Differential Distribution of Brain Metastases from Non-Small Cell Lung Cancer Based on Mutation Status
Abstract
:1. Introduction
2. Material and Methods
2.1. Clinical Data
2.2. Tumor Segmentation and Registration
2.3. Construction of Tumor Frequency Maps
2.4. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Tumor Characteristics
3.3. Distribution of GMD
3.4. Brain Regions with Higher Frequency of BM Than Random Distribution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoenmaekers, J.; Dingemans, A.C.; Hendriks, L.E.L. Brain imaging in early stage non-small cell lung cancer: Still a controversial topic? J. Thorac. Dis. 2018, 10 (Suppl. S18), S2168–S2171. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, H.; Shirato, H.; Tago, M.; Nakagawa, K.; Toyoda, T.; Hatano, K.; Kenjyo, M.; Oya, N.; Hirota, S.; Shioura, H.; et al. Stereotactic radiosurgery plus whole-brain radiation therapy vs stereotactic radiosurgery alone for treatment of brain metastases: A randomized controlled trial. JAMA 2006, 295, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Greenspoon, J.N.; Ellis, P.M.; Pond, G.; Caetano, S.; Broomfield, J.; Swaminath, A. Comparative survival in patients with brain metastases from non-small-cell lung cancer treated before and after implementation of radiosurgery. Curr. Oncol. 2017, 24, e146–e151. [Google Scholar] [CrossRef] [Green Version]
- Ernani, V.; Stinchcombe, T.E. Management of Brain Metastases in Non-Small-Cell Lung Cancer. J. Oncol. Pract. 2019, 15, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Hardesty, D.A.; Nakaji, P. The Current and Future Treatment of Brain Metastases. Front. Surg. 2016, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Yanagihara, T.K.; Lee, A.; Wang, T.J.C. Quantitative Analysis of the Spatial Distribution of Metastatic Brain Lesions. Tomogr. J. Imaging Res. 2017, 3, 16–22. [Google Scholar] [CrossRef]
- Lee, T.; Lee, B.; Choi, Y.L.; Han, J.; Ahn, M.J.; Um, S.W. Non-small Cell Lung Cancer with Concomitant EGFR, KRAS, and ALK Mutation: Clinicopathologic Features of 12 Cases. J. Pathol. Transl. Med. 2016, 50, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Takano, K.; Kinoshita, M.; Takagaki, M.; Sakai, M.; Tateishi, S.; Achiha, T.; Hirayama, R.; Nishino, K.; Uchida, J.; Kumagai, T.; et al. Different spatial distributions of brain metastases from lung cancer by histological subtype and mutation status of epidermal growth factor receptor. Neuro-Oncology 2016, 18, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Kyeong, S.; Cha, Y.J.; Ahn, S.G.; Suh, S.H.; Son, E.J.; Ahn, S.J. Subtypes of breast cancer show different spatial distributions of brain metastases. PLoS ONE 2017, 12, e0188542. [Google Scholar] [CrossRef] [Green Version]
