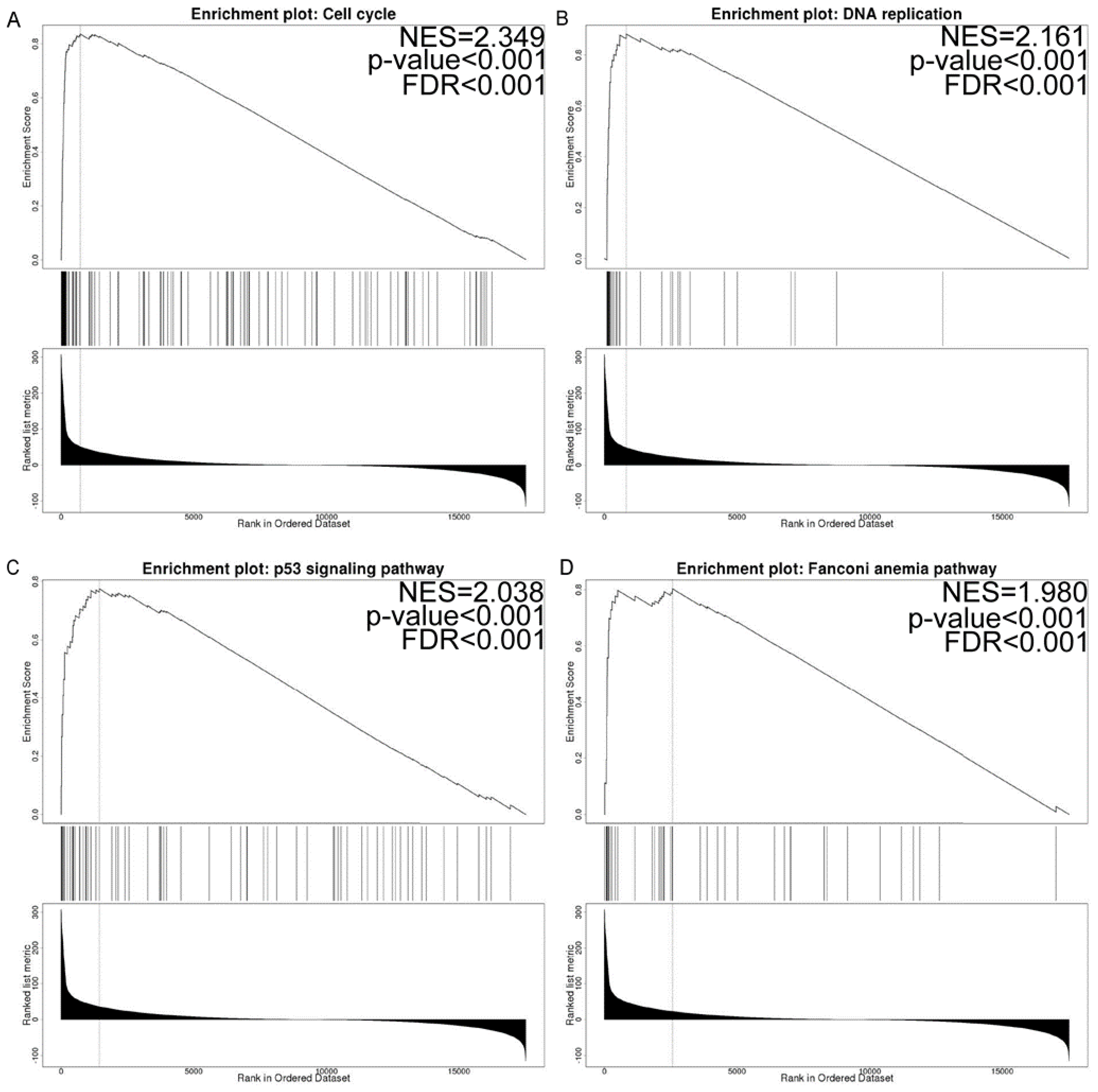
3.7. Co-Expressed Gene and Gene Set Enrichment Analysis
3.8. Candidate miRNA Prediction
3.9. Single-Cell Function Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Xu, S.; Tang, L.; Li, X.; Fan, F.; Liu, Z. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020, 476, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cabel, L.; Rosenblum, D.; Lerebours, F.; Brain, E.; Loirat, D.; Bergqvist, M.; Cottu, P.; Donnadieu, A.; Bethune, A.; Kiavue, N.; et al. Plasma thymidine kinase 1 activity and outcome of ER+ HER2- metastatic breast cancer patients treated with palbociclib and endocrine therapy. Breast Cancer Res. 2020, 22, 98. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, J.; Zhao, L.; He, H.; Zhao, P.; Peng, Z.; Liu, F.; Chen, J.; Wu, W.; Wang, G.; et al. Knockdown of Thymidine Kinase 1 Suppresses Cell Proliferation; Invasion; Migration; and Epithelial-Mesenchymal Transition in Thyroid Carcinoma Cells. Front. Oncol. 2020, 9, 1475. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Wang, H.; Li, L.; Pan, H.; Lu, L. Thymidine Kinase 1 Drives Skin Cutaneous Melanoma Malignant Progression and Metabolic Reprogramming. Front. Oncol. 2022, 12, 802807. [Google Scholar] [CrossRef]
- Zhu, X.; Shi, C.; Peng, Y.; Yin, L.; Tu, M.; Chen, Q.; Hou, C.; Li, Q.; Miao, Y. Thymidine kinase 1 silencing retards proliferative activity of pancreatic cancer cell via E2F1-TK1-P21 axis. Cell Prolif. 2018, 51, e12428. [Google Scholar] [CrossRef] [Green Version]
- Malvi, P.; Janostiak, R.; Nagarajan, A.; Cai, G.; Wajapeyee, N. Loss of thymidine kinase 1 inhibits lung cancer growth and metastatic attributes by reducing GDF15 expression. PLoS Genet. 2019, 15, e1008439. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Zhu, M.; Duan, J.; Wang, H.; Chen, J.; Xiao, Y.; Wang, Y.; Wang, J.; Yu, X.; Yang, H. Comprehensive Analysis of Immune-Related Prognosis of TK1 in Hepatocellular Carcinoma. Frontiers in oncology. Front. Oncol. 2022, 11, 786873. [Google Scholar] [CrossRef]
- Bitter, E.E.; Townsend, M.H.; Erickson, R.; Allen, C.; O’Neill, K.L. Thymidine kinase 1 through the ages: A comprehensive review. Cell Biosci. 2020, 10, 138. [Google Scholar] [CrossRef]
- Jagarlamudi, K.K.; Shaw, M. Thymidine kinase 1 as a tumor biomarker: Technical advances offer new potential to an old biomarker. Biomark. Med. 2018, 12, 1035–1048. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.; Zhou, X.; He, Y.; Guo, Q.; Shi, Q.; Eriksson, S.; Zhou, J.; He, E.; Skog, S. Thymidine kinase 1 expression in ovarian serous adenocarcinoma is superior to Ki-67: A new prognostic biomarker. Tumour Biol. 2017, 39, 1010428317706479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; He, C.; Li, L.; Lin, A.; Zheng, X.; He, E.; Skog, S. Nuclear TK1 expression is an independent prognostic factor for survival in pre-malignant and malignant lesions of the cervix. BMC Cancer 2013, 13, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gakis, G.; Hennenlotter, J.; Scharpf, M.; Hevler, J.; Schilling, D.; Kuehs, U.; Stenzl, A.; Schwentner, C. XPA-210: A new proliferation marker to characterize tumor biology and progression of renal cell carcinoma. World J. Urol. 2011, 29, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.; Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-Oncol. 