Machine Learning Models to Forecast Outcomes of Pituitary Surgery: A Systematic Review in Quality of Reporting and Current Evidence
Abstract
1. Introduction
2. Materials and Methods
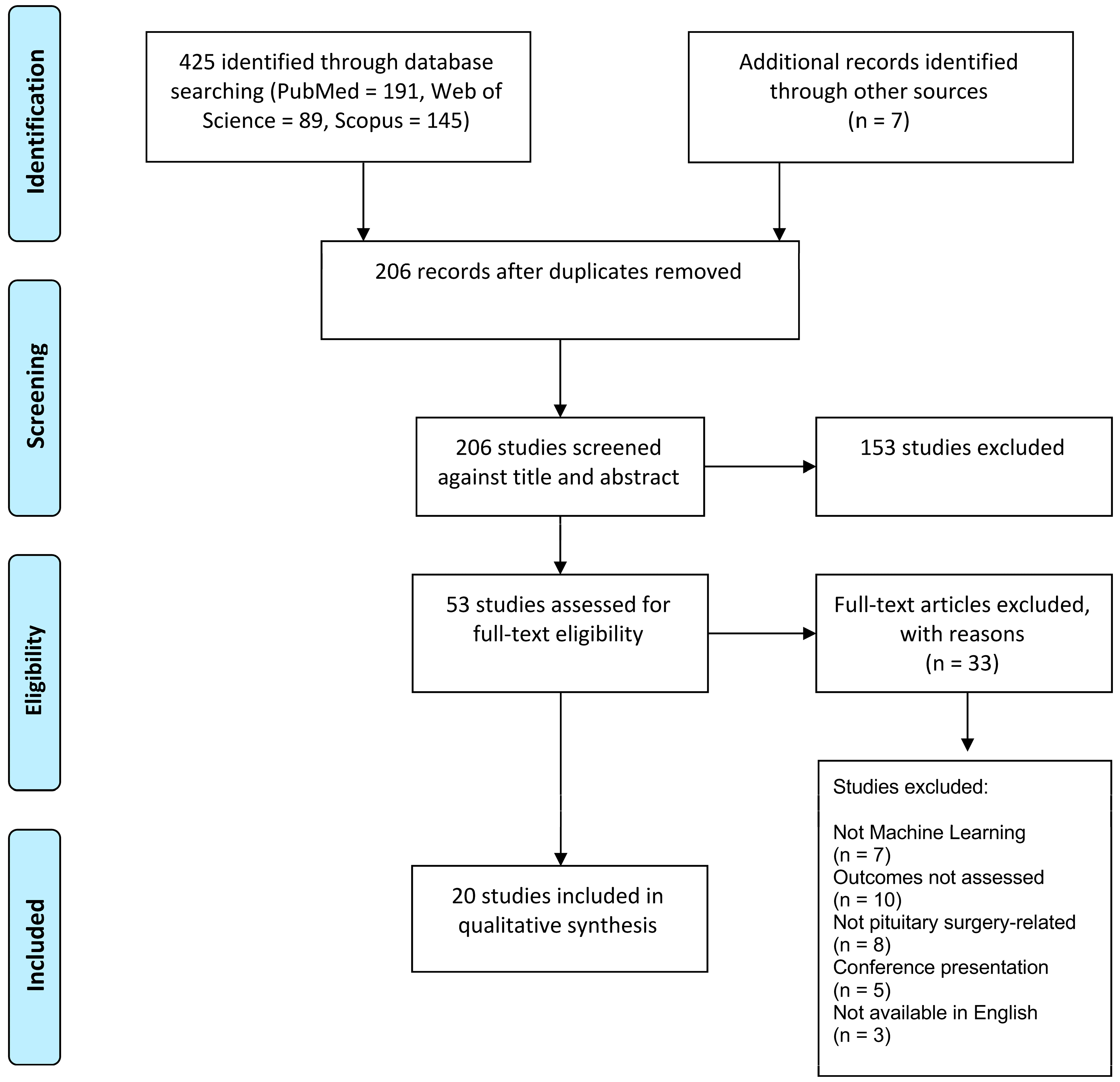
2.1. Literature Search and Studies Selection
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
2.4. Report Assessment
3. Results
3.1. Study Selection
3.2. Characteristics of Included Studies
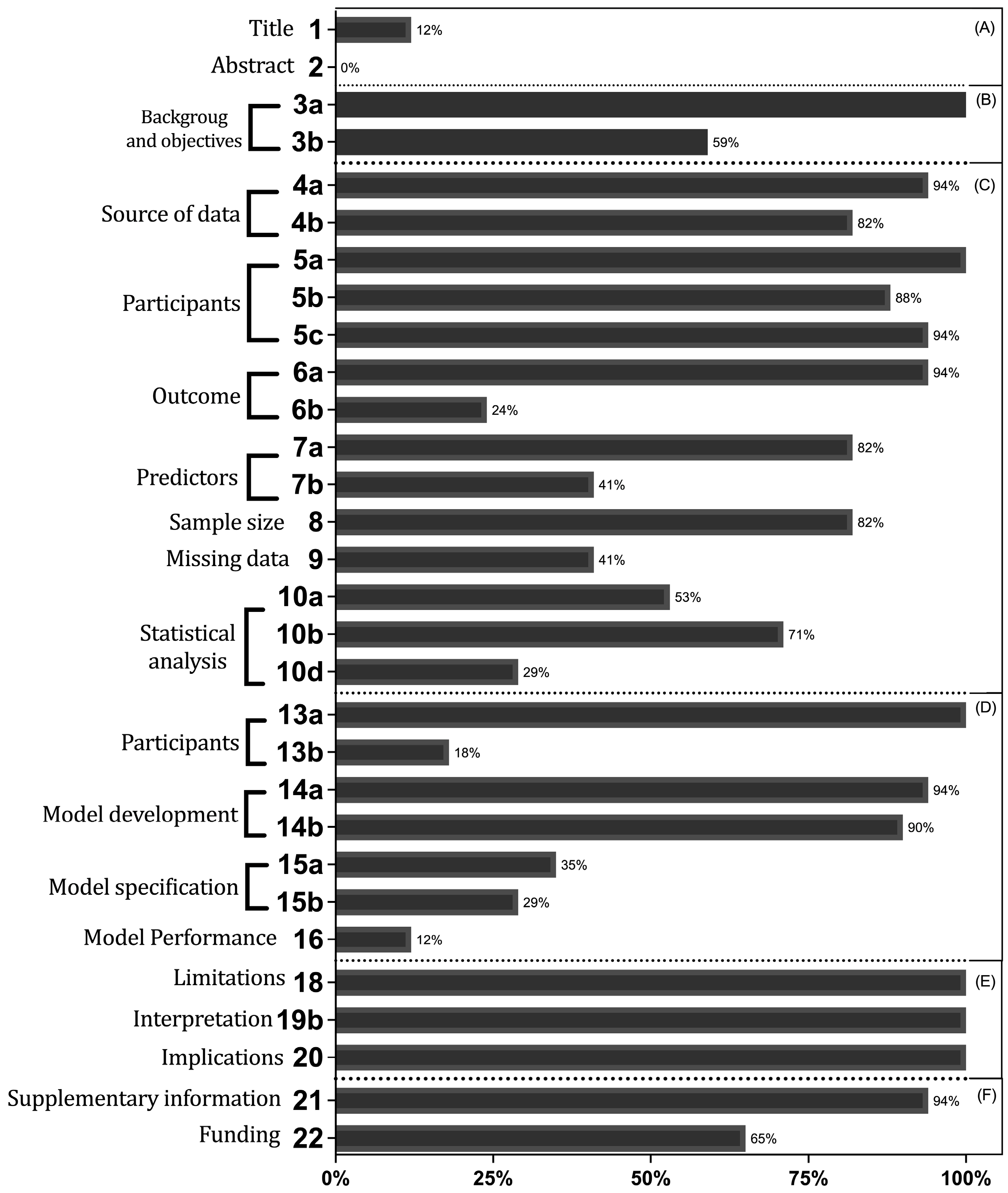
3.3. Report Assessment
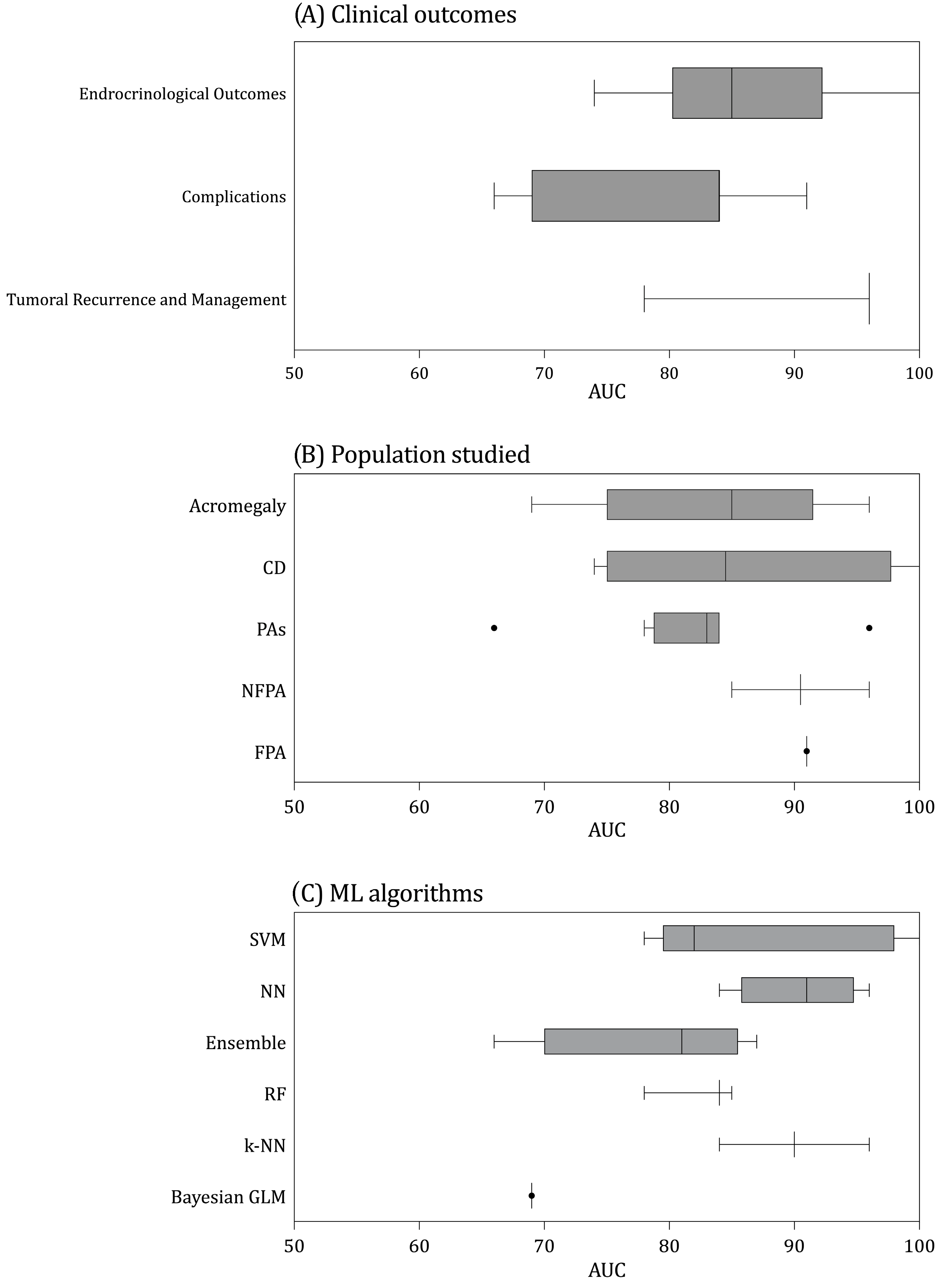
3.4. Models’ Assessment
3.5. Clinical Outcomes Predicted
3.5.1. Tumor Management and Recurrence
3.5.2. Endocrine Outcomes
3.5.3. Intra- and Post-Operative Complications
4. Discussion
4.1. Clinical Findings
4.2. Report Assessment
4.3. ML versus Traditional Statistical Methods
4.4. ML-Specific Reporting Guidelines
4.5. Future Perspectives
4.6. Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sivakumar, W.; Chamoun, R.; Nguyen, V.; Couldwell, W.T. Incidental Pituitary Adenomas. Neurosurg. Focus 2011, 31, E18. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, S.; Asa, S.L.; Couldwell, W.T.; Barr, C.E.; Dodge, W.E.; Vance, M.L.; McCutcheon, I.E. The Prevalence of Pituitary Adenomas: A Systematic Review. Cancer 2004, 101, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Liu, W.; Cao, P.; Zheng, Y.; Bu, Z.; Zhou, T. Endoscopic versus Microscopic Transsphenoidal Surgery in the Treatment of Pituitary Adenoma: A Systematic Review and Meta-Analysis. World Neurosurg. 2017, 101, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Gondim, J.A.; Almeida, J.P.C.; Albuquerque, L.A.F.; Schops, M.; Gomes, E.; Ferraz, T.; Sobreira, W.; Kretzmann, M.T. Endoscopic Endonasal Approach for Pituitary Adenoma: Surgical Complications in 301 Patients. Pituitary 2011, 14, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Gondim, J.A.; Schops, M.; de Almeida, J.P.C.; de Albuquerque, L.A.F.; Gomes, E.; Ferraz, T.; Barroso, F.A.C. Endoscopic Endonasal Transsphenoidal Surgery: Surgical Results of 228 Pituitary Adenomas Treated in a Pituitary Center. Pituitary 2010, 13, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.P.; Ruiz-Treviño, A.S.; Liang, B.; Omay, S.B.; Shetty, S.R.; Chen, Y.-N.; Anand, V.K.; Grover, K.; Christos, P.; Schwartz, T.H. Reoperation for Growth Hormone-Secreting Pituitary Adenomas: Report on an Endonasal Endoscopic Series with a Systematic Review and Meta-Analysis of the Literature. J. Neurosurg. 2018, 129, 404–416. [Google Scholar] [CrossRef]
- Asha, M.J.; Takami, H.; Velasquez, C.; Oswari, S.; Almeida, J.P.; Zadeh, G.; Gentili, F. Long-Term Outcomes of Transsphenoidal Surgery for Management of Growth Hormone-Secreting Adenomas: Single-Center Results. J. Neurosurg. 2019, 133, 1360–1370. [Google Scholar] [CrossRef]
- Johnson, K.W.; Torres Soto, J.; Glicksberg, B.S.; Shameer, K.; Miotto, R.; Ali, M.; Ashley, E.; Dudley, J.T. Artificial Intelligence in Cardiology. J. Am. Coll. Cardiol. 2018, 71, 2668–2679. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P. Artificial Intelligence in Nephrology: Core Concepts, Clinical Applications, and Perspectives. Am. J. Kidney Dis. 2019, 74, 803–810. [Google Scholar] [CrossRef]
- Hogarty, D.T.; Mackey, D.A.; Hewitt, A.W. Current State and Future Prospects of Artificial Intelligence in Ophthalmology: A Review. Clin. Experiment. Ophthalmol. 2019, 47, 128–139. [Google Scholar] [CrossRef]
- Shah, N.D.; Steyerberg, E.W.; Kent, D.M. Big Data and Predictive Analytics: Recalibrating Expectations. JAMA 2018, 320, 27–28. [Google Scholar] [CrossRef]
- Wilkinson, J.; Arnold, K.F.; Murray, E.J.; van Smeden, M.; Carr, K.; Sippy, R.; de Kamps, M.; Beam, A.; Konigorski, S.; Lippert, C.; et al. Time to Reality Check the Promises of Machine Learning-Powered Precision Medicine. Lancet Digit. Health 2020, 2, e677–e680. [Google Scholar] [CrossRef]
- Macyszyn, L.; Akbari, H.; Pisapia, J.M.; Da, X.; Attiah, M.; Pigrish, V.; Bi, Y.; Pal, S.; Davuluri, R.V.; Roccograndi, L.; et al. Imaging Patterns Predict Patient Survival and Molecular Subtype in Glioblastoma via Machine Learning Techniques. Neuro Oncol. 2016, 18, 417–425. [Google Scholar] [CrossRef]
- Oermann, E.K.; Rubinsteyn, A.; Ding, D.; Mascitelli, J.; Starke, R.M.; Bederson, J.B.; Kano, H.; Lunsford, L.D.; Sheehan, J.P.; Hammerbacher, J.; et al. Using a Machine Learning Approach to Predict Outcomes after Radiosurgery for Cerebral Arteriovenous Malformations. Sci. Rep. 2016, 6, 21161. [Google Scholar] [CrossRef]
- Abouzari, M.; Rashidi, A.; Zandi-Toghani, M.; Behzadi, M.; Asadollahi, M. Chronic Subdural Hematoma Outcome Prediction Using Logistic Regression and an Artificial Neural Network. Neurosurg. Rev. 2009, 32, 479–484. [Google Scholar] [CrossRef]
