Efficient Brain Age Prediction from 3D MRI Volumes Using 2D Projections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data
2.2. Two-Dimensional Projections
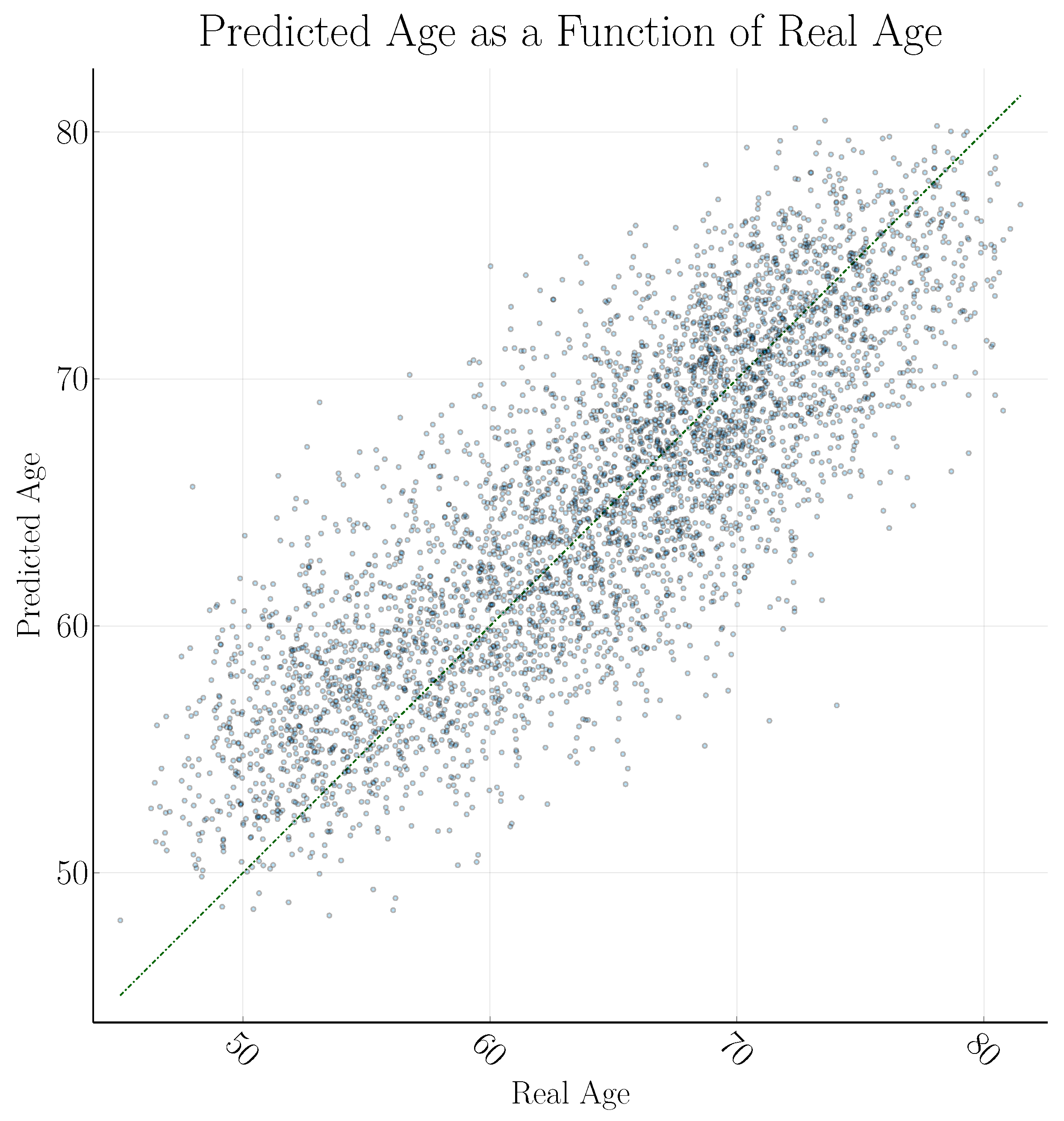
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bjørk, M.B.; Kvaal, S.I. CT and MR imaging used in age estimation: A systematic review. J. Forensic Odonto-Stomatol. 2018, 36, 14. [Google Scholar]
- Huang, T.W.; Chen, H.T.; Fujimoto, R.; Ito, K.; Wu, K.; Sato, K.; Taki, Y.; Fukuda, H.; Aoki, T. Age estimation from brain MRI images using deep learning. In Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI), Melbourne, Australia, 18–21 April 2017; pp. 849–852. [Google Scholar]
- Cole, J.H.; Poudel, R.P.; Tsagkrasoulis, D.; Caan, M.W.; Steves, C.; Spector, T.D.; Montana, G. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 2017, 163, 115–124. [Google Scholar] [CrossRef]
- Wang, J.; Knol, M.J.; Tiulpin, A.; Dubost, F.; de Bruijne, M.; Vernooij, M.W.; Adams, H.H.; Ikram, M.A.; Niessen, W.J.; Roshchupkin, G.V. Gray matter age prediction as a biomarker for risk of dementia. Proc. Natl. Acad. Sci. USA 2019, 116, 21213–21218. [Google Scholar] [CrossRef]
- Jónsson, B.A.; Bjornsdottir, G.; Thorgeirsson, T.; Ellingsen, L.M.; Walters, G.B.; Gudbjartsson, D.; Stefansson, H.; Stefansson, K.; Ulfarsson, M. Brain age prediction using deep learning uncovers associated sequence variants. Nat. Commun. 2019, 10, 5409. [Google Scholar] [CrossRef]
- Bashyam, V.M.; Erus, G.; Doshi, J.; Habes, M.; Nasrallah, I.M.; Truelove-Hill, M.; Srinivasan, D.; Mamourian, L.; Pomponio, R.; Fan, Y.; et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain 2020, 143, 2312–2324. [Google Scholar] [CrossRef]
- Peng, H.; Gong, W.; Beckmann, C.F.; Vedaldi, A.; Smith, S.M. Accurate brain age prediction with lightweight deep neural networks. Med. Image Anal. 2021, 68, 101871. [Google Scholar] [CrossRef]
- Bellantuono, L.; Marzano, L.; La Rocca, M.; Duncan, D.; Lombardi, A.; Maggipinto, T.; Monaco, A.; Tangaro, S.; Amoroso, N.; Bellotti, R. Predicting brain age with complex networks: From adolescence to adulthood. NeuroImage 2021, 225, 117458. [Google Scholar] [CrossRef]
- Gupta, U.; Lam, P.K.; Ver Steeg, G.; Thompson, P.M. Improved brain age estimation with slice-based set networks. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 840–844. [Google Scholar]
- Ning, K.; Duffy, B.A.; Franklin, M.; Matloff, W.; Zhao, L.; Arzouni, N.; Sun, F.; Toga, A.W. Improving brain age estimates with deep learning leads to identification of novel genetic factors associated with brain aging. Neurobiol. Aging 2021, 105, 199–204. [Google Scholar] [CrossRef]
- Dinsdale, N.K.; Bluemke, E.; Smith, S.M.; Arya, Z.; Vidaurre, D.; Jenkinson, M.; Namburete, A.I. Learning patterns of the ageing brain in MRI using deep convolutional networks. NeuroImage 2021, 224, 117401. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Burkett, B.J.; Min, H.K.; Senjem, M.L.; Lundt, E.S.; Botha, H.; Graff-Radford, J.; Barnard, L.R.; Gunter, J.L.; Schwarz, C.G.; et al. Deep learning-based brain age prediction in normal aging and dementia. Nat. Aging 2022, 2, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Pilli, R.; Goel, T.; Murugan, R.; Tanveer, M. Association of white matter volume with brain age classification using deep learning network and region wise analysis. Eng. Appl. Artif. Intell. 2023, 125, 106596. [Google Scholar] [CrossRef]
