Method Validation and Characterization of the Associated Uncertainty for Malondialdehyde Quantification in Exhaled Breath Condensate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents-Chemicals
2.2. Preparation of Standards and Procedural Blanks
2.3. EBC Samples and Quality Control (QC)
2.4. MDA Derivatization with DNPH
2.5. HPLC–MS/MS Analyses
2.6. Method Validation and Estimation of Its Expanded Uncertainty
2.7. Statistical Analysis
3. Results
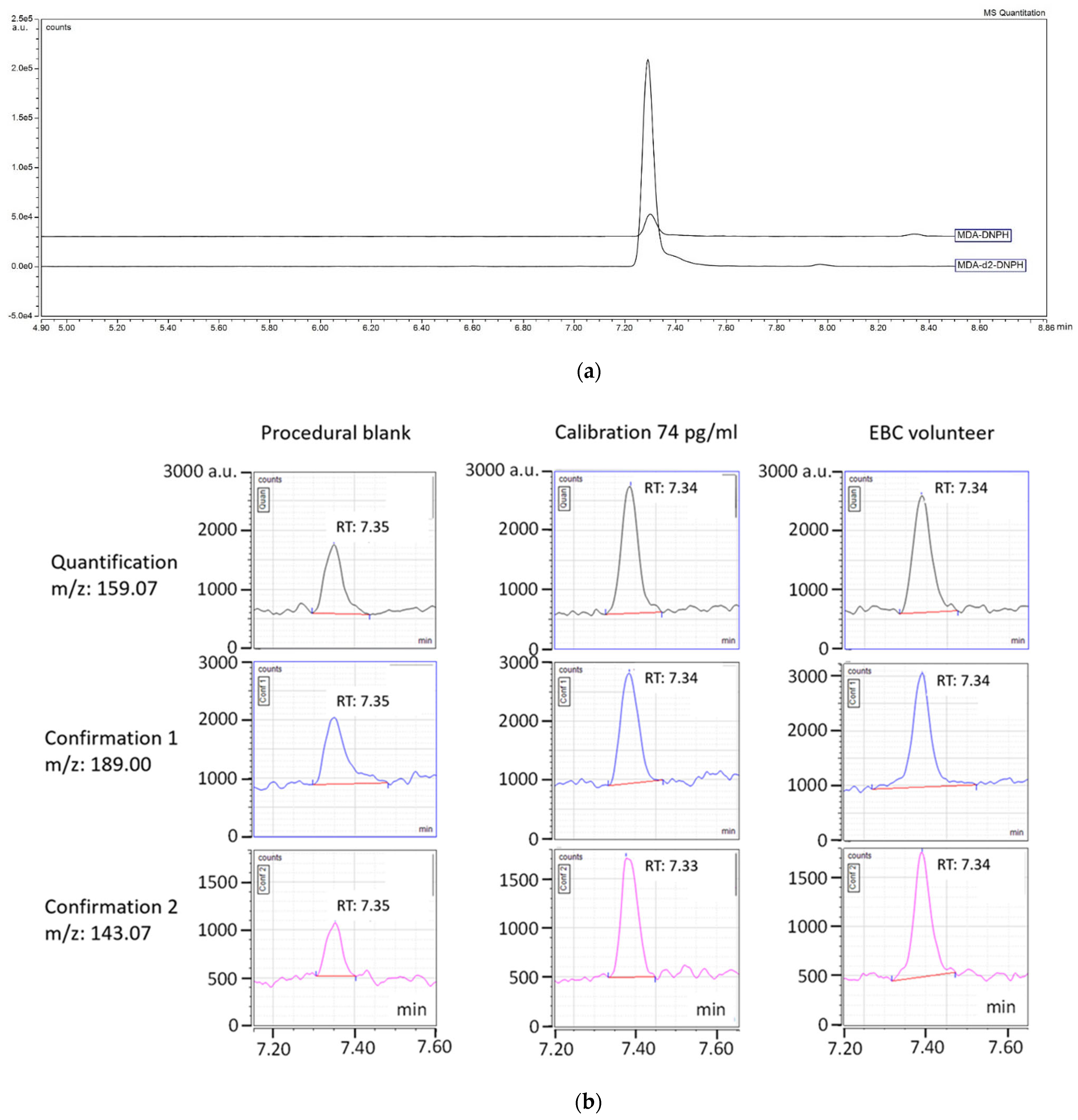
3.1. LC-MS/MS Analysis
3.2. Optimization of the DNPH Derivatization
3.3. Method Validation and Estimation of Uncertainty
3.4. Uncertainty
3.5. Levels of MDA in EBC of Healthy Adult Workers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Diseases, G.B.D.; Injuries, C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 4th ed.; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Pelclova, D.; Zdimal, V.; Kacer, P.; Zikova, N.; Komarc, M.; Fenclova, Z.; Vlckova, S.; Schwarz, J.; Makeš, O.; Syslova, K.; et al. Markers of lipid oxidative damage in the exhaled breath condensate of nano TiO2 production workers. Nanotoxicology 2017, 11, 52–63. [Google Scholar] [CrossRef]
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef]
- Kim, S.-S.; Gallaher, D.D.; Csallany, A.S. Lipophilic aldehydes and related carbonyl compounds in rat and human urine. Lipids 1999, 34, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Reitznerová, A.; Šuleková, M.; Nagy, J.; Marcinčák, S.; Semjon, B.; Čertík, M.; Klempová, T. Lipid Peroxidation Process in Meat and Meat Products: A Comparison Study of Malondialdehyde Determination between Modified 2-Thiobarbituric Acid Spectrophotometric Method and Reverse-Phase High-Performance Liquid Chromatography. Molecules 2017, 22, 1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilz, J.; Meineke, I.; Gleiter, C.H. Measurement of free and bound malondialdehyde in plasma by high-performance liquid chromatography as the 2,4-dinitrophenylhydrazine derivative. J. Chromatogr. B Biomed. Sci. Appl. 2000, 742, 315–325. [Google Scholar] [CrossRef]
- Esterbauer, H.; Cheeseman, K.H. Determination of aldehydic lipid peroxidation products: Malonaldehyde and 4-hydroxynonenal. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1990; Volume 186, pp. 407–421. [Google Scholar]
- Czauderna, M.; Kowalczyk, J.; Marounek, M. The simple and sensitive measurement of malondialdehyde in selected specimens of biological origin and some feed by reversed phase high performance liquid chromatography. J. Chromatogr. B 2011, 879, 2251–2258. [Google Scholar] [CrossRef] [PubMed]