- Senders, J.T.; Arnaout, O.; Karhade, A.V.; Dasenbrock, H.H.; Gormley, W.B.; Broekman, M.L.; Smith, T.R. Natural and Artificial Intelligence in Neurosurgery: A Systematic Review. Neurosurgery 2018, 83, 181–192. [Google Scholar] [CrossRef]
- Saha, A.; Tso, S.; Rabski, J.; Sadeghian, A.; Cusimano, M.D. Machine Learning Applications in Imaging Analysis for Patients with Pituitary Tumors: A Review of the Current Literature and Future Directions. Pituitary 2020, 23, 273–293. [Google Scholar] [CrossRef]
- Qiao, N.; Shen, M.; He, W.; He, M.; Zhang, Z.; Ye, H.; Li, Y.; Shou, X.; Li, S.; Jiang, C.; et al. Machine Learning in Predicting Early Remission in Patients after Surgical Treatment of Acromegaly: A Multicenter Study. Pituitary 2021, 24, 53–61. [Google Scholar] [CrossRef]
- Liu, X.; Cruz Rivera, S.; Moher, D.; Calvert, M.J.; Denniston, A.K.; SPIRIT-AI and CONSORT-AI Working Group. Reporting Guidelines for Clinical Trial Reports for Interventions Involving Artificial Intelligence: The CONSORT-AI Extension. Lancet Digit. Health 2020, 2, e537–e548. [Google Scholar] [CrossRef]
- Rivera, S.C.; Liu, X.; Chan, A.-W.; Denniston, A.K.; Calvert, M.J.; SPIRIT-AI and CONSORT-AI Working Group. Guidelines for Clinical Trial Protocols for Interventions Involving Artificial Intelligence: The SPIRIT-AI Extension. BMJ 2020, 370, m3210. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef]
- Heus, P.; Damen, J.A.A.G.; Pajouheshnia, R.; Scholten, R.J.P.M.; Reitsma, J.B.; Collins, G.S.; Altman, D.G.; Moons, K.G.M.; Hooft, L. Uniformity in Measuring Adherence to Reporting Guidelines: The Example of TRIPOD for Assessing Completeness of Reporting of Prediction Model Studies. BMJ Open 2019, 9, e025611. [Google Scholar] [CrossRef] [PubMed]
- Heus, P.; Damen, J.A.A.G.; Pajouheshnia, R.; Scholten, R.J.P.M.; Reitsma, J.B.; Collins, G.S.; Altman, D.G.; Moons, K.G.M.; Hooft, L. Poor Reporting of Multivariable Prediction Model Studies: Towards a Targeted Implementation Strategy of the TRIPOD Statement. BMC Med. 2018, 16, 120. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.; Liu, X.; Denniston, A.K. Reporting Guidelines for Artificial Intelligence in Healthcare Research. Clin. Experiment. Ophthalmol. 2021, 49, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Moons, K.G.M. Reporting of Artificial Intelligence Prediction Models. Lancet Lond. Engl. 2019, 393, 1577–1579. [Google Scholar] [CrossRef]
- Wang, W.; Kiik, M.; Peek, N.; Curcin, V.; Marshall, I.J.; Rudd, A.G.; Wang, Y.; Douiri, A.; Wolfe, C.D.; Bray, B. A Systematic Review of Machine Learning Models for Predicting Outcomes of Stroke with Structured Data. PLoS ONE 2020, 15, e0234722. [Google Scholar] [CrossRef]
- Staartjes, V.E.; Serra, C.; Muscas, G.; Maldaner, N.; Akeret, K.; van Niftrik, C.H.B.; Fierstra, J.; Holzmann, D.; Regli, L. Utility of Deep Neural Networks in Predicting Gross-Total Resection after Transsphenoidal Surgery for Pituitary Adenoma: A Pilot Study. Neurosurg. Focus 2018, 45, E12. [Google Scholar] [CrossRef]
- Hollon, T.C.; Parikh, A.; Pandian, B.; Tarpeh, J.; Orringer, D.A.; Barkan, A.L.; McKean, E.L.; Sullivan, S.E. A Machine Learning Approach to Predict Early Outcomes after Pituitary Adenoma Surgery. Neurosurg. Focus 2018, 45, E8. [Google Scholar] [CrossRef]
- Muhlestein, W.E.; Akagi, D.S.; McManus, A.R.; Chambless, L.B. Machine Learning Ensemble Models Predict Total Charges and Drivers of Cost for Transsphenoidal Surgery for Pituitary Tumor. J. Neurosurg. 2018, 131, 507–516. [Google Scholar] [CrossRef]
- Voglis, S.; van Niftrik, C.H.B.; Staartjes, V.E.; Brandi, G.; Tschopp, O.; Regli, L.; Serra, C. Feasibility of Machine Learning Based Predictive Modelling of Postoperative Hyponatremia after Pituitary Surgery. Pituitary 2020, 23, 543–551. [Google Scholar] [CrossRef]
- Zhang, S.; Song, G.; Zang, Y.; Jia, J.; Wang, C.; Li, C.; Tian, J.; Dong, D.; Zhang, Y. Non-Invasive Radiomics Approach Potentially Predicts Non-Functioning Pituitary Adenomas Subtypes before Surgery. Eur. Radiol. 2018, 28, 3692–3701. [Google Scholar] [CrossRef]
- Zhang, Y.; Ko, C.-C.; Chen, J.-H.; Chang, K.-T.; Chen, T.-Y.; Lim, S.-W.; Tsui, Y.-K.; Su, M.-Y. Radiomics Approach for Prediction of Recurrence in Non-Functioning Pituitary Macroadenomas. Front. Oncol. 2020, 10, 590083. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Huang, W.; Cheng, Y.; Teng, Y.; Zhang, L.; Xu, J. Machine Learning-Based Radiomics of the Optic Chiasm Predict Visual Outcome following Pituitary Adenoma Surgery. J. Pers. Med. 2021, 11, 991. [Google Scholar] [CrossRef]