- Tanveer, M.; Ganaie, M.; Beheshti, I.; Goel, T.; Ahmad, N.; Lai, K.T.; Huang, K.; Zhang, Y.D.; Del Ser, J.; Lin, C.T. Deep learning for brain age estimation: A systematic review. Inf. Fusion 2023, 96, 130–143. [Google Scholar] [CrossRef]
- Beheshti, I.; Ganaie, M.; Paliwal, V.; Rastogi, A.; Razzak, I.; Tanveer, M. Predicting brain age using machine learning algorithms: A comprehensive evaluation. IEEE J. Biomed. Health Inform. 2021, 26, 1432–1440. [Google Scholar] [CrossRef]
- Ganaie, M.; Tanveer, M.; Beheshti, I. Brain age prediction with improved least squares twin SVR. IEEE J. Biomed. Health Inform. 2022, 27, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Ganaie, M.; Tanveer, M.; Beheshti, I. Brain age prediction using improved twin SVR. Neural Comput. Appl. 2022, 1–11. [Google Scholar] [CrossRef]
- Cole, J.H.; Ritchie, S.J.; Bastin, M.E.; Hernández, V.; Muñoz Maniega, S.; Royle, N.; Corley, J.; Pattie, A.; Harris, S.E.; Zhang, Q.; et al. Brain age predicts mortality. Mol. Psychiatry 2018, 23, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Franke, K.; Gaser, C. Ten years of BrainAGE as a neuroimaging biomarker of brain aging: What insights have we gained? Front. Neurol. 2019, 10, 789. [Google Scholar] [CrossRef]
- Mei, X.; Liu, Z.; Robson, P.M.; Marinelli, B.; Huang, M.; Doshi, A.; Jacobi, A.; Cao, C.; Link, K.E.; Yang, T.; et al. RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning. Radiol. Artif. Intell. 2022, 4, e210315. [Google Scholar] [CrossRef]
- Langner, T.; Wikström, J.; Bjerner, T.; Ahlström, H.; Kullberg, J. Identifying morphological indicators of aging with neural networks on large-scale whole-body MRI. IEEE Trans. Med. Imaging 2019, 39, 1430–1437. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Alfaro-Almagro, F.; Jenkinson, M.; Bangerter, N.K.; Andersson, J.L.; Griffanti, L.; Douaud, G.; Sotiropoulos, S.N.; Jbabdi, S.; Hernandez-Fernandez, M.; Vallee, E.; et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 2018, 166, 400–424. [Google Scholar] [CrossRef]
- Littlejohns, T.J.; Holliday, J.; Gibson, L.M.; Garratt, S.; Oesingmann, N.; Alfaro-Almagro, F.; Bell, J.D.; Boultwood, C.; Collins, R.; Conroy, M.C.; et al. The UK Biobank imaging enhancement of 100,000 participants: Rationale, data collection, management and future directions. Nat. Commun. 2020, 11, 2624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Brady, M.; Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 2001, 20, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Bezanson, J.; Edelman, A.; Karpinski, S.; Shah, V.B. Julia: A fresh approach to numerical computing. SIAM Rev. 2017, 59, 65–98. [Google Scholar] [CrossRef]
- Innes, M. Flux: Elegant machine learning with Julia. J. Open Source Softw. 2018, 3, 602. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.E.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Li, X.; Chen, S.; Hu, X.; Yang, J. Understanding the disharmony between dropout and batch normalization by variance shift. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2682–2690. [Google Scholar]
