Detecting Cortical Thickness Changes in Epileptogenic Lesions Using Machine Learning
Abstract
1. Introduction
1.1. Logistic Regression (LR)
1.2. K-Nearest Neighbours (K-NN)
1.3. Support Vector Machine (SVM)
1.4. Naive Bayes
2. Literature Review
2.1. Morphometric Analysis Program (MAP) Strategies for FCD Detection
2.2. Temporal Lobe Epilepsy Applications Based on Machine Learning (ML)
2.2.1. Automated Detection of FCD Using ML
2.2.2. Automated Detection of MTS Using ML
2.3. Lesions Detection Based on Deep Learning (DL)
Automated Detection of FCD Using DL
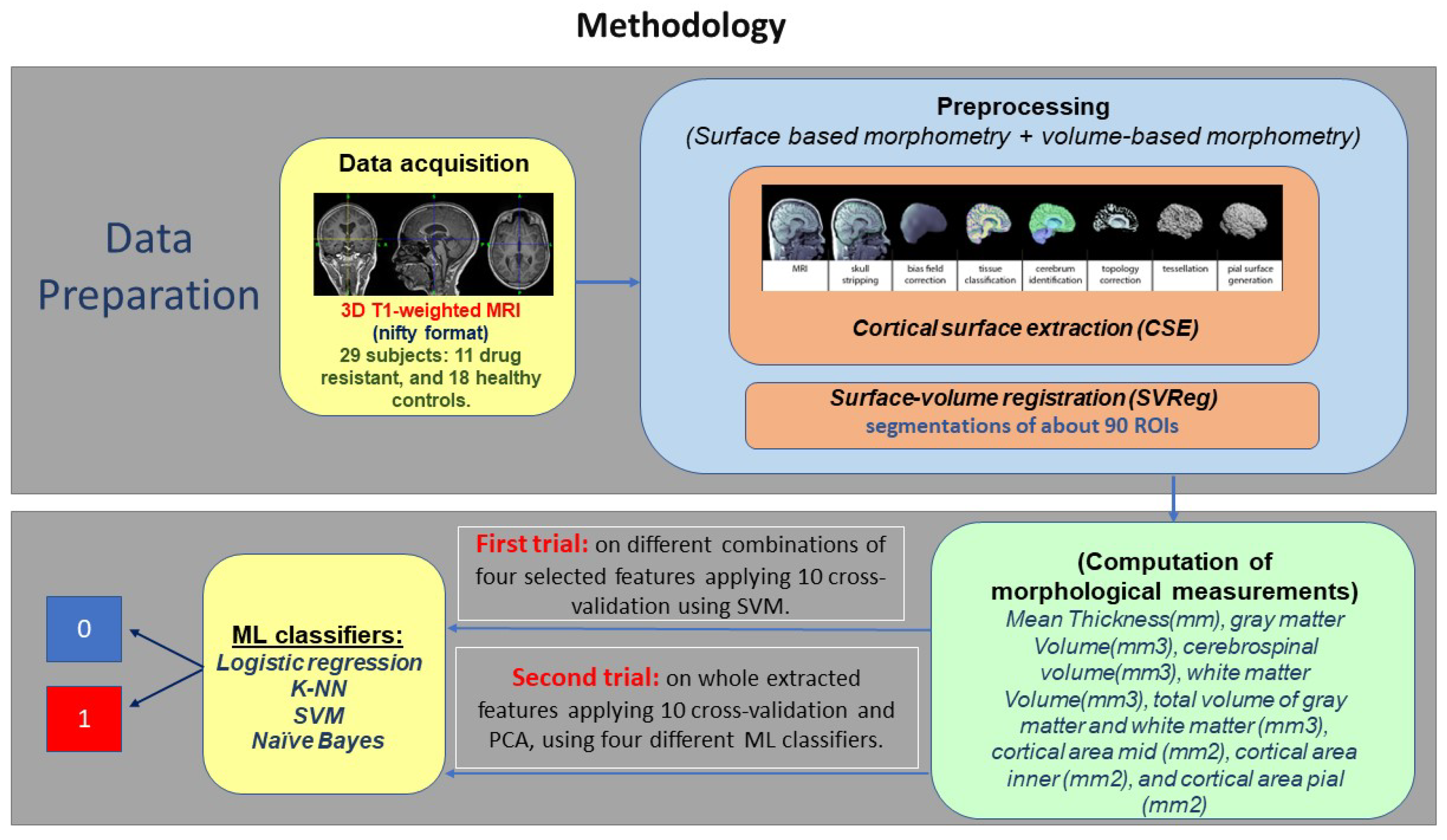
3. Materials and Methods
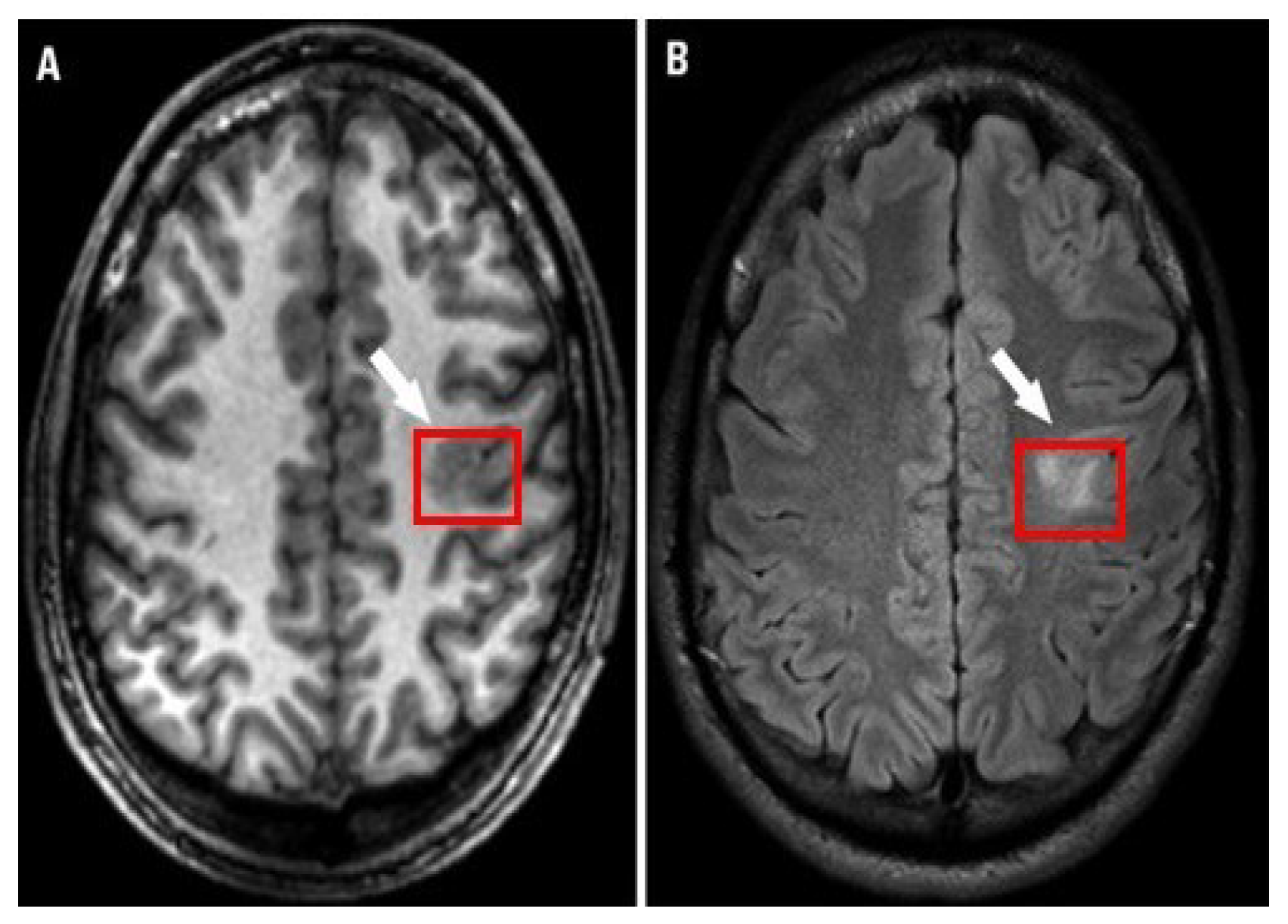
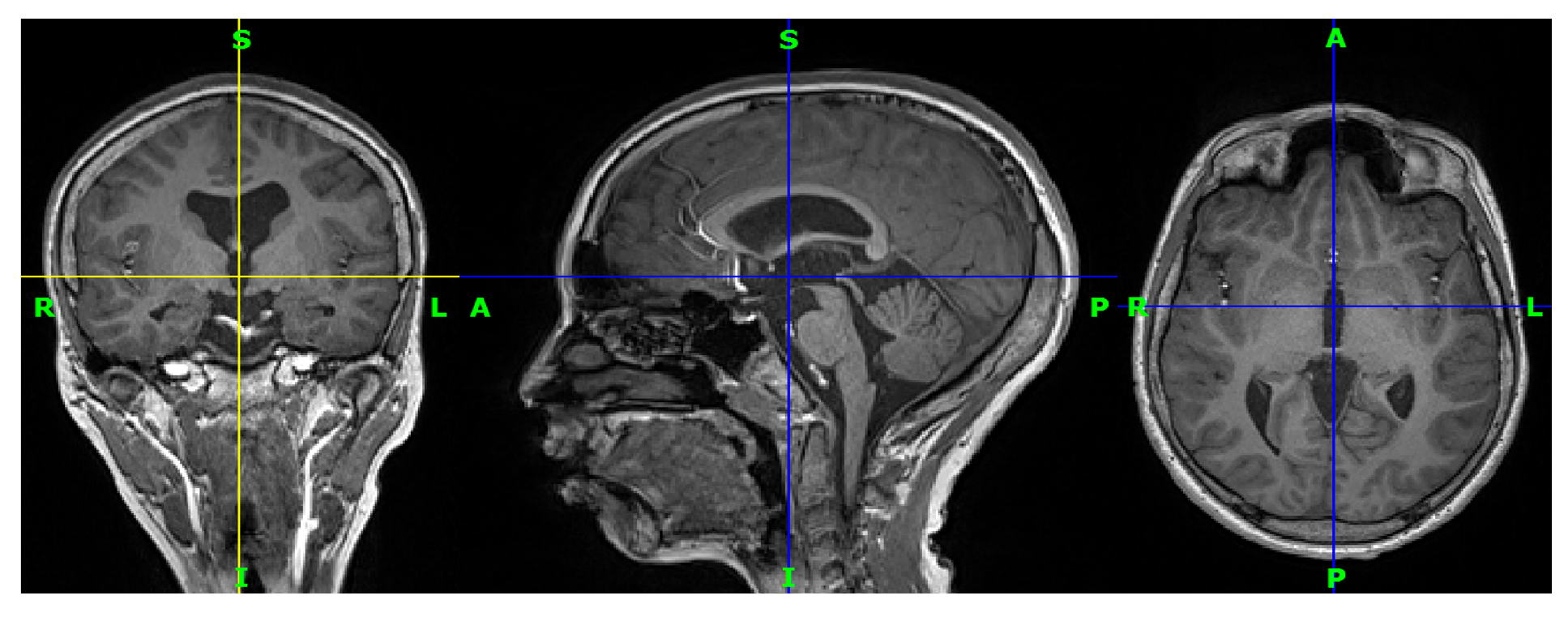
3.1. Data Acquisition
3.2. Preprocessing
3.3. Feature Extraction
3.4. ML Classifiers
3.4.1. First Trial on Four Commonly Selected Measurements
- Shuffle the data set at random.
- Sort the data into k groups.
- For every separate group:
- (a)
- As a holdout or test data set, use the group.
- (b)
- As a training data set, use the remaining groups.
- (c)
- Fit a model to the training data and test it on the test data.
- (d)
- Keep the evaluation score but throw out the model.
- Provide a brief summary of the model’s skill using the sample of model evaluation scores.
- C: a parameter for regularization.
- Kernel: a parameter that can be set to linear, rbf, or our own callable.
- Gamma: the rbf, poly, and sigmoid kernel parameter coefficient.