- Staartjes, V.E.; Zattra, C.M.; Akeret, K.; Maldaner, N.; Muscas, G.; Bas van Niftrik, C.H.; Fierstra, J.; Regli, L.; Serra, C. Neural Network-Based Identification of Patients at High Risk for Intraoperative Cerebrospinal Fluid Leaks in Endoscopic Pituitary Surgery. J. Neurosurg. 2019, 133, 329–335. [Google Scholar] [CrossRef]
- Fan, Y.; Jiang, S.; Hua, M.; Feng, S.; Feng, M.; Wang, R. Machine Learning-Based Radiomics Predicts Radiotherapeutic Response in Patients with Acromegaly. Front. Endocrinol. 2019, 10, 588. [Google Scholar] [CrossRef]
- Fan, Y.; Li, Y.; Li, Y.; Feng, S.; Bao, X.; Feng, M.; Wang, R. Development and Assessment of Machine Learning Algorithms for Predicting Remission after Transsphenoidal Surgery among Patients with Acromegaly. Endocrine 2020, 67, 412–422. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Kadioglu, P.; Polat Korkmaz, O.; Comunoglu, N.; Tanriover, N.; Kocer, N.; Islak, C.; Kizilkilic, O. Predicting Response to Somatostatin Analogues in Acromegaly: Machine Learning-Based High-Dimensional Quantitative Texture Analysis on T2-Weighted MRI. Eur. Radiol. 2019, 29, 2731–2739. [Google Scholar] [CrossRef]
- Zanier, O.; Zoli, M.; Staartjes, V.E.; Guaraldi, F.; Asioli, S.; Rustici, A.; Picciola, V.M.; Pasquini, E.; Faustini-Fustini, M.; Erlic, Z.; et al. Machine Learning-Based Clinical Outcome Prediction in Surgery for Acromegaly. Endocrine 2021, 75, 508–515. [Google Scholar] [CrossRef]
- Zoli, M.; Staartjes, V.E.; Guaraldi, F.; Friso, F.; Rustici, A.; Asioli, S.; Sollini, G.; Pasquini, E.; Regli, L.; Serra, C.; et al. Machine Learning-Based Prediction of Outcomes of the Endoscopic Endonasal Approach in Cushing Disease: Is the Future Coming? Neurosurg. Focus 2020, 48, E5. [Google Scholar] [CrossRef]
- Zhang, W.; Sun, M.; Fan, Y.; Wang, H.; Feng, M.; Zhou, S.; Wang, R. Machine Learning in Preoperative Prediction of Postoperative Immediate Remission of Histology-Positive Cushing’s Disease. Front. Endocrinol. 2021, 12, 635795. [Google Scholar] [CrossRef]
- Nadezhdina, E.Y.; Rebrova, O.Y.; Grigoriev, A.Y.; Ivaschenko, O.V.; Azizyan, V.N.; Melnichenko, G.A.; Dedov, I.I. Prediction of Recurrence and Remission within 3 Years in Patients with Cushing Disease after Successful Transnasal Adenomectomy. Pituitary 2019, 22, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, X.; Hong, X.; Liu, P.; Bao, X.; Yao, Y.; Xing, B.; Li, Y.; Huang, Y.; Zhu, H.; et al. Prediction of Recurrence after Transsphenoidal Surgery for Cushing’s Disease: The Use of Machine Learning Algorithms. Neuroendocrinology 2019, 108, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, H.; Feng, M.; Zhang, W.; Cao, L.; Ding, C.; Chen, H.; Wei, L.; Mu, S.; Pei, Z.; et al. Machine-Learning Prediction of Postoperative Pituitary Hormonal Outcomes in Nonfunctioning Pituitary Adenomas: A Multicenter Study. Front. Endocrinol. 2021, 12, 748725. [Google Scholar] [CrossRef]
- Machado, L.F.; Elias, P.C.L.; Moreira, A.C.; Dos Santos, A.C.; Murta Junior, L.O. MRI Radiomics for the Prediction of Recurrence in Patients with Clinically Non-Functioning Pituitary Macroadenomas. Comput. Biol. Med. 2020, 124, 103966. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Liu, Z.; Hou, B.; Li, L.; Liu, X.; Liu, Z.; Wang, R.; Lin, Y.; Feng, F.; Tian, J.; et al. Development and Validation of an MRI-Based Radiomic Signature for the Preoperative Prediction of Treatment Response in Patients with Invasive Functional Pituitary Adenoma. Eur. J. Radiol. 2019, 121, 108647. [Google Scholar] [CrossRef]
- Shahrestani, S.; Cardinal, T.; Micko, A.; Strickland, B.A.; Pangal, D.J.; Kugener, G.; Weiss, M.H.; Carmichael, J.; Zada, G. Neural Network Modeling for Prediction of Recurrence, Progression, and Hormonal Non-Remission in Patients following Resection of Functional Pituitary Adenomas. Pituitary 2021, 24, 523–529. [Google Scholar] [CrossRef]
- Qiao, N. A Systematic Review on Machine Learning in Sellar Region Diseases: Quality and Reporting Items. Endocr. Connect. 2019, 8, 952–960. [Google Scholar] [CrossRef]
- Agrawal, N.; Ioachimescu, A.G. Prognostic Factors of Biochemical Remission after Transsphenoidal Surgery for Acromegaly: A Structured Review. Pituitary 2020, 23, 582–594. [Google Scholar] [CrossRef]
- Fang, Y.; Pei, Z.; Chen, H.; Wang, R.; Feng, M.; Wei, L.; Li, J.; Zhang, H.; Wang, S. Diagnostic Value of Knosp Grade and Modified Knosp Grade for Cavernous Sinus Invasion in Pituitary Adenomas: A Systematic Review and Meta-Analysis. Pituitary 2021, 24, 457–464. [Google Scholar] [CrossRef]
- Knosp, E.; Steiner, E.; Kitz, K.; Matula, C. Pituitary Adenomas with Invasion of the Cavernous Sinus Space: A Magnetic Resonance Imaging Classification Compared with Surgical Findings. Neurosurgery 1993, 33, 610–617. [Google Scholar] [CrossRef]