- Chow, K.K.H.; Meola, A.; Chang, S.D. Commentary: Peritumoral Edema/Tumor Volume Ratio: A Strong Survival Predictor for Posterior Fossa Metastases. Neurosurgery 2019, 85, E18–E19. [Google Scholar] [CrossRef] [Green Version]
- Franchino, F.; Rudà, R.; Soffietti, R. Mechanisms and Therapy for Cancer Metastasis to the Brain. Front. Oncol. 2018, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Gardner, L.J.; Ward, M.; Andtbacka, R.H.; Boucher, K.M.; Bowen, G.M.; Bowles, T.L.; Cohen, A.L.; Grossmann, K.; Hitchcock, Y.J.; Holmen, S.L.; et al. Risk factors for development of melanoma brain metastasis and disease progression: A single-center retrospective analysis. Melanoma Res. 2017, 27, 477–484. [Google Scholar] [CrossRef]
- Palmer, J.D.; Trifiletti, D.M.; Gondi, V.; Chan, M.; Minniti, G.; Rusthoven, C.G.; Schild, S.E.; Mishra, M.V.; Bovi, J.; Williams, N.; et al. Multidisciplinary patient-centered management of brain metastases and future directions. Neuro-Oncol. Adv. 2020, 2, vdaa034. [Google Scholar] [CrossRef] [Green Version]
- Chang, E.L.; Hassenbusch, S.J., 3rd; Shiu, A.S.; Lang, F.F.; Allen, P.K.; Sawaya, R.; Maor, M.H. The role of tumor size in the radiosurgical management of patients with ambiguous brain metastases. Neurosurgery 2003, 53, 272–280, discussion 280–271. [Google Scholar] [CrossRef]
- Bennett, E.E.; Angelov, L.; Vogelbaum, M.A.; Barnett, G.H.; Chao, S.T.; Murphy, E.S.; Yu, J.S.; Suh, J.H.; Jia, X.; Stevens, G.H.; et al. The Prognostic Role of Tumor Volume in the Outcome of Patients with Single Brain Metastasis after Stereotactic Radiosurgery. World Neurosurg. 2017, 104, 229–238. [Google Scholar] [CrossRef]
- Calluaud, G.; Terrier, L.M.; Mathon, B.; Destrieux, C.; Velut, S.; François, P.; Zemmoura, I.; Amelot, A. Peritumoral Edema/Tumor Volume Ratio: A Strong Survival Predictor for Posterior Fossa Metastases. Neurosurgery 2019, 85, 117–125. [Google Scholar] [CrossRef]
- Chen, B.T.; Jin, T.; Ye, N.; Mambetsariev, I.; Daniel, E.; Wang, T.; Wong, C.W.; Rockne, R.C.; Colen, R.; Holodny, A.I.; et al. Radiomic prediction of mutation status based on MR imaging of lung cancer brain metastases. Magn. Reson. Imaging 2020, 69, 49–56. [Google Scholar] [CrossRef]