- Kadiiska, M.B.; Gladen, B.C.; Baird, D.D.; Germolec, D.; Graham, L.B.; Parker, C.E.; Nyska, A.; Wachsman, J.T.; Ames, B.N.; Basu, S.; et al. Biomarkers of oxidative stress study II: Are oxidation products of lipids, proteins, and DNA markers of CCl4 poisoning? Free Radic. Biol. Med. 2005, 38, 698–710. [Google Scholar] [CrossRef]
- Gönenç, A.; Ozkan, Y.; Torun, M.; Simşek, B. Plasma malondialdehyde (MDA) levels in breast and lung cancer patients. J. Clin. Pharm. Ther. 2001, 26, 141–144. [Google Scholar] [CrossRef]
- Corradi, M.; Folesani, G.; Andreoli, R.; Manini, P.; Bodini, A.; Piacentini, G.; Carraro, S.; Zanconato, S.; Baraldi, E. Aldehydes and glutathione in exhaled breath condensate of children with asthma exacerbation. Am. J. Respir. Crit. Care Med. 2003, 167, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Shin, J.H.; Hwang, J.H.; Baek, J.E.; Choi, B.S. Malondialdehyde and 3-nitrotyrosine in exhaled breath condensate in retired elderly coal miners with chronic obstructive pulmonary disease. Saf. Health Work 2014, 5, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Paliogiannis, P.; Fois, A.G.; Sotgia, S.; Mangoni, A.A.; Zinellu, E.; Pirina, P.; Carru, C.; Zinellu, A. Circulating malondialdehyde concentrations in patients with stable chronic obstructive pulmonary disease: A systematic review and meta-analysis. Biomark. Med. 2018, 12, 771–781. [Google Scholar] [CrossRef]
- Syslová, K.; Kacer, P.; Kuzma, M.; Najmanová, V.; Fenclová, Z.; Vlcková, S.; Lebedová, J.; Pelclová, D. Rapid and easy method for monitoring oxidative stress markers in body fluids of patients with asbestos or silica-induced lung diseases. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2009, 877, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, P.K.; Laitinen, A.; Venge, P. Biopsy markers of airway inflammation and remodelling. Respir. Med. 2000, 94, S9–S15. [Google Scholar] [CrossRef] [Green Version]
- Machado, R.F.; Laskowski, D.; Deffenderfer, O.; Burch, T.; Zheng, S.; Mazzone, P.J.; Mekhail, T.; Jennings, C.; Stoller, J.K.; Pyle, J.; et al. Detection of lung cancer by sensor array analyses of exhaled breath. Am. J. Respir. Crit. Care Med. 2005, 171, 1286–1291. [Google Scholar] [CrossRef] [Green Version]
- Janicka, M.; Kubica, P.; Kot-Wasik, A.; Kot, J.; Namieśnik, J. Sensitive determination of isoprostanes in exhaled breath condensate samples with use of liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 893–894, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Barnes, P.J. Analysis of exhaled breath condensate for monitoring airway inflammation. Trends Pharmacol. Sci. 2002, 23, 232–237. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Richardson, S.D.; Caughran, T.V.; Poiger, T.; Guo, Y.; Crumley, F.G. Application of DNPH Derivatization with LC/MS to the Identification of Polar Carbonyl Disinfection Byproducts in Drinking Water. Ozone Sci. Eng. 2000, 22, 653–675. [Google Scholar] [CrossRef]