- Hardy, J.; Vezina, J.L. Transsphenoidal Neurosurgery of Intracranial Neoplasm. Adv. Neurol. 1976, 15, 261–273. [Google Scholar]
- Mooney, M.A.; Hardesty, D.A.; Sheehy, J.P.; Bird, R.; Chapple, K.; White, W.L.; Little, A.S. Interrater and Intrarater Reliability of the Knosp Scale for Pituitary Adenoma Grading. J. Neurosurg. 2017, 126, 1714–1719. [Google Scholar] [CrossRef]
- Koçak, B.; Durmaz, E.Ş.; Ateş, E.; Kılıçkesmez, Ö. Radiomics with Artificial Intelligence: A Practical Guide for Beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Niu, J.; Zhang, S.; Ma, S.; Diao, J.; Zhou, W.; Tian, J.; Zang, Y.; Jia, W. Preoperative Prediction of Cavernous Sinus Invasion by Pituitary Adenomas Using a Radiomics Method Based on Magnetic Resonance Images. Eur. Radiol. 2019, 29, 1625–1634. [Google Scholar] [CrossRef]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the Performance of Prediction Models: A Framework for Traditional and Novel Measures. Epidemiol. Camb. Mass 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Li, T.; Convertino, G.; Wang, W.; Most, H.; Zajonc, T.; Tsai, Y.-H. Hypertuner: Visual Analytics for Hyperparameter Tuning by Professionals. In Proceedings of the Machine Learning from User Interaction for Visualization and Analytics Workshop at IEEE VIS, Berlin, Germany, October 2018. [Google Scholar]
- Luo, G. A Review of Automatic Selection Methods for Machine Learning Algorithms and Hyper-Parameter Values. Netw. Model. Anal. Health Inform. Bioinform. 2016, 5, 18. [Google Scholar] [CrossRef]
- Collins, G.S.; de Groot, J.A.; Dutton, S.; Omar, O.; Shanyinde, M.; Tajar, A.; Voysey, M.; Wharton, R.; Yu, L.-M.; Moons, K.G.; et al. External Validation of Multivariable Prediction Models: A Systematic Review of Methodological Conduct and Reporting. BMC Med. Res. Methodol. 2014, 14, 40. [Google Scholar] [CrossRef]
- Ramspek, C.L.; Jager, K.J.; Dekker, F.W.; Zoccali, C.; van Diepen, M. External Validation of Prognostic Models: What, Why, How, When and Where? Clin. Kidney J. 2021, 14, 49–58. [Google Scholar] [CrossRef]
- Knol, M.J.; Janssen, K.J.M.; Donders, A.R.T.; Egberts, A.C.G.; Heerdink, E.R.; Grobbee, D.E.; Moons, K.G.M.; Geerlings, M.I. Unpredictable Bias When Using the Missing Indicator Method or Complete Case Analysis for Missing Confounder Values: An Empirical Example. J. Clin. Epidemiol. 2010, 63, 728–736. [Google Scholar] [CrossRef]
- Kang, H. The Prevention and Handling of the Missing Data. Korean J. Anesthesiol. 2013, 64, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, E.; Ma, J.; Collins, G.S.; Steyerberg, E.W.; Verbakel, J.Y.; Van Calster, B. A Systematic Review Shows No Performance Benefit of Machine Learning over Logistic Regression for Clinical Prediction Models. J. Clin. Epidemiol. 2019, 110, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus Machine Learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Azimi, P.; Mohammadi, H.R.; Benzel, E.C.; Shahzadi, S.; Azhari, S.; Montazeri, A. Artificial Neural Networks in Neurosurgery. J. Neurol. Neurosurg. Psychiatry 2015, 86, 251–256. [Google Scholar] [CrossRef]
- Kitamura, F.C.; Marques, O. Trustworthiness of Artificial Intelligence Models in Radiology and the Role of Explainability. J. Am. Coll. Radiol. JACR 2021, 18, 1160–1162. [Google Scholar] [CrossRef]
- Goodman, B.; Flaxman, S. European Union Regulations on Algorithmic Decision-Making and a “Right to Explanation”. AI Mag. 2017, 38, 50–57. [Google Scholar] [CrossRef]
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Nair, B.; Vavilala, M.S.; Horibe, M.; Eisses, M.J.; Adams, T.; Liston, D.E.; Low, D.K.-W.; Newman, S.-F.; Kim, J.; et al. Explainable Machine-Learning Predictions for the Prevention of Hypoxaemia during Surgery. Nat. Biomed. Eng. 2018, 2, 749–760. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Moher, D.; Jones, A.; Lepage, L.; CONSORT Group (Consolidated Standards for Reporting of Trials). Use of the CONSORT Statement and Quality of Reports of Randomized Trials: A Comparative before-and-after Evaluation. JAMA 2001, 285, 1992–1995. [Google Scholar] [CrossRef]
- Turner, L.; Shamseer, L.; Altman, D.G.; Schulz, K.F.; Moher, D. Does Use of the CONSORT Statement Impact the Completeness of Reporting of Randomised Controlled Trials Published in Medical Journals? A Cochrane Review. Syst. Rev. 2012, 1, 60. [Google Scholar] [CrossRef]
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S.; PROBAST Group. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef]
| Term | Description |
|---|---|
| Artificial Intelligence | A broad area of computer applications with the ability to perform tasks that conventionally require human intelligence. |
