Over-Expression of Chorismate Mutase Enhances the Accumulation of Salicylic Acid, Lignin, and Antioxidants in Response to the White-Backed Planthopper in Rice Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Growth Conditions and Phenotypic Evaluation
2.2. Experimental Design
2.3. RNA Isolation and qRT-PCR
2.4. Measurement of Lignin Content
2.5. Quantification of Endogenous Salicylic Acid
2.6. Histological Staining
2.7. Antioxidant Enzyme Assay
- ΔA340 = A340/min (blank) − A340/min (sample);
- 6.22 = ƐmM for NADPH;
- DF = dilution factor of sample before adding to reaction;
- V = sample volume in mL.
- Sa = amount of MDA in unknown sample (nmole);
- Sv = sample volume added into each well (mL);
- D = sample dilution factor;
- C = concentration of MDA in the sample.
2.8. Amino Acid Isolation and Chlorophyll Content
3. Statistical Analysis
4. Results
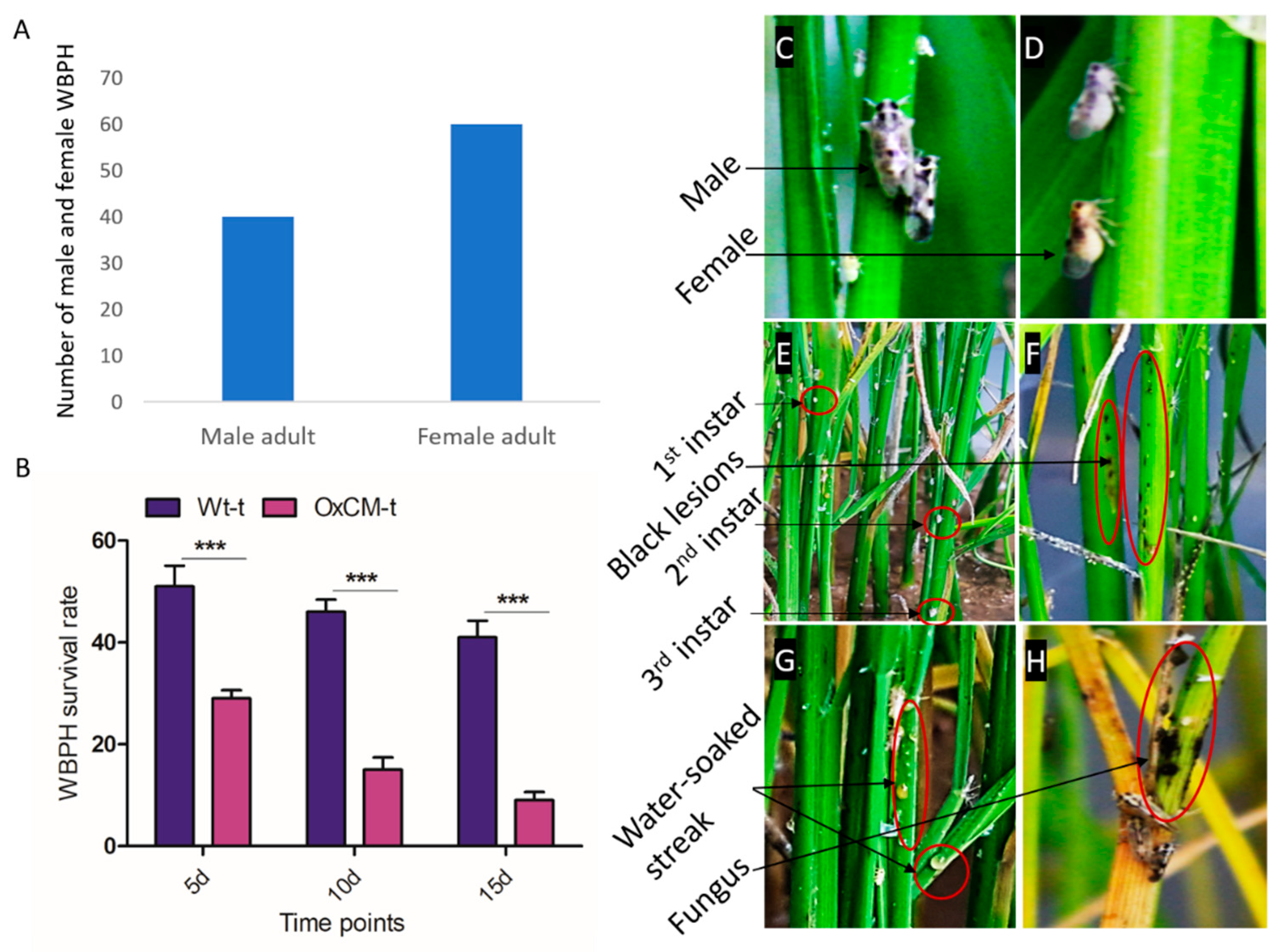
4.1. Characterization of Symptoms and Survival Rate of White-Backed Planthopper
4.2. White-Backed Planthopper Population Growth and the Resistance of Chorismate Mutase Transgenic Over-Expressor Plants
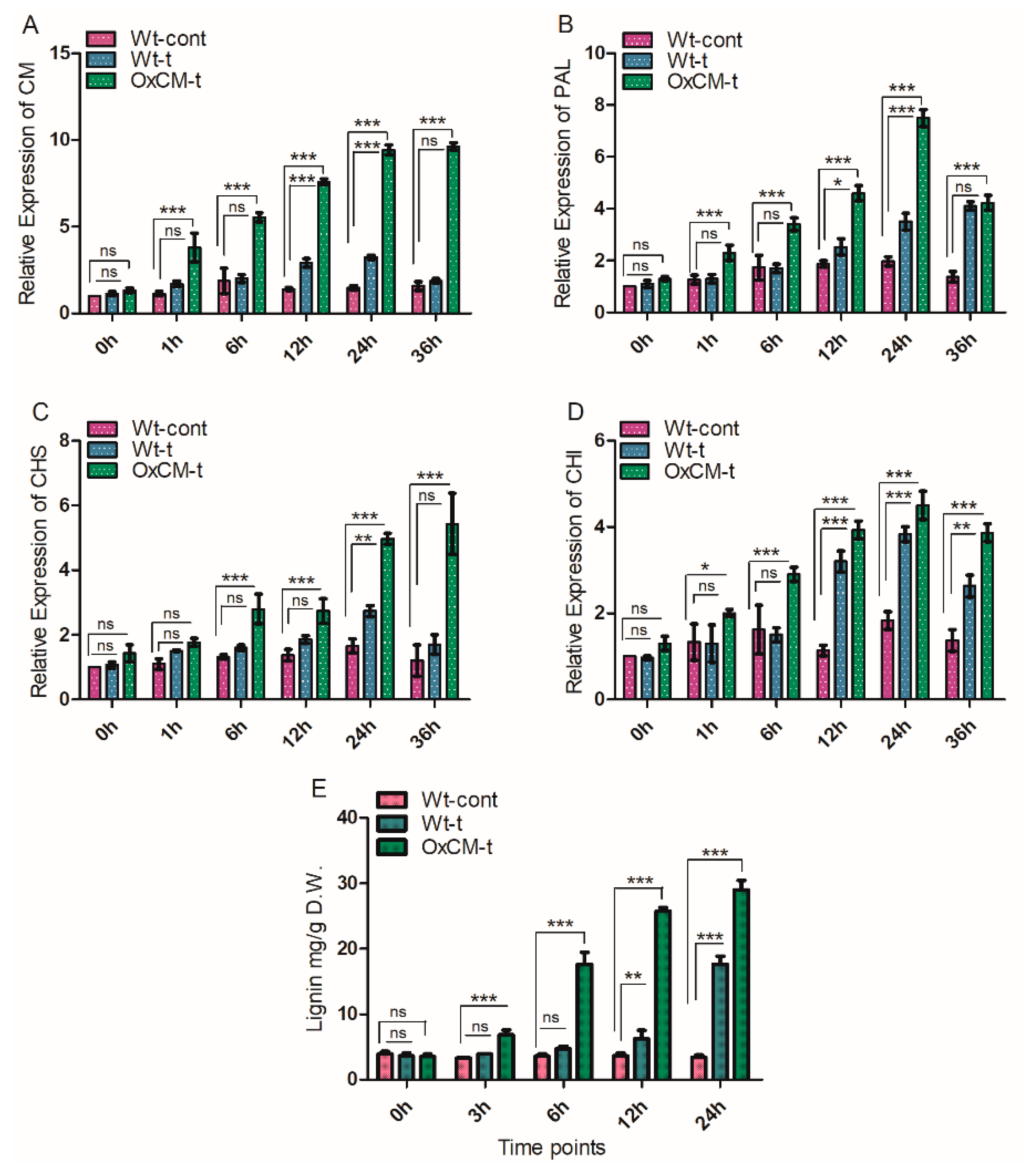
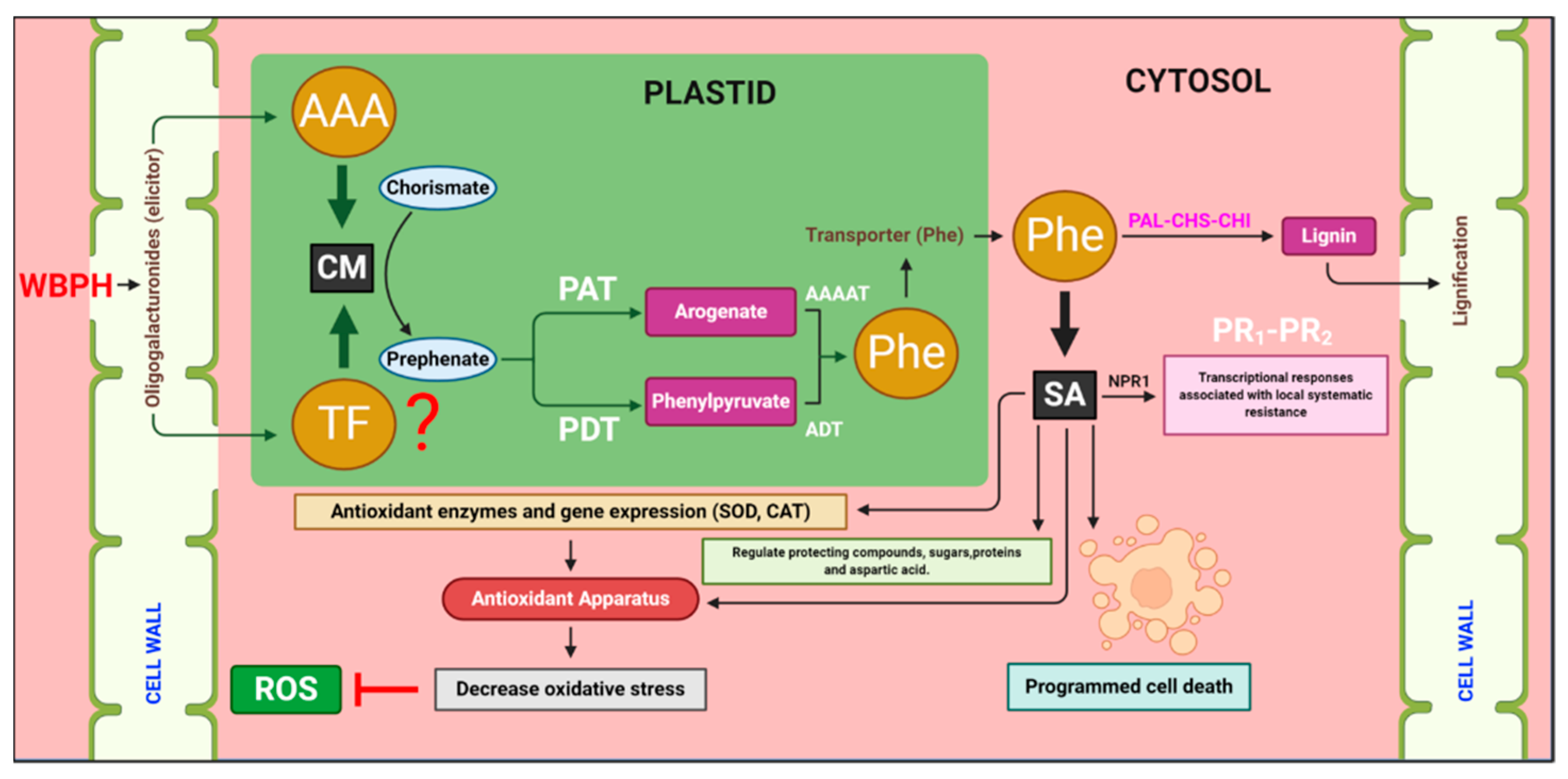
4.3. Chorismate Mutase Enhances Lignin Contents through the Induction of Downstream Genes
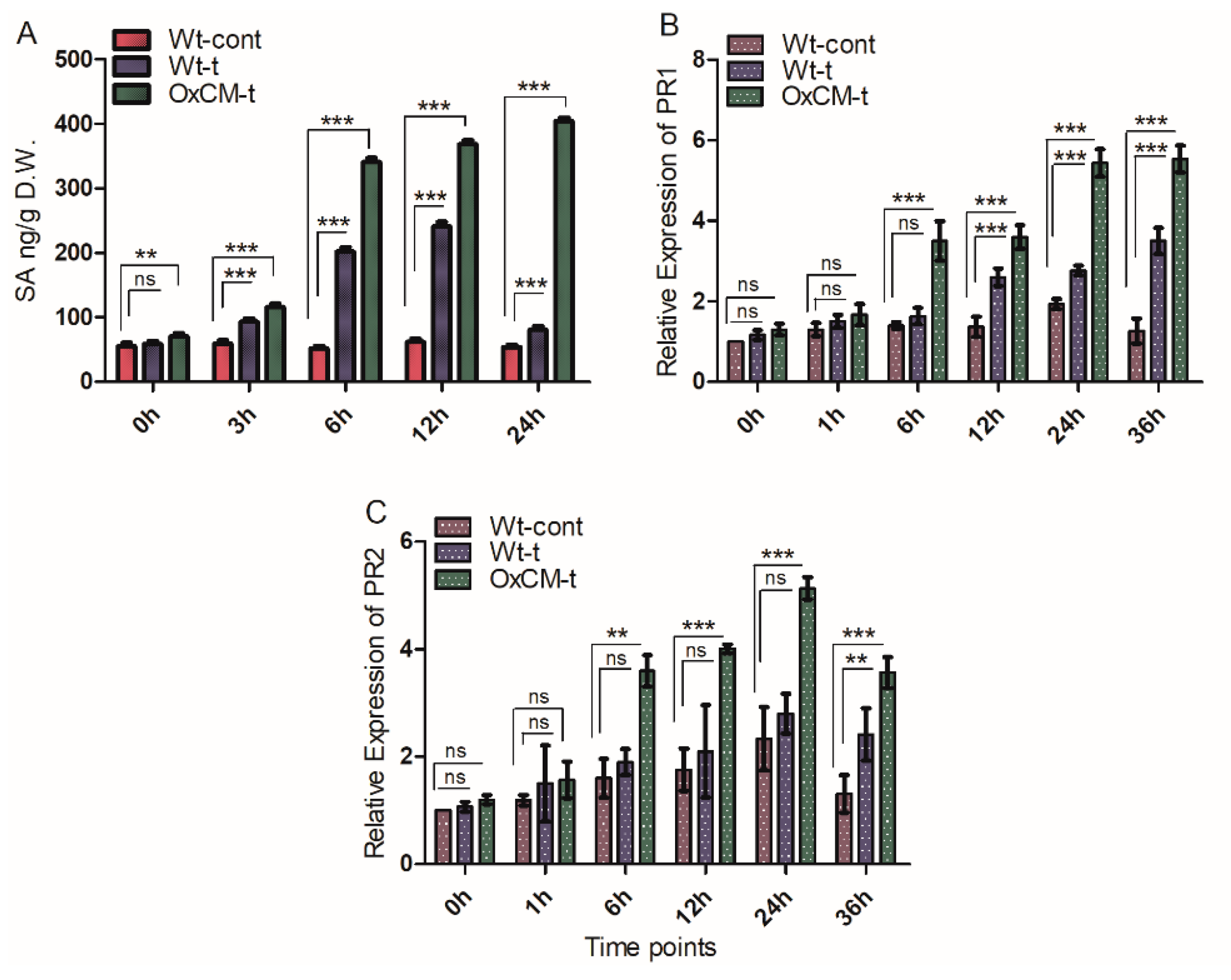
4.4. Salicylic Acid Stimulates PR1 and PR2 Genes in Response to White-Backed Planthopper Infestation
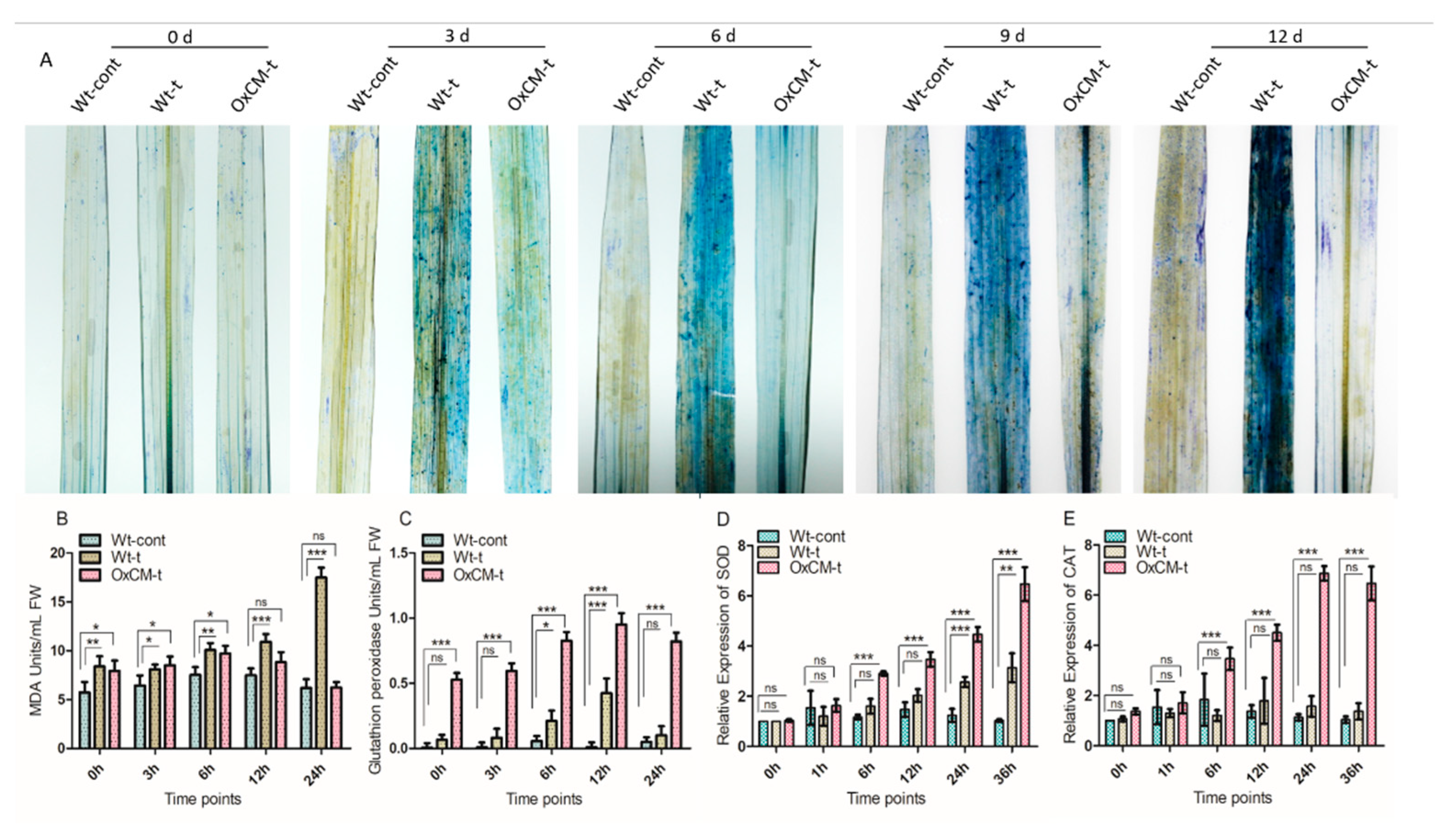
4.5. Hypersensitive Responses and Regulation of Antioxidant Apparatus
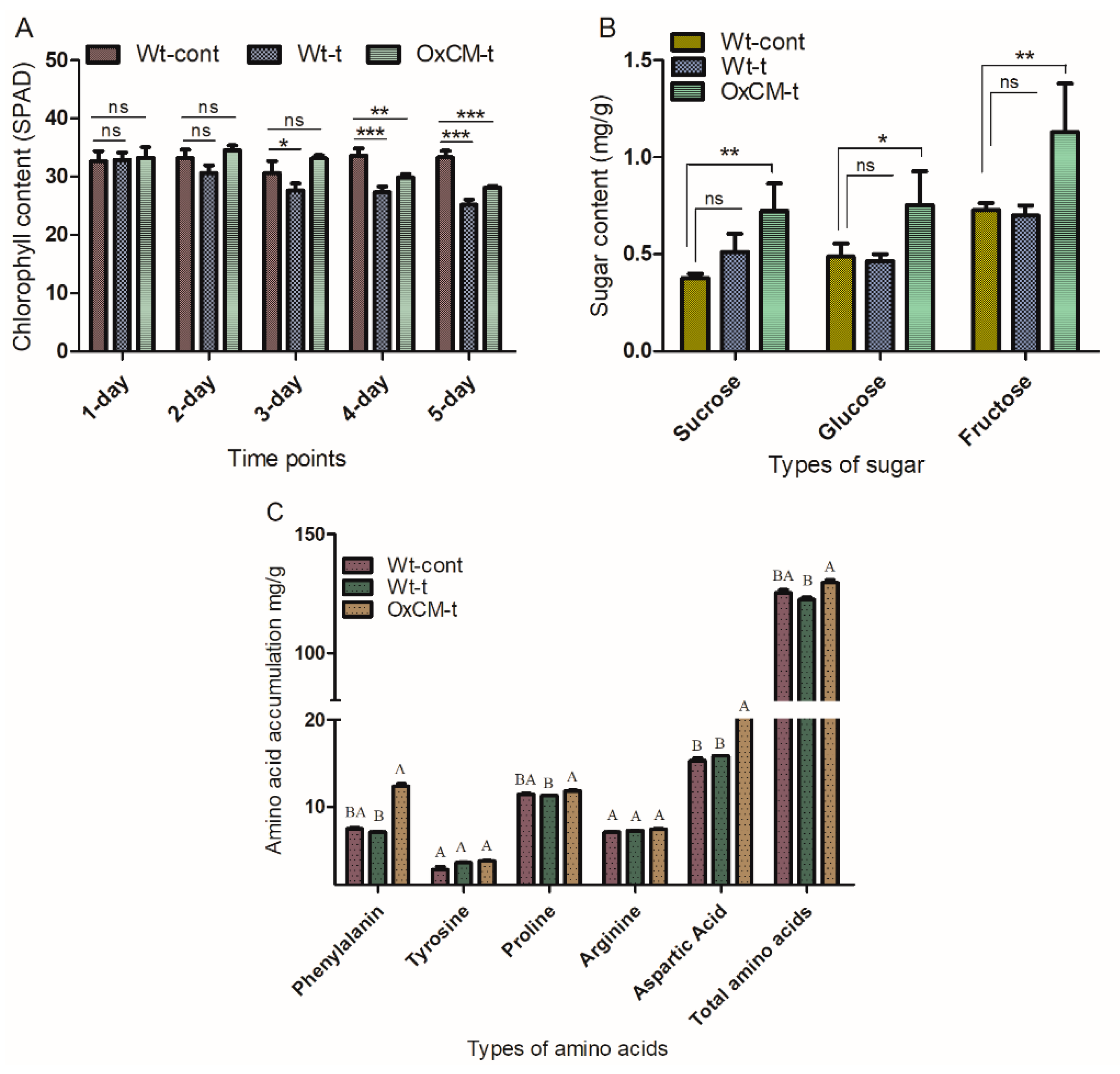