3.4.2. Second Trial on all Extracted Measurements
4. Results and Discussion
4.1. Evaluation Metrics
4.1.1. Accuracy
4.1.2. Precision
4.1.3. Recall
4.2. Results
5. Discussion
6. Conclusions
7. Recommendations and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barr, W.B.; Morrison, C. Handbook on the Neuropsychology of Epilepsy; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Kwan, P.; Brodie, M.J. Early identification of refractory epilepsy. N. Engl. J. Med. 2000, 342, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.T.; Vickrey, B.G.; Testa, F.M.; Levy, S.R.; Shinnar, S.; DiMario, F.; Smith, S. How long does it take for epilepsy to become intractable? A prospective investigation. Ann. Neurol. 2006, 60, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Blümcke, I.; Thom, M.; Aronica, E.; Armstrong, D.D.; Vinters, H.V.; Palmini, A.; Jacques, T.S.; Avanzini, G.; Barkovich, A.J.; Battaglia, G.; et al. The clinicopathologic spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc task force of the ILAE Diagnostic Methods Commission 1. Epilepsia 2011, 52, 158–174. [Google Scholar] [CrossRef] [PubMed]
- Tassi, L.; Colombo, N.; Garbelli, R.; Francione, S.; Lo Russo, G.; Mai, R.; Cardinale, F.; Cossu, M.; Ferrario, A.; Galli, C.; et al. Focal cortical dysplasia: Neuropathological subtypes, EEG, neuroimaging and surgical outcome. Brain 2002, 125, 1719–1732. [Google Scholar] [CrossRef]
- Taylor, D.; Falconer, M.; Bruton, C.; Corsellis, J. Focal dysplasia of the cerebral cortex in epilepsy. J. Neurol. Neurosurg. Psychiatry 1971, 34, 369–387. [Google Scholar] [CrossRef]
- Harvey, A.S.; Mandelstam, S.A.; Maixner, W.J.; Leventer, R.J.; Semmelroch, M.; MacGregor, D.; Kalnins, R.M.; Perchyonok, Y.; Fitt, G.J.; Barton, S.; et al. The surgically remediable syndrome of epilepsy associated with bottom-of-sulcus dysplasia. Neurology 2015, 84, 2021–2028. [Google Scholar] [CrossRef]
- Mellerio, C.; Labeyrie, M.A.; Chassoux, F.; Roca, P.; Alami, O.; Plat, M.; Naggara, O.; Devaux, B.; Meder, J.F.; Oppenheim, C. 3T MRI improves the detection of transmantle sign in type 2 focal cortical dysplasia. Epilepsia 2014, 55, 117–122. [Google Scholar] [CrossRef]
- Wagner, J.; Weber, B.; Urbach, H.; Elger, C.E.; Huppertz, H.J. Morphometric MRI analysis improves detection of focal cortical dysplasia type II. Brain 2011, 134, 2844–2854. [Google Scholar] [CrossRef]
- Andrade, C.S.; Leite, C.D.C. Malformations of cortical development: Current concepts and advanced neuroimaging review. Arq.-Neuro-Psiquiatr. 2011, 69, 130–138. [Google Scholar] [CrossRef]
- Wang, H.; Ahmed, S.N.; Mandai, M. Efficient detection of mesial temporal sclerosis using hippocampus and CSF features in MRI images. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Las Vegas, NV, USA, 4–7 March 2018; pp. 178–181. [Google Scholar]
- Barkovich, A.J.; Kuzniecky, R.I. Neuroimaging of focal malformations of cortical development. J. Clin. Neurophysiol. 1996, 13, 481–494. [Google Scholar] [CrossRef]
- Ganiler, O. Automated Detection of New Multiple Sclerosis Lesions in longitudinal Brain Magnetic Resonance Imaging. Ph.D. Thesis, Universitat de Girona, Girona, Spain, 2014. [Google Scholar]