| Machine Learning | Machine learning is an application of artificial intelligence (AI) that provides systems with the ability to automatically learn and improve from experience without being explicitly programmed. |
| Deep Learning | Is a subset of ML which is formally defined as computational models that are composed of multiple processing layers to learn representations of data with multiple levels of abstraction. |
| Supervised learning | A model that is trained based on inputs of data aiming at determining a target output, which are manually labeled a priori (e.g., diagnosis or prognosis). |
| Unsupervised learning | ML models that can perform tasks without being set with labels by a human (e.g., clustering data). |
| Structured data | Data that are pre-defined to be displayed in rows and columns (e.g., electronic medical records, administrative data). More qualitative form of data. |
| Unstructured data | Data without any predefined structure. More quantitative form of data (e.g., image analysis, text). |
| Missing values | Hyperparameters, which specify how a model learns, need to be set by the data scientist before training. They are perpetually improved (tuned) to find the model that performs best. |
| Single case analysis | Exclusion of a row with missing data among its features. |
| Feature | Data science term for predictor/independent variable. |
| Label/Target | Data science term for outcome/dependent variable. |
| Parameter | Inherent weights of a given model, which are set in the code of the algorithm. Define a search space as a grid of hyperparameter values and evaluate every position in the grid. |
| Hyperparameters | An ensemble of wights which define how a model learns. They are arbitrarily attributed, needing to be set by its developer to optimize its performance during and after training. |
| Overfitting | When a model performs well on the training data (seen patients) and performs poorly on the testing data (unseen patients). Regularization is often used to minimize overfitting and optimize generalizability of machine leaning algorithms |
| Discrimination | Describes the model’s ability to correctly identify from random pairs in which it was trained who will develop the target condition. Usually evaluated through the model’s AUC/C-statistic. |
| Area Under Curve/C-statistic | Most used discriminative statistic. An area of 1.0 represents a perfect test; an area of 0.5 represents a worthless test. It enables assessment of predictive ability, and identification of an optimal threshold to distinguish between classes. |
| Accuracy | Ratio between the total number of predictions that are correct. |
| Sensitivity/Recall | Proportion of true positives predictions. |
| Specificity | Proportion of correctly predicted true negatives which are correctly identified. |
| PPV/Precision | Proportion of correctly predicted true positives which are correctly identified |
| NPV | Proportion of correctly predicted negatives among all negative predictions. |
| F1 score | Composite metric defined as the harmonic mean between precision (or PPV) and recall (sensitivity). |
| Internal Validation | Assessment of a model’s performance with the same data or population, if prospective, used in the development process. |
| External Validation | Assessment of a model’s performance in a dataset which differs from the one used in its development geographically or temporally. |
| Cross Validation | Internal validation technique in which the dataset is randomly split into k-1 groups of similar size. Performance is evaluated in the remaining group with the whole process repeated n times; model performance is taken as average over n iterations. |
| Bootstrapping | Internal validation approach like cross validation but relying on random sampling with replacement; each sample is the same size as model development dataset |
| Split Sample | Internal validation approach in which the available development dataset is divided into two datasets: one to develop the model and the other to validate the model; division can be random or non-random. |
| Algorithm | Description |
|---|---|
| Neural Networks (NN or ANN) | Artificial neural networks are non-linear algorithms loosely inspired by human brain synapses. Convolutional neural networks, the most commonly applied, comprise input nodes, output nodes and intervening or hidden layers of nodes, which may number up to 100. Each node within a layer involves two or more inputs and applies an activation and weighting function to produce an output which serves as the input data for the next layer of nodes. |