4.6. White-Backed Planthoppers Alter Chlorophyll, Sugar and Amino Acid Contents
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Watanabe, T.; Kitagawa, H. Photosynthesis and translocation of assimilates in rice plants following phloem feeding by the planthopper Nilaparvata lugens (Homoptera: Delphacidae). J. Econ. Entomol. 2000, 93, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Ram, L.; Kumar, A.; Yadav, S.; Singh, B.; Kalkal, D. Biology of whitebacked plant hopper, Sogatella furcifera on basmati rice under agroclimatic condition of Haryana. Agric. Sci. Dig. Res. J. 2015, 35, 142–145. [Google Scholar] [CrossRef]
- Thompson, G.A.; Goggin, F.L. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 2006, 57, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Senthil-Nathan, S.; Kalaivani, K.; Choi, M.-Y.; Paik, C.-H. Effects of jasmonic acid-induced resistance in rice on the plant brownhopper, Nilaparvata lugens Stål (Homoptera: Delphacidae). Pestic. Biochem. Physiol. 2009, 95, 77–84. [Google Scholar] [CrossRef]
- Stael, S.; Kmiecik, P.; Willems, P.; Van Der Kelen, K.; Coll, N.S.; Teige, M.; Van Breusegem, F. Plant innate immunity–sunny side up? Trends Plant Sci. 2015, 20, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Yu, Z.; Meng, J.; Zhou, P.; Luo, T.; Zhang, J.; Wu, J.; Lou, Y. Rice phenolamindes reduce the survival of female adults of the white-backed planthopper Sogatella furcifera. Sci. Rep. 2020, 10, 5778. [Google Scholar] [CrossRef] [Green Version]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Van Loon, L.C.; Dicke, M. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant-Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Kaloshian, I.; Walling, L.L. Hemipterans as plant pathogens. Annu. Rev. Phytopathol. 2005, 43, 491–521. [Google Scholar] [CrossRef]
- De Ilarduya, O.M.; Xie, Q.; Kaloshian, I. Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant-Microbe Interact. 2003, 16, 699–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Dempsey, D.M.A.; Shah, J.; Klessig, D.F. Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 1999, 18, 547–575. [Google Scholar] [CrossRef]
- Abreu, M.E.; Munne-Bosch, S. Salicylic acid deficiency in NahG transgenic lines and sid2 mutants increases seed yield in the annual plant Arabidopsis thaliana. J. Exp. Bot. 2009, 60, 1261–1271. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [Green Version]
- Jannoey, P.; Channei, D.; Kotcharerk, J.; Pongprasert, W.; Nomura, M. Expression analysis of genes related to rice resistance against brown planthopper, Nilaparvata lugens. Rice Sci. 2017, 24, 163–172. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Handa, N.; Sharma, R.; Kaur, H.; Kohli, S.; Kumar, V.; Kaur, P. Lignins and abiotic stress: An overview. In Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment; Springer: Berlin/Heidelberg, Germany, 2014; pp. 267–296. [Google Scholar]
- Guo, W.; Jin, L.; Miao, Y.; He, X.; Hu, Q.; Guo, K.; Zhu, L.; Zhang, X. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Saboki Ebrahim, K.U.; Singh, B. Pathogenesis related (PR) proteins in plant defense mechanism. Sci. Against Microb. Pathog. 