- Wang, Z.I.; Alexopoulos, A.V.; Jones, S.E.; Jaisani, Z.; Najm, I.M.; Prayson, R.A. The pathology of magnetic-resonance-imaging-negative epilepsy. Mod. Pathol. 2013, 26, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Pail, M.; Mareček, R.; Hermanová, M.; Slaná, B.; Tyrlíková, I.; Kuba, R.; Brázdil, M. The role of voxel-based morphometry in the detection of cortical dysplasia within the temporal pole in patients with intractable mesial temporal lobe epilepsy. Epilepsia 2012, 53, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, H.J.; Grimm, C.; Fauser, S.; Kassubek, J.; Mader, I.; Hochmuth, A.; Spreer, J.; Schulze-Bonhage, A. Enhanced visualization of blurred gray–white matter junctions in focal cortical dysplasia by voxel-based 3D MRI analysis. Epilepsy Res. 2005, 67, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Wong-Kisiel, L.C.; Quiroga, D.F.T.; Kenney-Jung, D.L.; Witte, R.J.; Santana-Almansa, A.; Worrell, G.A.; Britton, J.; Brinkmann, B.H. Morphometric analysis on T1-weighted MRI complements visual MRI review in focal cortical dysplasia. Epilepsy Res. 2018, 140, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Dev, K.B.; Jogi, P.S.; Niyas, S.; Vinayagamani, S.; Kesavadas, C.; Rajan, J. Automatic detection and localization of focal cortical dysplasia lesions in MRI using fully convolutional neural network. Biomed. Signal Process. Control 2019, 52, 218–225. [Google Scholar]
- Wang, H.; Ahmed, S.N.; Mandal, M. Automated detection of focal cortical dysplasia using a deep convolutional neural network. Comput. Med. Imaging Graph. 2020, 79, 101662. [Google Scholar] [CrossRef]
- Fischl, B.; Dale, A.M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. USA 2000, 97, 11050–11055. [Google Scholar] [CrossRef]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 1999, 9, 179–194. [Google Scholar] [CrossRef]
- Kuperberg, G.R.; Broome, M.R.; McGuire, P.K.; David, A.S.; Eddy, M.; Ozawa, F.; Goff, D.; West, W.C.; Williams, S.C.; van der Kouwe, A.J.; et al. Regionally localized thinning of the cerebral cortex in schizophrenia. Arch. Gen. Psychiatry 2003, 60, 878–888. [Google Scholar] [CrossRef]
- Peterson, B.S.; Warner, V.; Bansal, R.; Zhu, H.; Hao, X.; Liu, J.; Durkin, K.; Adams, P.B.; Wickramaratne, P.; Weissman, M.M. Cortical thinning in persons at increased familial risk for major depression. Proc. Natl. Acad. Sci. USA 2009, 106, 6273–6278. [Google Scholar] [CrossRef]
- Westlye, L.T.; Walhovd, K.B.; Dale, A.M.; Espeseth, T.; Reinvang, I.; Raz, N.; Agartz, I.; Greve, D.N.; Fischl, B.; Fjell, A.M. Increased sensitivity to effects of normal aging and Alzheimer’s disease on cortical thickness by adjustment for local variability in gray/white contrast: A multi-sample MRI study. Neuroimage 2009, 47, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Frisoni, G.B.; Fox, N.C.; Jack, C.R.; Scheltens, P.; Thompson, P.M. The clinical use of structural MRI in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Lerch, J.P.; Evans, A.C. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage 2005, 24, 163–173. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, D.; Kabani, N.; Avis, D.; Evans, A.C. Automated 3-D extraction of inner and outer surfaces of cerebral cortex from MRI. NeuroImage 2000, 12, 340–356. [Google Scholar] [CrossRef]
- Salat, D.H.; Buckner, R.L.; Snyder, A.Z.; Greve, D.N.; Desikan, R.S.; Busa, E.; Morris, J.C.; Dale, A.M.; Fischl, B. Thinning of the cerebral cortex in aging. Cereb. Cortex 2004, 14, 721–730. [Google Scholar] [CrossRef]