- Martinez, M.P.; Kannan, K. Simultaneous Analysis of Seven Biomarkers of Oxidative Damage to Lipids, Proteins, and DNA in Urine. Environ. Sci. Technol. 2018, 52, 6647–6655. [Google Scholar] [CrossRef]
- Lärstad, M.; Ljungkvist, G.; Olin, A.C.; Torén, K. Determination of malondialdehyde in breath condensate by high-performance liquid chromatography with fluorescence detection. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2002, 766, 107–114. [Google Scholar] [CrossRef]
- Lykkesfeldt, J. Determination of Malondialdehyde as Dithiobarbituric Acid Adduct in Biological Samples by HPLC with Fluorescence Detection: Comparison with Ultraviolet-Visible Spectrophotometry. Clin. Chem. 2001, 47, 1725–1727. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Augusto, O.; Brigelius-Flohe, R.; Dennery, P.A.; Kalyanaraman, B.; Ischiropoulos, H.; Mann, G.E.; Radi, R.; Roberts, L.J., 2nd; Vina, J.; et al. Even free radicals should follow some rules: A guide to free radical research terminology and methodology. Free Radic. Biol. Med. 2015, 78, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Korchazhkina, O.; Exley, C.; Andrew Spencer, S. Measurement by reversed-phase high-performance liquid chromatography of malondialdehyde in normal human urine following derivatisation with 2,4-dinitrophenylhydrazine. J. Chromatogr. B 2003, 794, 353–362. [Google Scholar] [CrossRef]
- Andreoli, R.; Manini, P.; Corradi, M.; Mutti, A.; Niessen, W.M.A. Determination of patterns of biologically relevant aldehydes in exhaled breath condensate of healthy subjects by liquid chromatography/atmospheric chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 637–645. [Google Scholar] [CrossRef] [Green Version]
- Kartavenka, K.; Panuwet, P.; Greenwald, R.; Ehret, K.M.; D’Souza, P.E.; Barr, D.B.; Ryan, P.B. Quantification of malondialdehyde in exhaled breath condensate using pseudo two-dimensional ultra-performance liquid chromatography coupled with single quadrupole mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1105, 210–216. [Google Scholar] [CrossRef]
- Ellison, S.L.R.; Williams, A. Quantifying Uncertainty in Analytical Measurement. Eurachem-CITAC Guide CG4. 2012. Available online: https://www.eurachem.org/images/stories/Guides/pdf/QUAM2012_P1.pdf (accessed on 1 November 2020).