- Chen, B.T.; Jin, T.; Ye, N.; Mambetsariev, I.; Wang, T.; Wong, C.W.; Chen, Z.; Rockne, R.C.; Colen, R.R.; Holodny, A.I.; et al. Predicting Survival Duration With MRI Radiomics of Brain Metastases From Non-small Cell Lung Cancer. Front. Oncol. 2021, 11, 621088. [Google Scholar] [CrossRef]
- Ashburner, J.; Friston, K.J. Computing average shaped tissue probability templates. Neuroimage 2009, 45, 333–341. [Google Scholar] [CrossRef]
- Kwon, H.; Kim, J.W.; Park, M.; Kim, J.W.; Kim, M.; Suh, S.H.; Chang, Y.S.; Ahn, S.J.; Lee, J.-M. Brain Metastases From Lung Adenocarcinoma May Preferentially Involve the Distal Middle Cerebral Artery Territory and Cerebellum. Front. Oncol. 2020, 10, 1664. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fouad, A.; Pickren, J.W.; Lane, W.W. Central nervous system metastasis from breast carcinoma Autopsy study. Cancer 1983, 52, 2349–2354. [Google Scholar] [CrossRef] [PubMed]
- Bender, E.T.; Tomé, W.A. Distribution of brain metastases: Implications for non-uniform dose prescriptions. Br. J. Radiol. 2011, 84, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Huang, D.; Yang, X.; Zhang, Y.; Gang, F.; Du, X.B. Distribution of brain metastases: Low-risk metastasis areas may be avoided when treating with whole-brain radiotherapy. Cancer Imaging 2020, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Maccauro, G.; Spinelli, M.S.; Mauro, S.; Perisano, C.; Graci, C.; Rosa, M.A. Physiopathology of spine metastasis. Int. J. Surg. Oncol. 2011, 2011, 107969. [Google Scholar] [CrossRef] [Green Version]
- Fidler, I.J.; Yano, S.; Zhang, R.D.; Fujimaki, T.; Bucana, C.D. The seed and soil hypothesis: Vascularisation and brain metastases. Lancet Oncol. 2002, 3, 53–57. [Google Scholar] [CrossRef]
- Paget, S. The distribution of secondary growths in cancer of the breast. Lancet 1889, 133, 571–573. [Google Scholar] [CrossRef] [Green Version]
- Valiente, M.; Obenauf, A.C.; Jin, X.; Chen, Q.; Zhang, X.H.; Lee, D.J.; Chaft, J.E.; Kris, M.G.; Huse, J.T.; Brogi, E.; et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 2014, 156, 1002–1016. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Cossette, S.M.; Rarick, K.R.; Gershan, J.; Dwinell, M.B.; Harder, D.R.; Ramchandran, R. Astrocytes Directly Influence Tumor Cell Invasion and Metastasis In Vivo. PLoS ONE 2013, 8, e80933. [Google Scholar] [CrossRef] [Green Version]