- Koza, J.R.; Bennett, F.H.; Andre, D.; Keane, M.A. Automated design of both the topology and sizing of analog electrical circuits using genetic programming. In Artificial Intelligence in Design’96; Springer: Cham, Switzerland, 1996; pp. 151–170. [Google Scholar]
- Wright, R.E. Logistic Regression; American Psychological Association: Washington, DC, USA, 1995. [Google Scholar]
- Fix, E.; Hodges, J.L. Nonparametric Discrimination: Consistency Properties; Randolph Field, Texas, Project; California University Berkeley: Berkeley, CA, USA, 1951. [Google Scholar]
- Altman, N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Vishwanathan, S.; Murty, M.N. SSVM: A simple SVM algorithm. In Proceedings of the 2002 International Joint Conference on Neural Networks—IJCNN’02 (Cat. No. 02CH37290), Honolulu, HI, USA, 12–17 May 2002; Volume 3, pp. 2393–2398. [Google Scholar]
- Gurboga, S.; Sarp, G. Time Dependency Analysis of Automatic Lineament Extraction. Adv. Biomed. Eng. (ABE) 2012, 14, 405–409. [Google Scholar]
- Piryonesi, S.M.; El-Diraby, T.E. Role of data analytics in infrastructure asset management: Overcoming data size and quality problems. J. Transp. Eng. Part B Pavements 2020, 146, 04020022. [Google Scholar] [CrossRef]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach; Pearson Education, Inc.: Upper Saddle River, NJ, USA, 2002. [Google Scholar]
- Abbasi, B.; Goldenholz, D.M. Machine learning applications in epilepsy. Epilepsia 2019, 60, 2037–2047. [Google Scholar] [CrossRef]
- Wang, Z.I.; Jones, S.E.; Jaisani, Z.; Najm, I.M.; Prayson, R.A.; Burgess, R.C.; Krishnan, B.; Ristic, A.; Wong, C.H.; Bingaman, W.; et al. Voxel-based morphometric magnetic resonance imaging (MRI) postprocessing in MRI-negative epilepsies. Ann. Neurol. 2015, 77, 1060–1075. [Google Scholar] [CrossRef]
- Demerath, T.; Rubensdörfer, L.; Schwarzwald, R.; Schulze-Bonhage, A.; Altenmüller, D.M.; Kaller, C.; Kober, T.; Huppertz, H.J.; Urbach, H. Morphometric MRI analysis: Improved detection of focal cortical dysplasia using the MP2RAGE sequence. Am. J. Neuroradiol. 2020, 41, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- David, B.; Kröll-Seger, J.; Schuch, F.; Wagner, J.; Wellmer, J.; Woermann, F.; Oehl, B.; Van Paesschen, W.; Breyer, T.; Becker, A.; et al. External validation of automated focal cortical dysplasia detection using morphometric analysis. Epilepsia 2021, 62, 1005–1021. [Google Scholar] [CrossRef] [PubMed]
- Del Gaizo, J.; Mofrad, N.; Jensen, J.H.; Clark, D.; Glenn, R.; Helpern, J.; Bonilha, L. Using machine learning to classify temporal lobe epilepsy based on diffusion MRI. Brain Behav. 2017, 7, e00801. [Google Scholar] [CrossRef] [PubMed]
- Davoodi-Bojd, E.; Elisevich, K.V.; Schwalb, J.; Air, E.; Soltanian-Zadeh, H. TLE lateralization using whole brain structural connectivity. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1103–1106. [Google Scholar]
- Kamiya, K.; Amemiya, S.; Suzuki, Y.; Kunii, N.; Kawai, K.; Mori, H.; Kunimatsu, A.; Saito, N.; Aoki, S.; Ohtomo, K. Machine learning of DTI structural brain connectomes for lateralization of temporal lobe epilepsy. Magn. Reson. Med. Sci. 2015, 15, 121–129. [Google Scholar] [CrossRef]
- Sahebzamani, G.; Saffar, M.; Soltanian-Zadeh, H. Machine learning based analysis of structural MRI for epilepsy diagnosis. In Proceedings of the 2019 4th International Conference on Pattern Recognition and Image Analysis (IPRIA), Tehran, Iran, 6–7 March 2019; pp. 58–63. [Google Scholar]