| Support Vector Machine (SVM) | SVM is based on the idea of computing a hyperplane that best separates the features into different domains. Its objective is to find a decision boundary (the Hyperplane) that has the maximum separation degree between two nearer points of each class—i.e., the support vectors. Kernel functions are used when data are too non-linear functions; the algorithm can map examples to other dimensions and then operates on non-linear relationships by transforming low-dimensional input data into high-dimensional space. |
| k-Nearest Neighbors (k-NN) | The k-NN classing classes based on a distance criterion. The values of the distance from k (number of neighbors) in given distance between them and the subject of interest. This distance inputs-output is computed on comparing multidimensional vectors of feature values, defining the more similar ones as “neighbors”. |
| Decision Trees (DTs) | DT algorithms are architecture under a tree structure modeling approach with conditional control statements for establishing a framework of subsequent decisions. Its internal nodes represent ‘test’ on an attribute, branch represents the results of this test and “leaf” represents decision taken after computing all attributes. |
| Random Forest (RF) | RF is essentially an ensemble of DTs, although it differs from usual DTs by using randomly selected inputs or combinations of inputs at each node to grow each tree rather than a consistent set. That is intent yielding to avoid the overfitting usually present in deep DTs. Random distribution of inputs provides, when averaged, lower rates of error in the final output and reduced variance. |
| Study | Journal | Country | No. of Centers | Population | Time Interval | Source of Data | Sample Size |
|---|---|---|---|---|---|---|---|
| Fan et al., 2019 [35] | European Journal of Radiology | China | Single-Center | PAs | April 2012 May 2018 | Chart review | 163 |
| Fan et al., 2019 [45] | Frontiers in Endocrinology | China | Single-Center | Acromegaly | January 2008 and January 2016 | Case series | 57 |
| Fan et al., 2020 [36] | Endocrine | China | Single-Center | Acromegaly | 1983 to 2018 | Chart review | 668 |
| Fang et al., 2021 [43] | Frontiers in Endocrinology | China | Multicenter | NFPAs | 2015 to 2021 | Retrospective database | 215 |
| Hollon et al., 2018 [28] | Journal of Neurosurgery | USA | Single-Center | PAs | Not mentioned | Case series | 400 |
| Kocak et al., 2018 [37] | European Radiology | Istanbul | Single-Center | Acromegaly | January 2009 and December 2017 | Chart review | 47 |
| Liu et al., 2019 [42] | Neuroendocrinology | China | Single-Center | CD | January 2000 to December 2017 | Case series | 354 |
| Machado et al., 2020 [44] | Computes in Biology and Medicine | Brazil | Single-Center | NFPAs | Not mentioned | Electronical Clinical Records | 27 |
| Muhlestein et al., 2019 [29] | Journal of Neurosurgery | USA | National Inpatient Database | PAs | 2002 to 2011 | Administrative data | 15487 |
| Nadezhdina et al., 2019 [41] | Pituitary | Russia | Single-Center | CD | 2007 to 2014 | Chart review | 219 |
| Qiao et al., 2021 [47] | Pituitary | China | Multicenter | Acromegaly | 2010 to 2018 (D); 2019 (EV) | Prospective database | 833 (D); 52 (EV) |
| Shahrestani et al., 2021 [46] | Pituitary | USA | Single-Center | FPA | 1992 to 2019 | Chart review | 348 |
| Staartjes et al., 2018 [27] | Neurosurgical Focus | Switzerland | Single-Center | PAs | October 2012 onwards | Prospective clinical registry | 140 |
| Staartjes, et al., 2019 [34] | Journal of Neurosurgery | Switzerland | Single-Center | PAs | October 2012 onwards | Prospective clinical registry | 154 |
| Voglis et al., 2019 [30] | Pituitary | Switzerland | Single-Center | PAs | October 2012 to December 2019 | Case Series | 207 |
| Zanier et al., 2021 [38] | Endocrine | Switzerland | Multicenter | Acromegaly | August 1998 to January 2020 | Chart review | 307 (D); 40 (E) |
| Zhang et al., 2020 * [32] | Frontiers in Oncology | China | Single-Center | NFPAs | September 2010 to December 2017 | Chart review | 50 |
| Zhang et al., 2021 * [33] | Frontiers in Endocrinology | China | Single-Center | CD | February 2000 and September 2019 | Chart review | 1045 |
| Zhang et al., 2021 [40] | Journal of Personalized Medicine | China | Single-Centre | PAs | January 2017 to June 2019 | Chart review | 131 |
| Zoli et al., 2020 [39] | Neurosurgical Focus | Italy | Single-Center | CD | May 1998 to December 2017 | Case series | 151 |
| Study | ML Task | Outcome; Proportion | Software | Algorithm | AUC | Other Measures |
|---|---|---|---|---|---|---|
| Fan et al., 2019 [35] | Treatment response | Remission; 66 patients (33.7%) | MATLAB 2015b (Natick, MA, USA) | SVM | 0.81 | Acc: 74.5%; Sn: 61.3%; Sp 91.7%; PPV: 70.5%; NPV: 64.7% |