- Stoeckel, L.E.; Stewart, C.C.; Griffith, H.R.; Triebel, K.; Okonkwo, O.C.; Den Hollander, J.A.; Martin, R.C.; Belue, K.; Copeland, J.N.; Harrell, L.E.; et al. MRI volume of the medial frontal cortex predicts financial capacity in patients with mild Alzheimer’s disease. Brain Imaging Behav. 2013, 7, 282–292. [Google Scholar] [CrossRef] [Green Version]
- Euston, D.R.; Gruber, A.J.; McNaughton, B.L. The role of medial prefrontal cortex in memory and decision making. Neuron 2012, 76, 1057–1070. [Google Scholar] [CrossRef] [Green Version]
- Hwang, T.-L.; Close, T.P.; Grego, J.M.; Brannon, W.L.; Gonzales, F. Predilection of brain metastasis in gray and white matter junction and vascular border zones. Cancer Interdiscip. Int. J. Am. Cancer Soc. 1996, 77, 1551–1555. [Google Scholar] [CrossRef]
- Smith, F.O.; Yue, B.; Marzban, S.S.; Walls, B.L.; Carr, M.; Jackson, R.S.; Puleo, C.A.; Padhya, T.; Cruse, C.W.; Gonzalez, R.J.; et al. Both tumor depth and diameter are predictive of sentinel lymph node status and survival in Merkel cell carcinoma. Cancer 2015, 121, 3252–3260. [Google Scholar] [CrossRef] [Green Version]
- Spanberger, T.; Berghoff, A.S.; Dinhof, C.; Ilhan-Mutlu, A.; Magerle, M.; Hutterer, M.; Pichler, J.; Wöhrer, A.; Hackl, M.; Widhalm, G.; et al. Extent of peritumoral brain edema correlates with prognosis, tumoral growth pattern, HIF1a expression and angiogenic activity in patients with single brain metastases. Clin. Exp. Metastasis 2013, 30, 357–368. [Google Scholar] [CrossRef]

| EGFR (+) | ALK (+) | KRAS (+) | p-Value | |
|---|---|---|---|---|
| n = 75 | n = 21 | n = 15 | ||
| Age (years) | ||||
| Mean ± SD | 57.43 ± 12.09 | 53.81 ± 14.79 | 63.67 ± 6.40 | 0.09 |
| Gender | ||||
| Male | 24 (32%) | 8 (38.10%) | 5 (33.33%) | 0.83 |
| Female | 51 (68%) | 13 (61.90%) | 10 (66.67%) | |
| Race | ||||
| Caucasian | 34 (45.33%) | 13(61.90%) | 11 (73.33%) | 0.016 |
| Asian | 35 (46.67%) | 7 (33.33%) | 1 (6.67%) | |
| Other 1 | 6 (8%) | 1 (0.04%) | 3 (20%) | |
| History of Smoking | ||||
| Yes | 20 (26.67%) | 5 (23.80%) | 12 (80%) | <0.001 |
| No | 55 (73.33%) | 16 (76.19%) | 3 (20%) | |
| Histology | ||||
| Adenocarcinoma | 71 (94.67%) | 21 (100%) | 13 (86.67%) | 0.5 |
| Other 2 | 4 (5.33%) | 0 (0%) | 2 (13.33%) | |
| Number of tumors | 15.79 ± 35.24 | 9.07 ± 14.01 | 6.18 ± 7.41 | 0.34 |
| Tumor Size (cm3) | 0.51 ± 2.68 | 1.23 ± 3.38 | 2.83 ± 6.87 | <0.0001 |
| Median Tumor Depth (mm) | 7.9 | 8.7 | 9.1 | 0.044 |
| Edema/Tumor Volume Ratio | 2.38 ± 6.13 | 5.66 ± 21.15 | 5.08 ± 8.70 | <0.0001 |