- Ganji, Z.; Hakak, M.A.; Zamanpour, S.A.; Zare, H. Automatic Detection of Focal Cortical Dysplasia Type II in MRI: Is the Application of Surface-Based Morphometry and Machine Learning Promising? Front. Hum. Neurosci. 2021, 15, 608285. [Google Scholar] [CrossRef]
- Feng, C.; Zhao, H.; Tian, M.; Lu, M.; Wen, J. Detecting focal cortical dysplasia lesions from FLAIR-negative images based on cortical thickness. Biomed. Eng. Online 2020, 19, 13. [Google Scholar] [CrossRef]
- Gill, R.S.; Hong, S.J.; Fadaie, F.; Caldairou, B.; Bernhardt, B.; Bernasconi, N.; Bernasconi, A. Automated detection of epileptogenic cortical malformations using multimodal MRI. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2017; pp. 349–356. [Google Scholar]
- El Azami, M.; Hammers, A.; Jung, J.; Costes, N.; Bouet, R.; Lartizien, C. Detection of lesions underlying intractable epilepsy on T1-weighted MRI as an outlier detection problem. PLoS ONE 2016, 11, e0161498. [Google Scholar] [CrossRef]
- Jin, B.; Krishnan, B.; Adler, S.; Wagstyl, K.; Hu, W.; Jones, S.; Najm, I.; Alexopoulos, A.; Zhang, K.; Zhang, J.; et al. Automated detection of focal cortical dysplasia type II with surface-based magnetic resonance imaging postprocessing and machine learning. Epilepsia 2018, 59, 982–992. [Google Scholar] [CrossRef]
- Ahmed, B.; Brodley, C.E.; Blackmon, K.E.; Kuzniecky, R.; Barash, G.; Carlson, C.; Quinn, B.T.; Doyle, W.; French, J.; Devinsky, O.; et al. Cortical feature analysis and machine learning improves detection of “MRI-negative” focal cortical dysplasia. Epilepsy Behav. 2015, 48, 21–28. [Google Scholar] [CrossRef]
- Adler, S.; Wagstyl, K.; Gunny, R.; Ronan, L.; Carmichael, D.; Cross, J.H.; Fletcher, P.C.; Baldeweg, T. Novel surface features for automated detection of focal cortical dysplasias in paediatric epilepsy. NeuroImage Clin. 2017, 14, 18–27. [Google Scholar] [CrossRef]
- Mo, J.J.; Zhang, J.G.; Li, W.L.; Chen, C.; Zhou, N.J.; Hu, W.H.; Zhang, C.; Wang, Y.; Wang, X.; Liu, C.; et al. Clinical value of machine learning in the automated detection of focal cortical dysplasia using quantitative multimodal surface-based features. Front. Neurosci. 2019, 12, 1008. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Chung, C.K. Electrophysiological resting-state biomarker for diagnosing mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsy Res. 2017, 129, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Rudie, J.D.; Colby, J.B.; Salamon, N. Machine learning classification of mesial temporal sclerosis in epilepsy patients. Epilepsy Res. 2015, 117, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Qu, G.; Yuan, Q. Epileptogenic region detection based on deep CNN with transfer learning. In Proceedings of the 2019 IEEE 11th International Conference on Advanced Infocomm Technology (ICAIT), Jinan, China, 18–20 October 2019; pp. 73–77. [Google Scholar]
- Chen, X.; Qian, T.; Maréchal, B.; Zhang, G.; Yu, T.; Ren, Z.; Ni, D.; Liu, C.; Fu, Y.; Chen, N.; et al. Quantitative volume-based morphometry in focal cortical dysplasia: A pilot study for lesion localization at the individual level. Eur. J. Radiol. 2018, 105, 240–245. [Google Scholar] [CrossRef]