- Chen, J.-L.; Huang, Y.-J.; Pan, C.-H.; Hu, C.-W.; Chao, M.-R. Determination of urinary malondialdehyde by isotope dilution LC-MS/MS with automated solid-phase extraction: A cautionary note on derivatization optimization. Free Radic. Biol. Med. 2011, 51, 1823–1829. [Google Scholar] [CrossRef]
- Guseva Canu, I.; Hemmendinger, M.; Sauvain, J.J.; Suarez, G.; Hopf, N.B.; Pralong, J.A.; Ben Rayana, T.; Besançon, S.; Sakthithasan, K.; Jouannique, V.; et al. Respiratory Disease Occupational Biomonitoring Collaborative Project (ROBoCoP): A longitudinal pilot study and implementation research in the Parisian transport company. J. Occup. Med. Toxicol. 2021, 16, 22. [Google Scholar] [CrossRef]
- Horváth, I.; Barnes, P.J.; Loukides, S.; Sterk, P.J.; Högman, M.; Olin, A.C.; Amann, A.; Antus, B.; Baraldi, E.; Bikov, A.; et al. A European Respiratory Society technical standard: Exhaled biomarkers in lung disease. Eur. Respir. J. 2017, 49, 1600965. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration US. Bioanalytical Method Validation Guidance for Industry. US Dep. Health Hum. Serv. 2018, 44, 1–41. [Google Scholar]
- Gu, H.; Liu, G.; Wang, J.; Aubry, A.F.; Arnold, M.E. Selecting the correct weighting factors for linear and quadratic calibration curves with least-squares regression algorithm in bioanalytical LC-MS/MS assays and impacts of using incorrect weighting factors on curve stability, data quality, and assay performance. Anal. Chem. 2014, 86, 8959–8966. [Google Scholar] [CrossRef]
- Hornung, R.W.; Reed, L.D. Estimation of Average Concentration in the Presence of Nondetectable Values. Appl. Occup. Environ. Hyg. 1990, 5, 46–51. [Google Scholar] [CrossRef]
- Chen, I.C.; Bertke, S.J.; Curwin, B.D. Quantile regression for exposure data with repeated measures in the presence of non-detects. J. Expo. Sci. Environ. Epidemiol. 2021, 31, 1–10. [Google Scholar] [CrossRef]
- Manini, P.; Andreoli, R.; Sforza, S.; Dall’Asta, C.; Galaverna, G.; Mutti, A.; Niessen, W.M.A. Evaluation of Alternate Isotope-Coded Derivatization Assay (AIDA) in the LC–MS/MS analysis of aldehydes in exhaled breath condensate. J. Chromatogr. B 2010, 878, 2616–2622. [Google Scholar] [CrossRef] [PubMed]
- Casimirri, E.; Stendardo, M.; Bonci, M.; Andreoli, R.; Bottazzi, B.; Leone, R.; Schito, M.; Vaccari, A.; Papi, A.; Contoli, M.; et al. Biomarkers of oxidative-stress and inflammation in exhaled breath condensate from hospital cleaners. Biomarkers 2016, 21, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Pelclova, D.; Zdimal, V.; Kacer, P.; Fenclova, Z.; Vlckova, S.; Syslova, K.; Navratil, T.; Schwarz, J.; Zikova, N.; Barosova, H.; et al. Oxidative stress markers are elevated in exhaled breath condensate of workers exposed to nanoparticles during iron oxide pigment production. J. Breath Res. 2016, 10, 016004. [Google Scholar] [CrossRef]
- Mendonça, R.; Gning, O.; Di Cesaré, C.; Lachat, L.; Bennett, N.C.; Helfenstein, F.; Glauser, G. Sensitive and selective quantification of free and total malondialdehyde in plasma using UHPLC-HRMS. J. Lipid Res. 2017, 58, 1924–1931. [Google Scholar] [CrossRef] [Green Version]