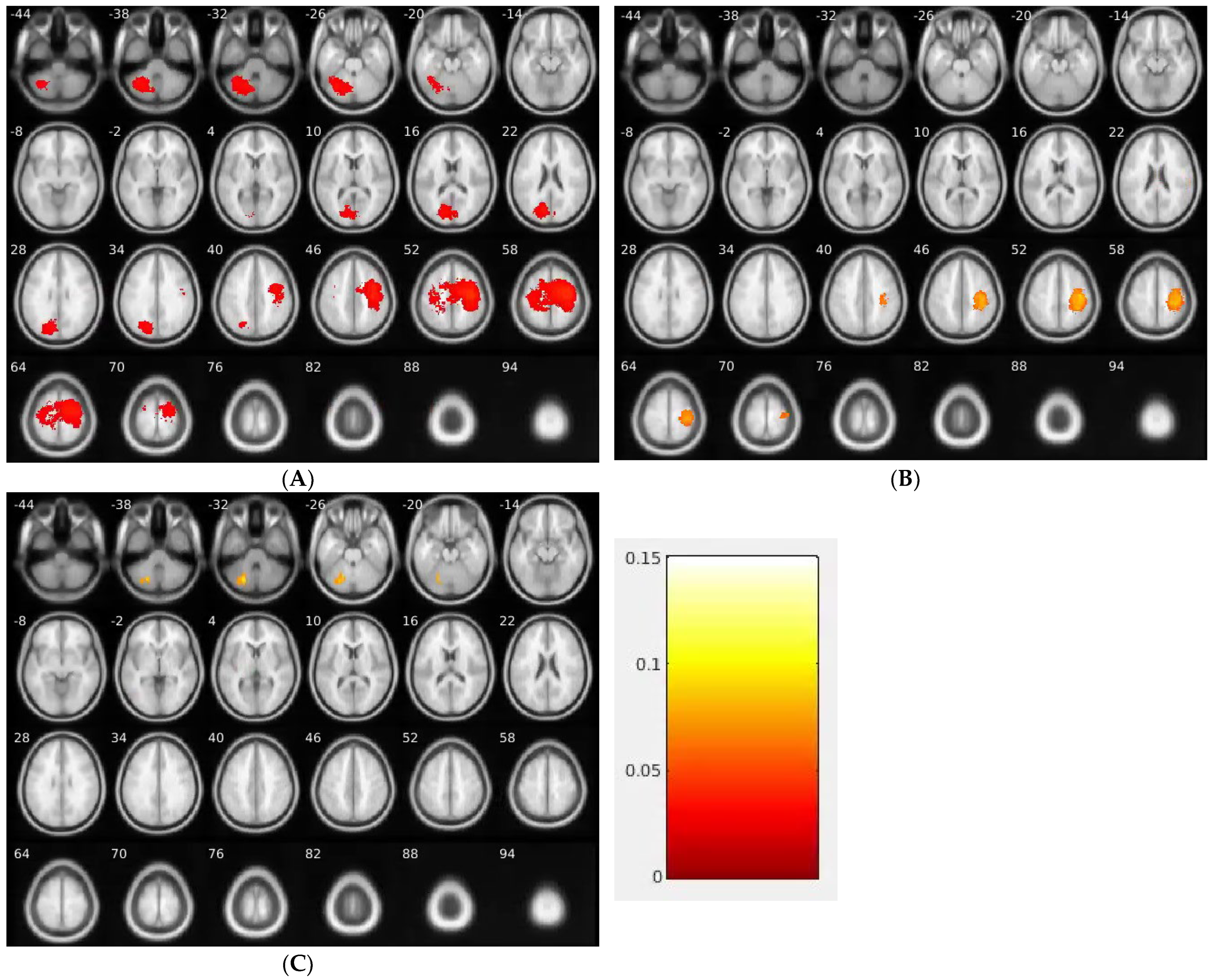
| Group | Center (MNI) | Cluster Size | p Value | Brain Region |
|---|---|---|---|---|
| EGFR | -28.5_-63.0_-33.0 | 6060 | 0.0002 | L Cerebellum, Anterior Lobe, Culmen |
| -30.0_-61.5_-40.5 | 6060 | 0.0002 | L Cerebellum, Posterior Lobe, Cerebellar Tonsil | |
| -18.0_-66.0_-28.5 | 6060 | 0.0002 | L Cerebellum, Posterior Lobe, Declive | |
| -16.5_-61.5_-31.5 | 6060 | 0.0002 | L Cerebellum, Anterior Lobe, Dentate | |
| -13.5_-73.5_31.5 | 5433 | 0.0006 | L Occipital Lobe, Cuneus | |
| -12.0_-76.5_21.0 | 5433 | 0.0006 | L Occipital Lobe, Cuneus (BA 18) | |
| -9.0_-75.0_21.0 | 5433 | 0.0006 | L Parietal Lobe, Precuneus (BA 31) | |
| -15.0_-69.0_24.0 | 5433 | 0.0006 | L Parietal Lobe, Precuneus | |
| 36.0_-16.5_49.5 | 23,128 | 0.0001 | R Frontal Lobe, Precentral Gyrus | |
| 28.5_-19.5_63.0 | 23,128 | 0.0001 | R Frontal Lobe, Precentral Gyrus (BA 6) | |
| ALK | 28.5_-79.5_13.5 | 1576 | 0.022 | R Occipital Lobe, Middle Occipital Gyrus |
| 25.5_-66.0_10.5 | 1576 | 0.022 | R Limbic Lobe, Posterior Cingulate | |
| 21.0_-78.0_21.0 | 1576 | 0.022 | R Parietal Lobe, Precuneus (BA 18) | |
| 37.5_-16.5_51.0 | 8814 | 0.0001 | R Frontal Lobe, Precentral Gyrus (BA 4) | |
| 40.5_-19.5_54.0 | 8814 | 0.0001 | R Frontal Lobe, Precentral Gyrus | |
| 37.5_-31.5_43.5 | 8814 | 0.0001 | R Parietal Lobe, Inferior Parietal Lobule (BA 40) | |
| 40.5_-33.0_45.0 | 8814 | 0.0001 | R Parietal Lobe, Postcentral Gyrus | |
| KRAS | -24.0_-72.0_-30.0 | 1496 | 0.0278 | L Cerebellum, Posterior Lobe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.T.; Jin, T.; Ye, N.; Chen, S.W.; Rockne, R.C.; Yoon, S.; Mambetsariev, I.; Daniel, E.; Salgia, R. Differential Distribution of Brain Metastases from Non-Small Cell Lung Cancer Based on Mutation Status. Brain Sci. 2023, 13, 1057. https://doi.org/10.3390/brainsci13071057
Chen BT, Jin T, Ye N, Chen SW, Rockne RC, Yoon S, Mambetsariev I, Daniel E, Salgia R. Differential Distribution of Brain Metastases from Non-Small Cell Lung Cancer Based on Mutation Status. Brain Sciences. 2023; 13(7):1057. https://doi.org/10.3390/brainsci13071057
Chicago/Turabian StyleChen, Bihong T., Taihao Jin, Ningrong Ye, Sean W. Chen, Russell C. Rockne, Stephanie Yoon, Isa Mambetsariev, Ebenezer Daniel, and Ravi Salgia. 2023. "Differential Distribution of Brain Metastases from Non-Small Cell Lung Cancer Based on Mutation Status" Brain Sciences 13, no. 7: 1057. https://doi.org/10.3390/brainsci13071057