- Aliev, R.; Kondrateva, E.; Sharaev, M.; Bronov, O.; Marinets, A.; Subbotin, S.; Bernstein, A.; Burnaev, E. Convolutional neural networks for automatic detection of Focal Cortical Dysplasia. In Proceedings of the International Conference on Cognitive Sciences, Moscow, Russia, 10–16 October 2020; Springer: Cham, Switzerland, 2020; pp. 582–588. [Google Scholar]
- Feng, C.; Zhao, H.; Li, Y.; Wen, J. Automatic localization and segmentation of focal cortical dysplasia in FLAIR-negative patients using a convolutional neural network. J. Appl. Clin. Med. Phys. 2020, 21, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Shattuck, D.W.; Leahy, R.M. BrainSuite: An automated cortical surface identification tool. Med. Image Anal. 2002, 6, 129–142. [Google Scholar] [CrossRef]
- Prieto, P.B. Las 5 Diferencias Entre Cerebro y Cerebelo. Available online: https://medicoplus.com/neurologia/diferencias-cerebro-cerebelo (accessed on 16 January 2023).
- Brownlee, J. Machine Learning Mastery with Weka; 2019; Volume 1. Available online: https://machinelearningmastery.com/machine-learning-mastery-weka/ (accessed on 16 January 2023).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Geron, A. Hands-On Machine Learning with Scikit-Learn & Tensorflow; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2017; p. 564. [Google Scholar]


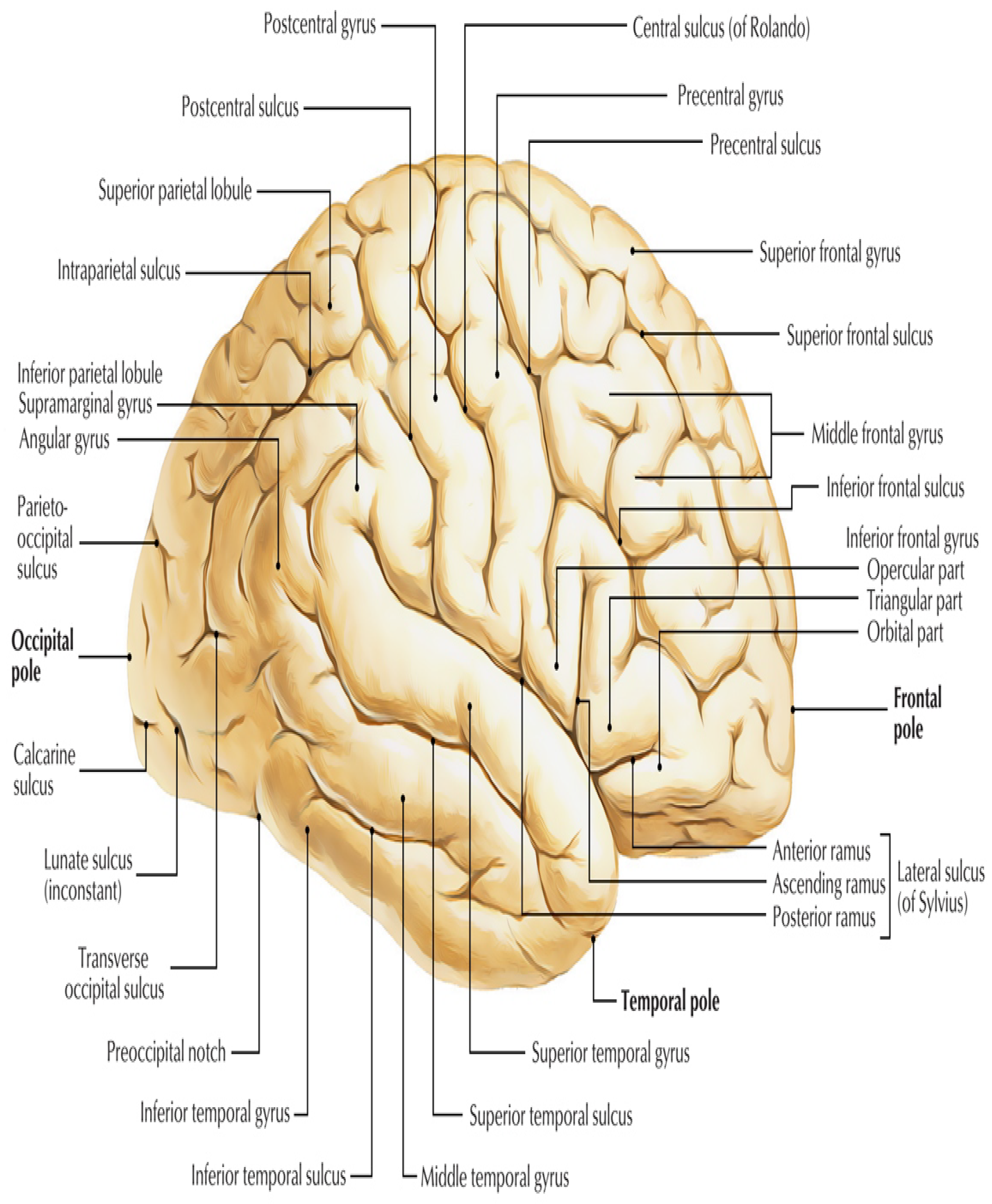
| ROI_ID | Name of Region | ROI_ID | Name of Region | ROI | Name of Region |
|---|---|---|---|---|---|
| 120 | R. superior frontal gyrus | 226 | R. angular gyrus | 500 | R. Insula |
| 121 | L. superior frontal gyrus | 227 | L. angular gyrus | 501 | L. Insula |
| 130 | R. middle frontal gyrus | 228 | R. superior parietal gyrus | 612 | R. caudate nucleus |
| 131 | L. middle frontal gyrus | 229 | L. superior parietal gyrus | 613 | L. caudate nucleus |
| 142 | R. pars opercularis | 242 | R. pre-cuneus | 614 | R. putamen |
| 143 | L. pars opercularis | 243 | L. pre-cuneus | 615 | L. putamen |
| 144 | R. pars triangularis | 310 | R. temporal pole | 616 | R. globus pallidus |
| 145 | L. pars triangularis | 311 | L. temporal pole | 617 | L. globus pallidus |
| 146 | R. pars orbitalis | 322 | R. superior temporal gyrus | 620 | R. nucleus accumbens |
| 147 | L. pars orbitalis | 323 | L. superior temporal gyrus | 621 | L. nucleus accumbens |
| 150 | R. pre-central gyrus | 324 | R. transverse temporal gyrus | 630 | R. claustrum |
| 151 | L. pre-central gyrus | 325 | L. transverse temporal gyrus | 631 | L. claustrum |
| 162 | R. transvers frontal gyrus | 326 | R. middle temporal gyrus | 640 | R. thalamus |
| 163 | L. transvers frontal gyrus | 327 | L. middle temporal gyrus | 641 | L. thalamus |
| 164 | R. gyrus rectus | 328 | R. inferior temporal gyrus | 650 | R. basal forebrain |
| 165 | L. gyrus rectus | 329 | L. inferior temporal gyrus | 651 | L. basal forebrain |
| 166 | R. middle orbito-frontal gyrus | 330 | R. fusiforme gyrus | 660 | R. lateral geniculate nucleus |
| 167 | L. middle orbito-frontal gyrus | 331 | L. fusiforme gyrus | 661 | L. lateral geniculate nucleus |
| 168 | R. anterior orbito-frontal gyrus | 342 | R. parahippocampal gyrus | 662 | R. medial geniculate nucleus |
| 169 | L. anterior orbito-frontal gyrus | 343 | L. parahippocampal gyrus | 663 | L. medial geniculate nucleus |
| 170 | R. posterior orbito-frontal gyrus | 344 | R. hippocampus | 670 | R. superior colliculus |
| 171 | L. posterior orbito-frontal gyrus | 345 | L. hippocampus | 671 | L. superior colliculus |
| 172 | R. lateral orbitofrontal gyrus | 346 | R. amygdala | 680 | R. inferior colliculus |