- Bloice, M.D.; Stocker, C.; Holzinger, A. Augmentor: An Image Augmentation Library for Machine Learning. J. Open Source Softw. 2017, 2, 432. [Google Scholar] [CrossRef]
- Fry, A.; Littlejohns, T.J.; Sudlow, C.; Doherty, N.; Adamska, L.; Sprosen, T.; Collins, R.; Allen, N.E. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 2017, 186, 1026–1034. [Google Scholar] [CrossRef] [PubMed]





| Paper/Settings | Approach | N Subjects | Test Accuracy | Parameters | Training Time |
|---|---|---|---|---|---|
| Huang et al., 2017 [2] | 2D slices | 600 | 4.00 MAE | - | 12 h |
| Cole et al., 2017 [3] | 3D CNN | 1601 | 4.16 MAE | 889,960 | 72–332 h |
| Wang et al., 2019 [4] | 3D CNN | 3688 | 4.45 MAE | - | 30 h |
| Jonsson et al., 2019 [5] | 3D CNN | 809 | 3.39 MAE | - | 48 h |
| Bashyam et al., 2020 [6] | 2D slices | 9383 | 3.70 MAE | - | 10 h |
| Peng et al., 2021 [7] | 3D CNN | 12,949 | 2.14 MAE | 3 million | 130 h |
| Bellantuono et al., 2021 [8] | Dense | 800 | 2.19 MAE | - | - |
| Gupta et al., 2021 [9] | 2D slices | 7312 | 2.82 MAE | 998,625 | 6.75 h |
| Ning et al., 2021 [10] | 3D CNN | 13,598 | 2.70 MAE | - | 96 h |
| Dinsdale et al., 2021 [11] | 3D CNN | 12,802 | 2.90 MAE | - | - |
| Lee et al., 2022 [12] | 3D CNN | 1805 | 3.49 MAE | 70,183,073 | 24 h |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.55 (4.49) | 2,009,261 | 22 min (3 h 53 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.51 (4.43) | 2,009,261 | 24 min (3 h 30 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.53 (4.44) | 2,009,369 | 24 min (3 h 26 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.46 (4.38) | 827,841 | 25 min (4 h 36 min) |
| Dropout between dense | |||||
| 0.3 dropout rate | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.70 (4.66) | 2,009,261 | 22 min (3 h 12 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.67 (4.62) | 2,009,261 | 27 min (4 h 27 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.56 (4.47) | 2,009,369 | 27 min (3 h 32 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.63 (4.56) | 827,841 | 28 min (4 h 23 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| trained with augmentation | |||||
| Ours, 3 mean channels | 2D proj | 20,324 1 | 3.44 (4.31) | 2,009,261 | > 3 days 2 |
| Ours, 3 std channels | 2D proj | 20,324 1 | 3.40 (4.33) | 2,009,261 | > 3 days 2 |
| Ours, all 6 channels | 2D proj | 20,324 1 | 3.47 (4.40) | 2,009,369 | > 3 days 2 |
| Ours, all 6 channels, iso | 2D proj | 20,324 1 | 3.85 (4.80) | 827,841 | > 3 days 2 |
| Settings | Approach | N Subjects | Test Accuracy | Parameters | Training Time |
|---|---|---|---|---|---|
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| trained using only | |||||
| 2000 subjects | |||||
| Ours, 3 mean channels | 2D proj | 2000 | 4.05 (5.09) | 2,009,261 | 18 min (22 min) |
| Ours, 3 std channels | 2D proj | 2000 | 4.01 (5.08) | 2,009,261 | 20 min (22 min) |
| Ours, all 6 channels | 2D proj | 2000 | 4.06 (5.13) | 2,009,369 | 7 min (22 min) |
| Ours, all 6 channels, iso | 2D proj | 2000 | 4.13 (5.18) | 827,841 | 8 min (27 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| trained using only | |||||