2017, 19, 139–141. [Google Scholar] [CrossRef] [Green Version]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pandey, A.; Chinnaiyan, A.M. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gill, B.J.; Pisapia, D.J.; Malone, H.R.; Goldstein, H.; Lei, L.; Sonabend, A.; Yun, J.; Samanamud, J.; Sims, J.S.; Banu, M.; et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 12550–12555. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017, 45, W130–W137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Zhang, K.N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Wang, P.; Yan, Q.; Liao, B.; Zhao, L.; Xiong, S.; Wang, J.; Zou, D.; Pan, J.; Wu, L.; Deng, Y.; et al. The HIF1α/HIF2α-miR210-3p network regulates glioblastoma cell proliferation, dedifferentiation and chemoresistance through EGF under hypoxic conditions. Cell Death Dis. 2020, 11, 992. [Google Scholar] [CrossRef]
- Griesinger, A.M.; Josephson, R.J.; Donson, A.M.; Levy, J.M.M.; Amani, V.; Birks, D.K.; Hoffman, L.M.; Furtek, S.L.; Reigan, P.; Handler, M.H.; et al. Interleukin-6/STAT3 Pathway Signaling Drives an Inflammatory Phenotype in Group A Ependymoma. Cancer Immunol. Res. 2015, 3, 1165–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Hui, A.M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gravendeel, L.A.; Kouwenhoven, M.C.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.; Kloosterhof, N.K.; et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef] [Green Version]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, Y.; Zhao, Y.; Liu, T.; Wang, Z.; Zhang, N.; Dai, Z.; Wu, W.; Cao, H.; Feng, S.; et al. PTX3 mediates the infiltration, migration, and inflammation-resolving-polarization of macrophages in glioblastoma. CNS Neurosci. Ther. 2022, 28, 1748–1766. [Google Scholar] [CrossRef]
- Alshamrani, A.A. Roles of microRNAs in Ovarian Cancer Tumorigenesis: Two Decades Later, What Have We Learned? Front. Oncol. 2020, 10, 1084. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhu, W.; Jiang, J. miR-150-5p suppresses the stem cell-like characteristics of glioma cells by targeting the Wnt/β-catenin signaling pathway. Cell Biol. Int. 2020, 44, 1156–1167. [Google Scholar] [CrossRef]
- Tian, W.; Wu, W.; Li, X.; Rui, X.; Wu, Y. MiRNA-139-3p inhibits the proliferation, invasion, and migration of human glioma cells by targeting MDA-9/syntenin. Biochem. Biophys. Res. Commun. 2019, 508, 295–301. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Han, J.J.; Fan, S.Q.; Yang, W.; Zhang, Y.B.; Xu, T.J.; Xu, G.M. miR-132 weakens proliferation and invasion of glioma cells via the inhibition of Gli1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1971–1978. [Google Scholar] [CrossRef]
- Chiang, K.C.; Lai, C.Y.; Chiou, H.L.; Lin, C.L.; Chen, Y.S.; Kao, S.H.; Hsieh, Y.H. Timosaponin AIII inhibits metastasis of renal carcinoma cells through suppressing cathepsin C expression by AKT/miR-129-5p axis. J. Cell. Physiol. 2019, 234, 13332–13341. [Google Scholar] [CrossRef]