2011, 2, 1043–1054. [Google Scholar]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Wang, L.; He, Y.; Liang, J.; Tao, R. Expression of defense genes and activities of antioxidant enzymes in rice resistance to rice stripe virus and small brown planthopper. Plant Physiol. Biochem. 2011, 49, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhu, L.; He, G. Differential gene expression in response to brown planthopper feeding in rice. J. Plant Physiol. 2004, 161, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Zhu, L.; He, G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant 2013, 6, 621–634. [Google Scholar] [CrossRef] [Green Version]
- Jan, R.; Khan, M.A.; Asaf, S.; Lee, I.-J.; Bae, J.-S.; Kim, K.-M. Overexpression of OsCM alleviates BLB stress via phytohormonal accumulation and transcriptional modulation of defense-related genes in Oryza sativa. Sci. Rep. 2020, 10, 19520. [Google Scholar] [CrossRef]
- Jan, R.; Khan, M.A.; Asaf, S.; Lee, I.-J.; Kim, K.-M. Overexpression of OsF 3 H modulates WBPH stress by alteration of phenylpropanoid pathway at a transcriptomic and metabolomic level in Oryza sativa. Sci. Rep. 2020, 10, 14685. [Google Scholar] [CrossRef]
- Zheng, M.; Chen, J.; Shi, Y.; Li, Y.; Yin, Y.; Yang, D.; Luo, Y.; Pang, D.; Xu, X.; Li, W.; et al. Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci. Rep. 2017, 7, 41805. [Google Scholar] [CrossRef] [PubMed]
- Koch, E.; Slusarenko, A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 1990, 2, 437–445. [Google Scholar] [PubMed] [Green Version]