| Fan et al., 2019 [45] | Radiotherapeutic response | radiotherapy response; 25 patients (78.1%) | ITK-SNAP 3.8 (Philadelphia, PA, USA); Python 3.0 (Wilmington, NC, USA), PyRadiomic library | SVM | 0.96 | Acc: 91%; Sn: 90%; Sp: 92%; PPV: 935; NPV 0.885 |
| Fan et al., 2020 [36] | Remission of acromegaly after surgery | Acromegaly remission; 349 patients (52.2%) | Python 2.7 (Wilmington, NC, USA) | GBDT | 0.81 | Acc: 79%; Sn: 81%; Sn: 78%; PPV: 81%; NPV: 77% |
| Fang et al., 2021 [43] | Postoperative hypofunction, new postoperative hypofunction, and hormonal recovery | hormone level normalization; 21 patients (64.7%) | R 4.0.4 (Vienna, Austria); Python 3.9 (Wilmington, NC, USA) | RF | 0.85 | AUC-PR: 0.52 |
| Hollon et al., 2018 [28] | Poor early postoperative outcome | Poor early postoperative outcome; 124 (31%) | R 4.0.4 (Vienna, Austria), caret package; Python 3.2 (Wilmington, NC, USA), SciPy 0.19.1 library | RF | 0.84 | Acc: 85%; Sn: 56%; Sp: 94.7%; PPV: 77.8%; NPV: 86.6% |
| Kocak et al., 2018 [37] | Response to somatostatin analogues | Responsive; 24 patients responsive (51%) | WEKA 3.8.2 (Waikato, New Zeland) | k-NN | 0.85 | Acc: 85.1% |
| Liu et al., 2019 [42] | Immediate CD remission | CD recurrence; 46 patients (13.0%) | Python 2.7 (Wilmington, NC, USA) | RF | 0.78 | Acc: 87%; Sn: 71.7%; Sp: 58.4% |
| Machado et al., 2020 [44] | Tumor recurrence | Tumor recurrence; 10 (37%) | Python 3.0 (Wilmington, NC, USA), Scikit-learn library | k-NN | 0.96 | Acc: 96.3%; Sp: 100%; Sn: 91.7% |
| Muhlestein et al., 2019 [29] | Hospital total charges, Postoperative complications | Postoperative complications; 3365 inpatients (25%) | Python 2.7 (Wilmington, NC, USA), SciPy 0.17 library; DataRobot 3.0 (Boston, MA, USA) | GBDT | 0.66 | RMSLE: 0.446; Holdout: 0.455 |
| Nadezhdina et al., 2019 [41] | Remission/Recurrence of CD | Remission; 172 patients (78.5%) | IBM SPSS 18 (Armonk, NY, USA) | NN | 0.91 | Acc: 92%; Sn: 75%; Sp 97%; PPV: 85%; NPV: 93% |
| Qiao et al., 2021 [47] | Acromegaly endocrine remission | Remission; 434 patients (52.1%) | R version 3.4.3 (Vienna, Austria); Python version 3.6 (Wilmington, NC, USA) | GBM | 0.87 | Acc: 80.3%; Sn 90.5%; Sp 69.6% |
| Shahrestani et al., 2021 [46] | Suboptimal outcomes | Suboptimal outcomes; 81 patients (23.3%) | Python 2.7 (Wilmington, NC, USA), PyRadiomics, Scikit-learn libraries | NN | 0.91 | Acc: 87.1%; Sn: 89.5%; Sp: 76.9%; PPV: 94.4%; NPV: 62.5% |
| Staartjes et al., 2018 [27] | GTR | GTR; 95 patients (68%) | R 3.4.4 (Vienna, Austria), TensorFlow, Keras | NN | 0.96 | Acc: 91%; Sn: 94%; Sp 89% |
| Staartjes, et al., 2019 [34] | Risk level of intraoperative CSF Leak | CSF leak; 45 patients (29%) | R 3.5.1 (Vienna, Austria); TensorFlow (Mountain View, CA, USA), Keras | NN | 0.84 | Sn: 83%; Sp: 89%; PPV: 71%; NPV: 94%; F1 score: 0.77 |
| Voglis et al., 2019 [30] | Post-operative hyponatremia | Post-operative hyponatremia; 44 patients (22%) | R 3.6.2 (Vienna, Austria), caret package | Boosted GLM | 0.84 | Acc:78.4%; Sn: 81.4%; Sp: 77.5%; F1 Score: 62.1%; NPV: 93.9%; PPV: 50% |
| Zanier et al., 2021 [38] | GTR, Biochemical remission, or CSF leak | CSF leak; 38 patients (12.5%) | R 4.0.2 (Vienna, Austria) | Bayesian GLM | 0.69 | Acc: 60%; Sn: 71%; Sp: 59%; PPV: 19%; 93%; Calibration intercept: −1.77; calibration slope: 0.39 |
| Zhang et al., 2020 * [32] | NFPA recurrence | Tumor recurrence; 28 patients (56%) | MATLAB 2018b (Natick, MA, USA) | SVM | 0.78 | Acc: 82% |
| Zhang et al., 2021 * [33] | Postoperative Immediate Remission of CD | CD remission; 766 patients (73.3%) | R Studio 1.2 (Vienna, Austria); IBM SPSS 23 (Armonk, NY, USA); Python 3.6 (Wilmington, NC, USA), Scikit-learn library | Stacking | 0.74 | Acc: 72% |
| Zhang et al., 2021 [40] | Visual field recovery following pituitary adenoma surgery | Visual field recovery; 79 patients (60.3%) | ITK-SNAP (Philadelphia, PA, USA); R 3.6.3 (Vienna, Austria) | SVM | 0.82 | Acc: 70%; Sn: 65%; Sp: 80%; PPV: 70%; NPV: 80% |
| Zoli et al., 2020 [39] | GTR, postsurgical remission, and long-term control of disease | GTR; 137 patients (91%) | R 3.5.2 (Vienna, Austria) | SVM | 1.00 | Acc: 100%; Sn: 100%; Sp: 100%; PPV: 100%; NPV: 100%; F1 score: 100%; Brier score: 0.097 |
| Study | Demographics | Medical History | Tumor Morphology and Behavior | Endocrine Parameters | Surgical Aspects | Histological |
|---|---|---|---|---|---|---|
| Fan et al., 2019 [35] | Age; sex | NA | Diagnosis type of tumor; KG | NA | NA | NA |
| Fan et al., 2019 [45] | NA | NA | KG; tumor consistency; tumor volume; | Random GH; IGF-1 standard deviation score; GH inhibition ratio; | NA | P53 |