- Beyer, H.; John, W. Exploratory Data Analysis. Addison-Wesley Publishing Company Reading, Mass—Menlo Park, Cal., London, Amsterdam, Don Mills, Ontario, Sydney 1977, XVI, 688 S. Biom. J. 1981, 23, 413–414. [Google Scholar] [CrossRef]
- Sanagi, M.M.; Ling, S.L.; Nasir, Z.; Hermawan, D.; Ibrahim, W.A.; Abu Naim, A. Comparison of signal-to-noise, blank determination, and linear regression methods for the estimation of detection and quantification limits for volatile organic compounds by gas chromatography. J. AOAC Int. 2009, 92, 1833–1838. [Google Scholar] [CrossRef] [Green Version]
- Claeson, K.; Thorsén, G.; Karlberg, B. Methyl malondialdehyde as an internal standard for the determination of malondialdehyde. J. Chromatogr. B Biomed. Sci. Appl. 2001, 751, 315–323. [Google Scholar] [CrossRef]
- Cighetti, G.; Allevi, P.; Anastasia, L.; Bortone, L.; Paroni, R. Use of methyl malondialdehyde as an internal standard for malondialdehyde detection: Validation by isotope-dilution gas chromatography-mass spectrometry. Clin. Chem. 2002, 48, 2266–2269. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, S.; Ando, M.; Aoyagi, S. Isomerization of aldehyde-2,4-dinitrophenylhydrazone derivatives and validation of high-performance liquid chromatographic analysis. J. Chromatogr. A 2003, 996, 95–102. [Google Scholar] [CrossRef]
- Corradi, M.; Pignatti, P.; Manini, P.; Andreoli, R.; Goldoni, M.; Poppa, M.; Moscato, G.; Balbi, B.; Mutti, A. Comparison between exhaled and sputum oxidative stress biomarkers in chronic airway inflammation. Eur. Respir. J. 2004, 24, 1011–1017. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Moral, M.-P.; Kannan, K. How stable is oxidative stress level? An observational study of intra- and inter-individual variability in urinary oxidative stress biomarkers of DNA, proteins, and lipids in healthy individuals. Environ. Int. 2019, 123, 382–389. [Google Scholar] [CrossRef]




| Method Characteristics | Expected Performance | Observed Value | Conclusion |
|---|---|---|---|
| Calibration function (n = 12) | - | - | Linear |
| Concentration range (SD) 1 | |||
| Error for SD 1 = 74 pg·mL−1 (n = 6) | 20% | 11.1 ± 7.4% | Verified |
| Error for SD 2 = 148 pg·mL−1 (n = 6) | 10% | 5.9 ± 3.5% | Verified |
| Error for SD 3 = 370 pg·mL−1 (n = 6) | 10% | 2.2 ± 1.1% | Verified |
| Error for SD 4 = 740 pg·mL−1 (n = 6) | 10% | 2.7 ± 2.2% | Verified |
| Error for SD 5 = 1110 pg·mL−1 (n = 6) | 10% | 2.2 ± 1.4% | Verified |
| Error for SD 6 = 1480 pg·mL−1 (n = 6) | 10% | 2.2 ± 1.4% | Verified |
| Error for SD 7 = 2220 pg·mL−1 (n = 6) | 10% | 3.0 ± 4.5% | Verified |
| LOQ | 211 pg·mL−1 | ||
| Maximum acceptable deviation | 20% | 13.1% 2 | Verified |
| Recovery | |||
| EBC not spiked (n = 12) | 98.63 pg·mL−1 | ||
| EBC level 1 spiked with 12 pg (n = 15) | 211.2 pg·mL−1 | 209.1 ± 39.9 pg·mL−1 | |
| EBC level 2 spiked with 24 pg (n = 15) | 349.3 pg·mL−1 | 371.4 ± 40.8 pg·mL−1 | |
| EBC level 3 spiked with 62 pg (n = 15) | 642.2 pg·mL−1 | 651.2 ± 49.6 pg·mL−1 | |
| EBC level 4 spiked with 124 pg (n = 15) | 1180.9 pg·mL−1 | 1161.4 ± 60.4 pg·mL−1 | |
| EBC level 5 spiked with 248 pg (n = 15) | 2258.3 pg·mL−1 | 2070.4 ± 175.7 pg·mL−1 | |
| Recovery EBC spiked level 1 | 90–110% | 92.4 ± 13.0% | Verified |
| Recovery EBC spiked level 2 | 90–110% | 106.5 ± 2.8% | Verified |