- Bilal, S.; Shahzad, R.; Imran, M.; Jan, R.; Kim, K.M.; Lee, I.-J. Synergistic association of endophytic fungi enhances Glycine max L. resilience to combined abiotic stresses: Heavy metals, high temperature and drought stress. Ind. Crop. Prod. 2020, 143, 111931. [Google Scholar] [CrossRef]
- Jan, R.; Khan, M.A.; Asaf, S.; Lee, I.-J.; Kim, K.M. Metal resistant endophytic bacteria reduces cadmium, nickel toxicity, and enhances expression of metal stress related genes with improved growth of Oryza Sativa, via regulating its antioxidant machinery and endogenous hormones. Plants 2019, 8, 363. [Google Scholar] [CrossRef] [Green Version]
- Pavlík, M.; Pavlíková, D.; Zemanová, V.; Hnilička, F.; Urbanová, V.; Száková, J. Trace elements present in airborne particulate matter—Stressors of plant metabolism. Ecotoxicol. Environ. Saf. 2012, 79, 101–107. [Google Scholar] [CrossRef]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant-Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Weng, J.K.; Chapple, C. Improvement of biomass through lignin modification. Plant J. 2008, 54, 569–581. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Kourelis, J.; Van Der Hoorn, R.A. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renganayaki, K.; Fritz, A.K.; Sadasivam, S.; Pammi, S.; Harrington, S.E.; McCouch, S.R.; Kumar, S.M.; Reddy, A.S. Mapping and Progress toward Map-Based Cloning of Brown Planthopper Biotype-4 Resistance Gene Introgressed from Oryza officinalis into Cultivated Rice, O. sativa. Crop Sci. 2002, 42, 2112–2117. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Wang, Z.; Zhang, K.; Tian, X.; Yin, Y.; Zhao, X.; Shen, A.; Gao, C.F. Status of insecticide resistance of the whitebacked planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Fla. Entomol. 2013, 96, 948–956. [Google Scholar] [CrossRef]
- Yamasaki, M.; Yoshimura, A.; Yasui, H. Genetic basis of ovicidal response to whitebacked planthopper (Sogatella furcifera Horváth) in rice (Oryza sativa L.). Mol. Breed. 2003, 12, 133–143. [Google Scholar] [CrossRef]
- Park, J.-R.; Yun, S.; Jan, R.; Kim, K.-M. Screening and Identification of Brown Planthopper Resistance Genes OsCM9 in Rice. Agronomy 2020, 10, 1865. [Google Scholar] [CrossRef]
- Ferrari, S.; Galletti, R.; Denoux, C.; De Lorenzo, G.; Ausubel, F.M.; Dewdney, J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007, 144, 367–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhang, X.; Wu, W.; Chen, Z.; Gu, H.; Qu, L.J. Overexpression of the wounding-responsive gene AtMYB15 activates the shikimate pathway in Arabidopsis. J. Integr. Plant Biol. 2006, 48, 1084–1095. [Google Scholar] [CrossRef]
- Alvarez, A.; Montesano, M.; Schmelz, E.; Ponce de León, I. Activation of shikimate, phenylpropanoid, oxylipins, and auxin pathways in Pectobacterium carotovorum elicitors-treated moss. Front. Plant Sci. 2016, 7, 328. [Google Scholar] [CrossRef] [Green Version]
- Jwa, N.-S.; Agrawal, G.K.; Rakwal, R.; Park, C.-H.; Agrawal, V.P. Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. 