| 6376 subjects | |||||
| Ours, 3 mean channels | 2D proj | 6376 | 3.75 (4.74) | 2,009,261 | 7 min (58 min) |
| Ours, 3 std channels | 2D proj | 6376 | 3.73 (4.72) | 2,009,261 | 4 min (58 min) |
| Ours, all 6 channels | 2D proj | 6376 | 3.73 (4.73) | 2,009,369 | 50 min (1 h 7 min) |
| Ours, all 6 channels, iso | 2D proj | 6376 | 3.77 (4.75) | 827,841 | 53 min (1 h 16 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| half as many filters | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.61 (4.51) | 505,037 | 37 min (2 h 40 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.61 (4.57) | 505,037 | 43 min (3 h 3 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.49 (4.40) | 505,091 | 17 min (3 h 10 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.49 (4.39) | 209,167 | 40 min (4 h 52 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| twice as many filters | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.45 (4.39) | 8,015,333 | 25 min (4 h 51 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.45 (4.37) | 8,015,333 | 23 min (4 h 52 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.40 (4.30) | 8,015,549 | 23 min (4 h 55 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.42 (4.33) | 3,293,773 | 19 min (5 h 39 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| with 19 convolution layers | |||||
| per stack rather than 13 | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.56 (4.50) | 2,599,697 | 37 min (4 h 24 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.49 (4.40) | 2,599,697 | 50 min (4 h 39 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.40 (4.28) | 2,599,805 | 31 min (4 h 43 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.37 (4.26) | 1,024,653 | 60 min (5 h 44 min) |
| Dropout between conv | |||||
| 0.2 dropout rate | |||||
| with 25 convolution layers | |||||
| per stack rather than 13 | |||||
| Ours, 3 mean channels | 2D proj | 20,324 | 3.49 (4.41) | 3,189,985 | 1 h 22 min (5 h 29 min) |
| Ours, 3 std channels | 2D proj | 20,324 | 3.47 (4.38) | 3,189,985 | 1 h 20 min (5 h 27 min) |
| Ours, all 6 channels | 2D proj | 20,324 | 3.50 (4.47) | 3,190,093 | 1 h 37 min (5 h 46 min) |
| Ours, all 6 channels, iso | 2D proj | 20,324 | 3.48 (4.38) | 1,221,465 | 1 h 14 min (7 h 26 min) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jönemo, J.; Akbar, M.U.; Kämpe, R.; Hamilton, J.P.; Eklund, A. Efficient Brain Age Prediction from 3D MRI Volumes Using 2D Projections. Brain Sci. 2023, 13, 1329. https://doi.org/10.3390/brainsci13091329
Jönemo J, Akbar MU, Kämpe R, Hamilton JP, Eklund A. Efficient Brain Age Prediction from 3D MRI Volumes Using 2D Projections. Brain Sciences. 2023; 13(9):1329. https://doi.org/10.3390/brainsci13091329
Chicago/Turabian StyleJönemo, Johan, Muhammad Usman Akbar, Robin Kämpe, J. Paul Hamilton, and Anders Eklund. 2023. "Efficient Brain Age Prediction from 3D MRI Volumes Using 2D Projections" Brain Sciences 13, no. 9: 1329. https://doi.org/10.3390/brainsci13091329
APA StyleJönemo, J., Akbar, M. U., Kämpe, R., Hamilton, J. P., & Eklund, A. (2023). Efficient Brain Age Prediction from 3D MRI Volumes Using 2D Projections. Brain Sciences, 13(9), 1329. https://doi.org/10.3390/brainsci13091329