| Recovery EBC spiked level 3 | 90–110% | 101.4 ± 4.0% | Verified |
| Recovery EBC spiked level 4 | 90–110% | 98.3 ± 0.9% | Verified |
| Recovery EBC spiked level 5 | 90–110% | 93.5 ± 7.3% | Verified |
| Repeatability | |||
| EBC spiked level 1 (n = 15) | 20% | 9.4% | Verified |
| EBC spiked level 2 (n = 15) | 15% | 7.9% | Verified |
| EBC spiked level 3 (n = 15) | 15% | 4.8% | Verified |
| EBC spiked level 4 (n = 15) | 15% | 4.3% | Verified |
| EBC spiked level 5 (n = 15) | 15% | 2.7% | Verified |
| Inter-day precision | |||
| EBC spiked level 1 (n = 15) | 20% | 19.6% | Verified |
| EBC spiked level 2 (n = 15) | 15% | 8.9% | Verified |
| EBC spiked level 3 (n = 15) | 15% | 7.0% | Verified |
| EBC spiked level 4 (n = 15) | 15% | 3.4% | Verified |
| EBC spiked level 5 (n = 15) | 15% | 9.5% | Verified |
| Professional Category 1 | Professional Category 2 | Professional Category 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Participant N° | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| MDA mean concentration (pg/mL) (number of replicates) | 130 a (19) | 255 (19) | 393 (20) | 75 a (18) | 110 a (18) | 170 a (17) | <LOD (18) | <LOD (18) | 155 a (17) |
| Minimum (pg/mL) | 78 a | <LOD | 95 a | <LOD | <LOD | 115 a | <LOD | <LOD | 85 a |
| Maximum (pg/mL) | 339 | 597 | 886 | 154 a | 236 | 345 | 141a | 229 | 419 |
| Intra-individual variability (%) (number of replicates) | 46 (19) | 60 (19) | 68 (20) | 39 (18) | 35 (18) | 33 (17) | 47 (18) | 85 (18) | 49 (17) |
| Inter-individual variability (%)b | 51 | 41 | 57 | ||||||
| Reference | Technique | EBC Volume (µL) | LOD (pg/mL) | LOQ (pg/mL) | Intermediate Precision (%) | Recovery (%) | N a | EBC Levels (pg/mL) |
|---|---|---|---|---|---|---|---|---|
| This study | LC-ESI-MS/MS | 125 | 70 | 211 | 9 | 92 | 164 | 115 (<LOD-886) b |
| [29] | LC-ESI-MS | 100 | 42 | 126 | 12 | 92 | 205 | 491 (294–711) c |
| [28] | LC-APCI-MS/MS | 100 | 72 | 216 | 7 | n.a. | 12 | 806 (77–1685) b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hemmendinger, M.; Sauvain, J.-J.; Hopf, N.B.; Wild, P.; Suárez, G.; Guseva Canu, I. Method Validation and Characterization of the Associated Uncertainty for Malondialdehyde Quantification in Exhaled Breath Condensate. Antioxidants 2021, 10, 1661. https://doi.org/10.3390/antiox10111661
Hemmendinger M, Sauvain J-J, Hopf NB, Wild P, Suárez G, Guseva Canu I. Method Validation and Characterization of the Associated Uncertainty for Malondialdehyde Quantification in Exhaled Breath Condensate. Antioxidants. 2021; 10(11):1661. https://doi.org/10.3390/antiox10111661
Chicago/Turabian StyleHemmendinger, Maud, Jean-Jacques Sauvain, Nancy B. Hopf, Pascal Wild, Guillaume Suárez, and Irina Guseva Canu. 2021. "Method Validation and Characterization of the Associated Uncertainty for Malondialdehyde Quantification in Exhaled Breath Condensate" Antioxidants 10, no. 11: 1661. https://doi.org/10.3390/antiox10111661
APA StyleHemmendinger, M., Sauvain, J. -J., Hopf, N. B., Wild, P., Suárez, G., & Guseva Canu, I. (2021). Method Validation and Characterization of the Associated Uncertainty for Malondialdehyde Quantification in Exhaled Breath Condensate. Antioxidants, 10(11), 1661. https://doi.org/10.3390/antiox10111661