2001, 286, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yu, J.; Bai, J.; Zhu, Z.; Wang, X. Induced defense responses in rice plants against small brown planthopper infestation. Crop J. 2014, 2, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Fuzhang, D. Research progress on application of superoxide dismutases in tobacco. J. Anhui Agric. Sci. 2008, 36, 1897. [Google Scholar]
- Wang, Y.; Wang, X.; Yuan, H.; Chen, R.; Zhu, L.; He, R.; He, G. Responses of two contrasting genotypes of rice to brown planthopper. Mol. Plant-Microbe Interact. 2008, 21, 122–132. [Google Scholar] [CrossRef]
- Verslues, P.E.; Juenger, T.E. Drought, metabolites, and Arabidopsis natural variation: A promising combination for understanding adaptation to water-limited environments. Curr. Opin. Plant Biol. 2011, 14, 240–245. [Google Scholar] [CrossRef]
- Yokota, A.; Kawasaki, S.; Iwano, M.; Nakamura, C.; Miyake, C.; Akashi, K. Citrulline and DRIP-1 protein (ArgE homologue) in drought tolerance of wild watermelon. Ann. Bot. 2002, 89, 825–832. [Google Scholar] [CrossRef] [Green Version]
- Akashi, K.; Miyake, C.; Yokota, A. Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Lett. 2001, 508, 438–442. [Google Scholar] [CrossRef] [Green Version]

| S/No | Gene | Forward Primers | Reverse Primers | Accession No |
|---|---|---|---|---|
| 1 | CM | ATGGCGGCGGCGATGATTCTCTCCTGCA | TCAGGCATTGCAAGTTCGAATCCTAACAAG | XM_015793648.2 |
| 2 | PAL | GCAACCCCAGCTTGGACTAT | TCACACTCTCGAAATGCTC | XM_015769639 |
| 3 | CHS | AGGGAAGAATGGGGACTGAT | TGCCTCGAACTAGCATTCCT | X91811.1 |
| 4 | CHI | AGCTCCTGAAGGCGGAAT | GATTTTCACGCGGACACC | XM_015772801 |
| 5 | PR1 | GGAAGTACGGCGAGAACATC | GGTCGTACCACTGCTTCTCC | XM_015792496.2 |