| 173 | L. lateral orbitofrontal gyrus | 347 | L. amygdala | 681 | L. inferior colliculus |
| 182 | R. paracentral lobule | 422 | R. superior occipital gyrus | 690 | R. mamillary body |
| 183 | L. paracentral lobule | 423 | L. superior occipital gyrus | 691 | L. mamillary body |
| 184 | R. cingulate gyrus | 424 | R. middle occipital gyrus | 701 | L. Ventricular System |
| 185 | L. cingulate gyrus | 425 | L. middle occipital gyrus | 720 | R. lateral ventricle |
| 186 | R. subcallosal gyrus | 442 | R. inferior occipital gyrus | 740 | third ventricle |
| 187 | L. subcallosal gyrus | 443 | L. inferior occipital gyrus | 800 | Brainstem |
| 222 | R. post-central gyrus | 444 | R. lingual gyrus | 900 | Cerebellum |
| 223 | L. post-central gyrus | 445 | L. lingual gyrus | 2000 | White matter (cerebrum) |
| 224 | R. supramarginal gyrus | 446 | R. cuneus | ||
| 225 | L. supramarginal gyrus | 447 | L. cuneus |
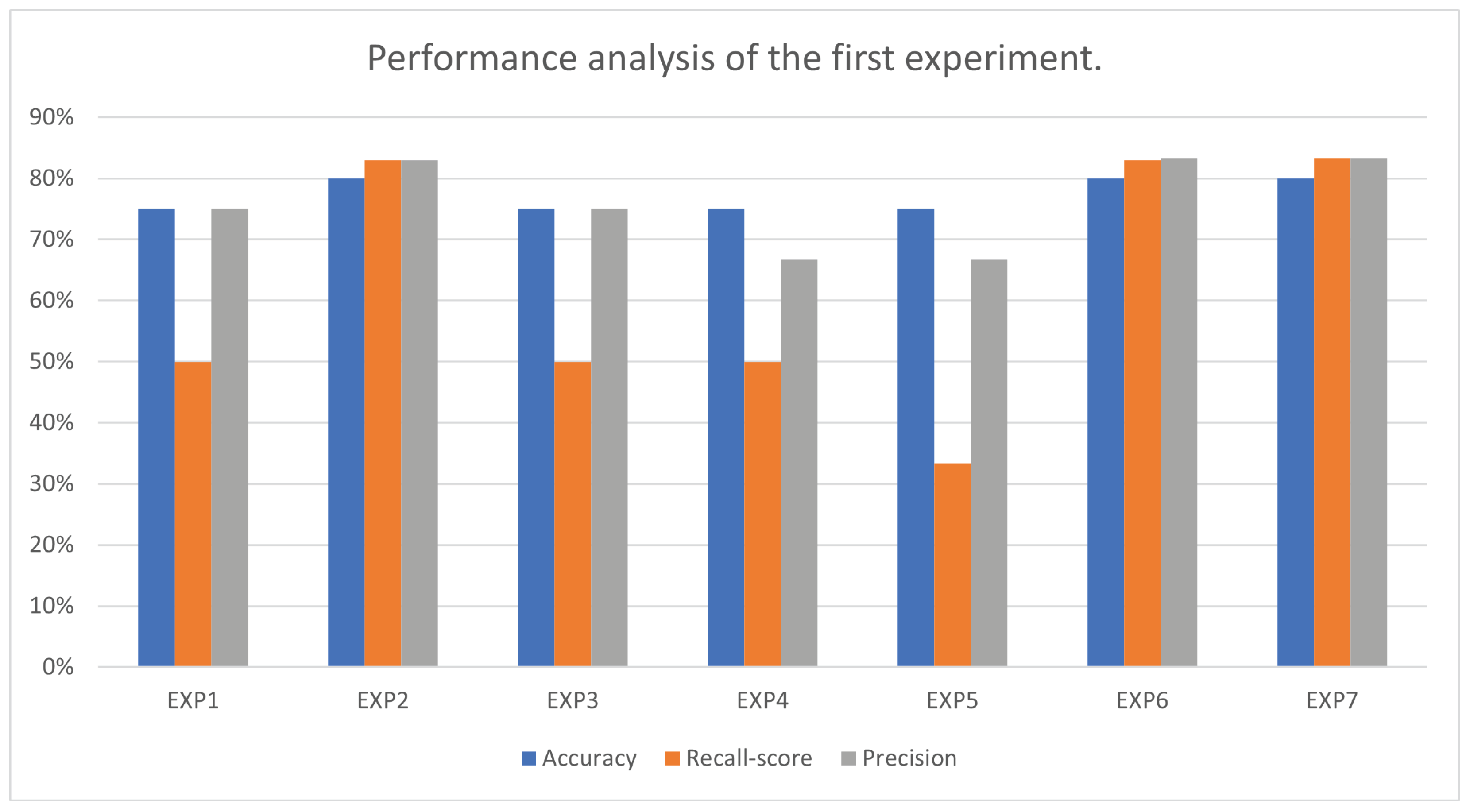
| Experiment No. | Accuracy | Best Parameters for Accuracy | Recall Score | Precision |
|---|---|---|---|---|
| EXP1 | 75.00% | ‘C’: 0.25 ‘kernel’: ‘linear’ | 50.00% | 75.00% |
| EXP2 | 80.00% | ‘C’: 0.25 ‘kernel’: ‘linear’ | 83.00% | 83.00% |
| EXP3 | 75.00% | ‘C’: 0.25 ‘gamma’: 0.1 ‘kernel’: ‘rbf’ | 50.00% | 75.00% |
| EXP4 | 75.00% | ‘C’: 0.25 ‘kernel’: ‘linear’ | 50.00% | 66.66% |
| EXP5 | 75.00% | ‘C’: 0.25 ‘gamma’: 0.1 ‘kernel’: ‘rbf’ | 33.33% | 66.66% |
| EXP6 | 80.00% | ‘C’: 0.25 ‘kernel’: ‘linear’ | 83.33% | 83.33% |
| EXP7 | 80.00% | ‘C’: 0.25 ‘kernel’: ‘linear’ | 83.33% | 83.33% |
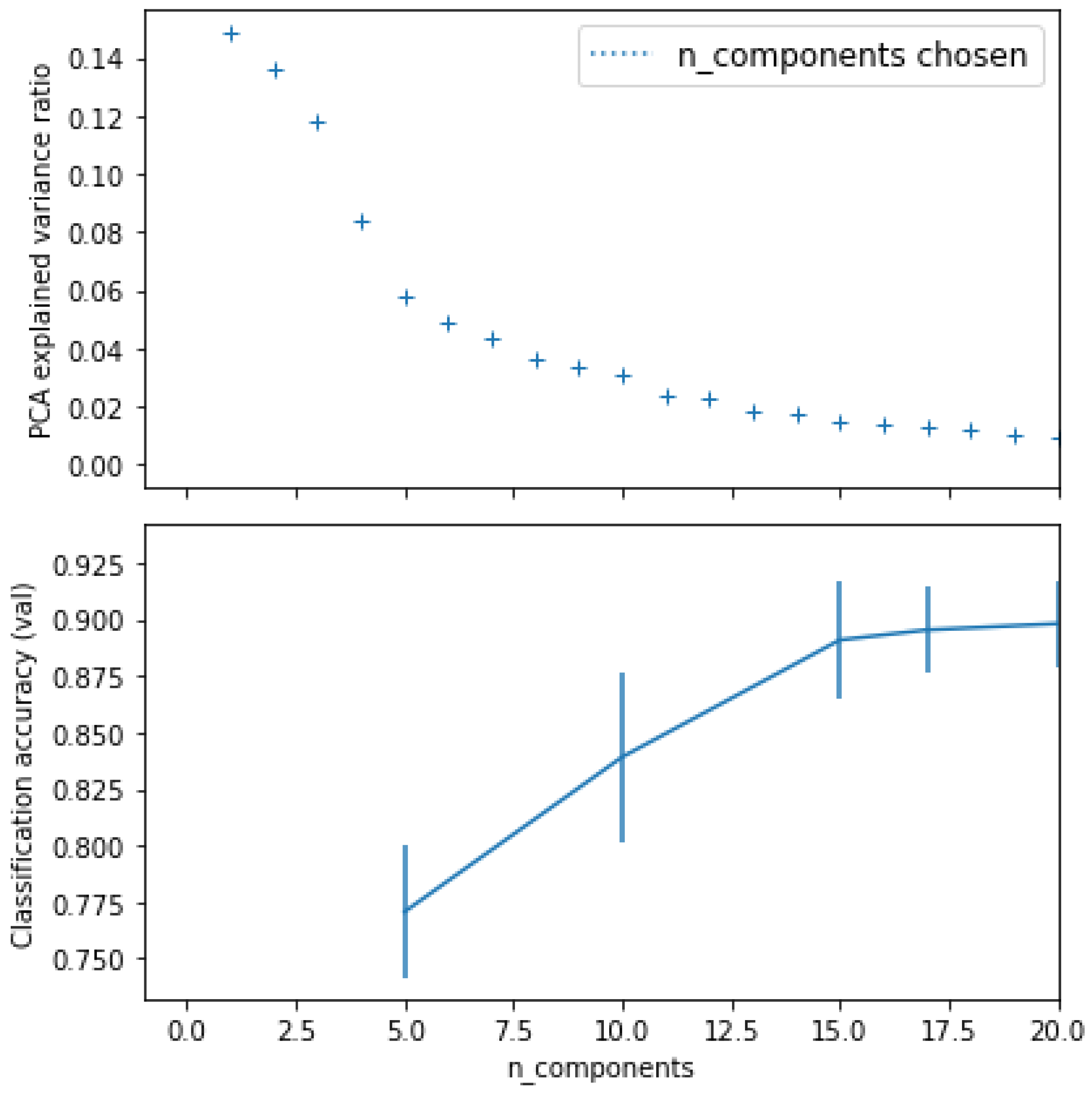
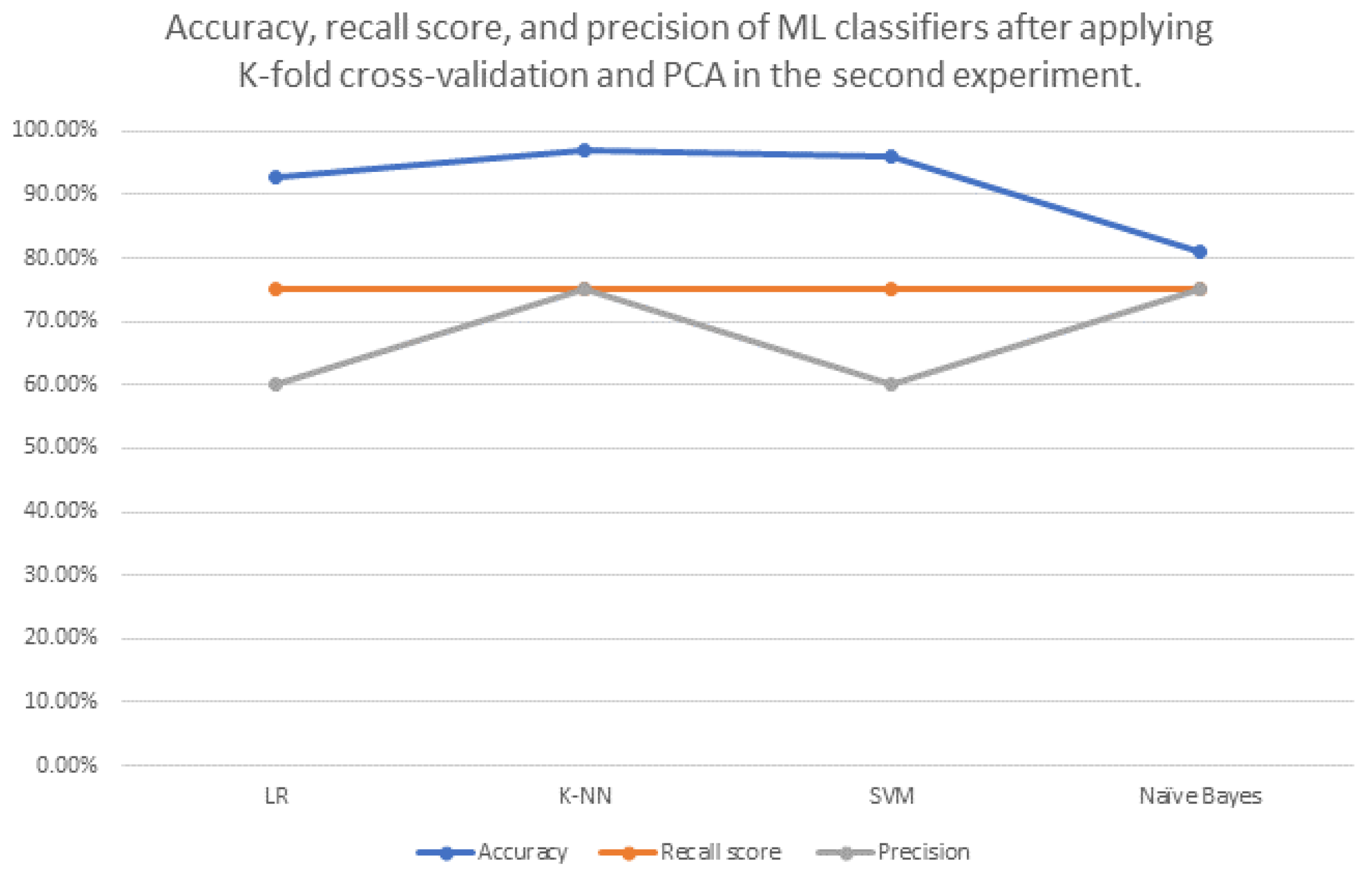
| ML Classifier Name | Best Parameters for Accuracy (If Exist) | Accuracy before Applying K-Fold Cross Validation and PCA | Accuracy after Applying K-Fold Cross Validation Only | Accuracy after Applying PCA Only | Accuracy after Applying K-Fold Cross Validation and PCA | Recall Score | Precis Ion |
|---|---|---|---|---|---|---|---|
| Logistic Regression (LR) | - | 66.67% | 90.00% | (n_components = 20) 66.67% | 92.82% | 75.00% | 60.00% |
| K-Nearest Neighbours (K-NN) | metric: ‘minkowski’, and n_neighbours = 5 | 77.78% | 55.00% | (n_components = 20) 77.78% | 97.11% | 75.00% | 75.00% |
| Support vector machine (SVM) | C = 0.25, and kernel: ‘linear’ | 66.67% | 90.00% | (n_components = 20) 66.67% | 96.05% | 75.00% | 60.00% |
| Naive Bayes “GaussianNB” | - | 77.78% | 70.00% | (n_components = 20) 77.78% | 81.14% | 75.00% | 75.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzony, S.; Moria, K.; Alghamdi, J. Detecting Cortical Thickness Changes in Epileptogenic Lesions Using Machine Learning. Brain Sci. 2023, 13, 487. https://doi.org/10.3390/brainsci13030487
Azzony S, Moria K, Alghamdi J. Detecting Cortical Thickness Changes in Epileptogenic Lesions Using Machine Learning. Brain Sciences. 2023; 13(3):487. https://doi.org/10.3390/brainsci13030487
Chicago/Turabian StyleAzzony, Sumayya, Kawthar Moria, and Jamaan Alghamdi. 2023. "Detecting Cortical Thickness Changes in Epileptogenic Lesions Using Machine Learning" Brain Sciences 13, no. 3: 487. https://doi.org/10.3390/brainsci13030487
APA StyleAzzony, S., Moria, K., & Alghamdi, J. (2023). Detecting Cortical Thickness Changes in Epileptogenic Lesions Using Machine Learning. Brain Sciences, 13(3), 487. https://doi.org/10.3390/brainsci13030487