| Fan et al., 2020 [36] | Age | Hypertension, ophthalmic disorders, maximal tumor diameter | KG | GH, IGF-1, nadir GH, | NA | NA |
| Fang et al., 2021 [43] | NA | NA | NA | Preoperative hormone levels (SH, FSH, LH, PRL, ACTH | NA | NA |
| Hollon et al., 2018 [28] | Age; gender; race; BMI | Prior cardiovascular, renal, pulmonary or hepatic disease; prior TSS, prior craniotomy; current blood thinners intake; prior visual deficit; use of postoperative desmopressin | Tumor type | Postoperative sodium low; postoperative sodium elevated; diabetes insipidus | NA | NA |
| Kocak et al., 2018 [37] | NA | NA | NA | NA | NA | NA |
| Liu et al., 2019 [42] | Age | Disease course; | NA | Postoperative levels of morning ACTH (nadir), morning serum cortisol (nadir), 24 h UFC; preoperative levels of morning ACTH, and serum cortisol | NA | NA |
| Machado et al., 2020 [44] | NA | NA | NA | NA | NA | NA |
| Muhlestein et al., 2019 [29] | NA | NA | NA | NA | NA | NA |
| Nadezhdina et al., 2019 [41] | Sex; age | Duration of disease (months) | Type of tumor; | Postoperative morning levels of ACTH and cortisol | NA | NA |
| Qiao et al., 2021 [47] | Age; gender; BMI | TSS; specific pharmacotherapy; radiotherapy | Tumor dimensions; KG; clivus invasiveness; intraoperative cavernous sinus invasion; tumor texture; presence of pseudocapsule; | Serum random GH; serum IGF-1 level; preoperative hypopituitarism; preoperative diabetes insipidus | Surgeons’ experience (based on annual pituitary operations performed surgical approach; total resection or subtotal | NA |
| Shahrestani et al., 2021 [46] | NA | Hospital LOS (days); prior craniotomy; preoperative visual deficit not improved after surgery; | NA | Transient diabetes Insipidus; low cortisol axis low GH axis; panhypopituitarism; acromegaly | NA | MIB-1/Ki-67 labeling index |
| Staartjes et al., 2018 [27] | Sex; age | TSS | KG; HG; tumor invasiveness; ICD at the C6, C4 horizontal, and C4 vertical segments; R ratio between maximum adenoma diameter and ICD C4 horizontal segment; adenoma secretory status, volume, and diameters in 3 axes | NA | NA | NA |
| Staartjes, et al., 2019 [34] | Sex; age | TSS | KG; HG; tumor invasiveness; ICD at the C6, C4 horizontal, and C4 vertical segments; R ratio between maximum adenoma diameter and ICD C4 horizontal segment; adenoma secretory status, volume, and diameters in 3 axes | NA | Targeted level of resection | NA |
| Voglis et al., 2019 [30] | Sex; Age; weight; height; BMI | TSS; | KG; HG | Hypofunctional ACTH and GNRH in preoperative levels; prior diabetes insipidus; preoperative levels of potassium, sodium, cortisol, IGF-1, fT3, fT4, TSH, LH, FSH, and PRL | NA | NA |
| Zanier et al., 2021 [38] | Age; gender | TSS | KG; HG; tumoral size | NA | NA | NA |
| Zhang et al., 2020 * [32] | NA | NA | NA | NA | NA | NA |
| Zhang et al., 2021 * [33] | NA | NA | NA | NA | NA | NA |
| Zhang et al., 2021 [40] | NA | TSS | Cavernous sinus invasion on preoperative MRI; tumor size | Preoperative ACTH | NA | NA |
| Zoli et al., 2020 [39] | Age; sex | TSS; specific pharmacotherapy; radiotherapy | Tumor size; HG; KG; bony tumor or cavernous invasiveness | Preoperative hypopituitarism; preoperative diabetes insipidus | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rech, M.M.; de Macedo Filho, L.; White, A.J.; Perez-Vega, C.; Samson, S.L.; Chaichana, K.L.; Olomu, O.U.; Quinones-Hinojosa, A.; Almeida, J.P. Machine Learning Models to Forecast Outcomes of Pituitary Surgery: A Systematic Review in Quality of Reporting and Current Evidence. Brain Sci. 2023, 13, 495. https://doi.org/10.3390/brainsci13030495
Rech MM, de Macedo Filho L, White AJ, Perez-Vega C, Samson SL, Chaichana KL, Olomu OU, Quinones-Hinojosa A, Almeida JP. Machine Learning Models to Forecast Outcomes of Pituitary Surgery: A Systematic Review in Quality of Reporting and Current Evidence. Brain Sciences. 2023; 13(3):495. https://doi.org/10.3390/brainsci13030495
Chicago/Turabian StyleRech, Matheus M., Leonardo de Macedo Filho, Alexandra J. White, Carlos Perez-Vega, Susan L. Samson, Kaisorn L. Chaichana, Osarenoma U. Olomu, Alfredo Quinones-Hinojosa, and Joao Paulo Almeida. 2023. "Machine Learning Models to Forecast Outcomes of Pituitary Surgery: A Systematic Review in Quality of Reporting and Current Evidence" Brain Sciences 13, no. 3: 495. https://doi.org/10.3390/brainsci13030495
APA StyleRech, M. M., de Macedo Filho, L., White, A. J., Perez-Vega, C., Samson, S. L., Chaichana, K. L., Olomu, O. U., Quinones-Hinojosa, A., & Almeida, J. P. (2023). Machine Learning Models to Forecast Outcomes of Pituitary Surgery: A Systematic Review in Quality of Reporting and Current Evidence. Brain Sciences, 13(3), 495. https://doi.org/10.3390/brainsci13030495