| 6 | PR2 | TGCTATGTTCGACGAGAACG | GTTGAACAGCCCAAAGTGCT | XM_015766957.2 |
| 7 | SOD | CTTGATGCCCTGGAACCTTA | GCCAGACCCCAAAAGTGATA | KY752532.1 |
| 8 | CAT | CCACCACAACAACCACTACGACGG | CCAACGACTCATCACACTGG | KY752529.1 |
| 9 | Actin | ATCACCATCGGAGCAGAAAG | AAAAGATGGCTGGAAGAGCA | XM_015785964.2 |
| GPx Assay Buffer (μL) | NADPH Assay Reagent (μL) | Enzyme (0.25 unit/mL) (μL) | Sample (μL) | 30 mM t-Bu-OOH (μL) | |
|---|---|---|---|---|---|
| Blink | 940 | 50 | --- | --- | 10 |
| Positive control | 900 | 50 | 50 | --- | 10 |
| Sample | 900 | 50 | --- | 50 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jan, R.; Khan, M.A.; Asaf, S.; Lubna; Lee, I.-J.; Kim, K.-M. Over-Expression of Chorismate Mutase Enhances the Accumulation of Salicylic Acid, Lignin, and Antioxidants in Response to the White-Backed Planthopper in Rice Plants. Antioxidants 2021, 10, 1680. https://doi.org/10.3390/antiox10111680
Jan R, Khan MA, Asaf S, Lubna, Lee I-J, Kim K-M. Over-Expression of Chorismate Mutase Enhances the Accumulation of Salicylic Acid, Lignin, and Antioxidants in Response to the White-Backed Planthopper in Rice Plants. Antioxidants. 2021; 10(11):1680. https://doi.org/10.3390/antiox10111680
Chicago/Turabian StyleJan, Rahmatullah, Muhammad Aaqil Khan, Sajjad Asaf, Lubna, In-Jung Lee, and Kyung-Min Kim. 2021. "Over-Expression of Chorismate Mutase Enhances the Accumulation of Salicylic Acid, Lignin, and Antioxidants in Response to the White-Backed Planthopper in Rice Plants" Antioxidants 10, no. 11: 1680. https://doi.org/10.3390/antiox10111680