- Ersahin, T.; Tuncbag, N.; Cetin-Atalay, R. The PI3K/AKT/mTOR interactive pathway. Mol. Biosyst. 2015, 11, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dai, W.; Song, J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem. Biophys. Res. Commun. 2016, 470, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xiao, Y.F.; Zhang, J.W.; Xie, R.; Hu, C.J.; Tang, B.; Wang, S.M.; Wu, Y.Y.; Hao, N.B.; Yang, S.M. miR-1182 attenuates gastric cancer proliferation and metastasis by targeting the open reading frame of hTERT. Cancer Lett. 2015, 360, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiang, W.; Liu, T.; Lv, J.; Guan, J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem. Biophys. Res. Commun. 2019, 510, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiao, L.; Pan, D.; Hu, L. ASF1B enhances migration and invasion of lung cancers cell via regulating the P53-mediated epithelial-mesenchymal transformation (EMT) signaling pathway. Neoplasma 2022, 69, 361–369. [Google Scholar] [CrossRef]
- Jiang, X.; Shi, Y.; Chen, X.; Xu, H.; Huang, X.; Li, L.; Pu, J. The N6-Methylandenosine-Related Gene BIRC5 as a Prognostic Biomarker Correlated with Cell Migration and Immune Cell Infiltrates in Low Grade Glioma. Front. Mol. Biosci. 2022, 9, 773662. [Google Scholar] [CrossRef]
- Sun, H.; Yang, B.; Zhang, H.; Song, J.; Zhang, Y.; Xing, J.; Yang, Z.; Wei, C.; Xu, T.; Yu, Z.; et al. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int. J. Biol. Sci. 2019, 15, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Lei, Y.; Li, J.K.; Du, W.X.; Li, R.G.; Yang, J.; Li, J.; Li, F.; Tan, H.B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Dong, S.; Shen, J.; Yu, H.; Zhou, J.; Li, J.; Ma, H.; He, E.; Skog, S. Serum TK1 is a more reliable marker than CEA and AFP for cancer screening in a study of 56,286 people. Cancer Biomark. 2016, 16, 529–536. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.H.; Huang, S.Q.; Wang, Y.; Yang, A.Z.; Wen, J.; Xu, X.H.; Chen, Y.; Chen, Q.B.; Wang, Y.H.; He, E.; et al. Serological thymidine kinase 1 is a biomarker for early detection of tumours—A health screening study on 35,365 people, using a sensitive chemiluminescent dot blot assay. Sensors 2011, 11, 11064–11080. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, C.; Wang, P.; Liao, B.; Gong, S.; Wu, N. Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches. Brain Sci. 2023, 13, 230. https://doi.org/10.3390/brainsci13020230
Shao C, Wang P, Liao B, Gong S, Wu N. Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches. Brain Sciences. 2023; 13(2):230. https://doi.org/10.3390/brainsci13020230
Chicago/Turabian StyleShao, Chuan, Pan Wang, Bin Liao, Sheng Gong, and Nan Wu. 2023. "Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches" Brain Sciences 13, no. 2: 230. https://doi.org/10.3390/brainsci13020230
APA StyleShao, C., Wang, P., Liao, B., Gong, S., & Wu, N. (2023). Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches. Brain Sciences, 13(2), 230. https://doi.org/10.3390/brainsci13020230





