Effects of the Quinone Oxidoreductase WrbA on Escherichia coli Biofilm Formation and Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Escherichia Coli Strain and Preparation of the wrbA-mutant
2.2. E. coli Planktonic Growth
2.3. E. coli Biofilm Growth
2.4. Level of Oxidative Stress within the Biofilm
2.5. Biofilm Catalase Activities
2.6. Biofilm Susceptibility Assay
2.7. E. coli Crude Extracts
2.8. NADH-Dependent Oxidoreductase Activities
2.9. Purification of WrbA
2.10. Statistical Analysis
3. Results and Discussion
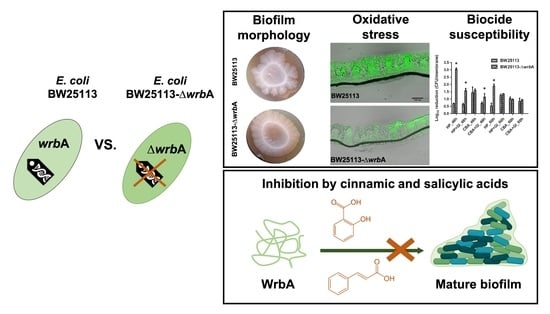
3.1. WrbA Is Not Essential for the Planktonic Growth but It Affects the Biofilm Growth
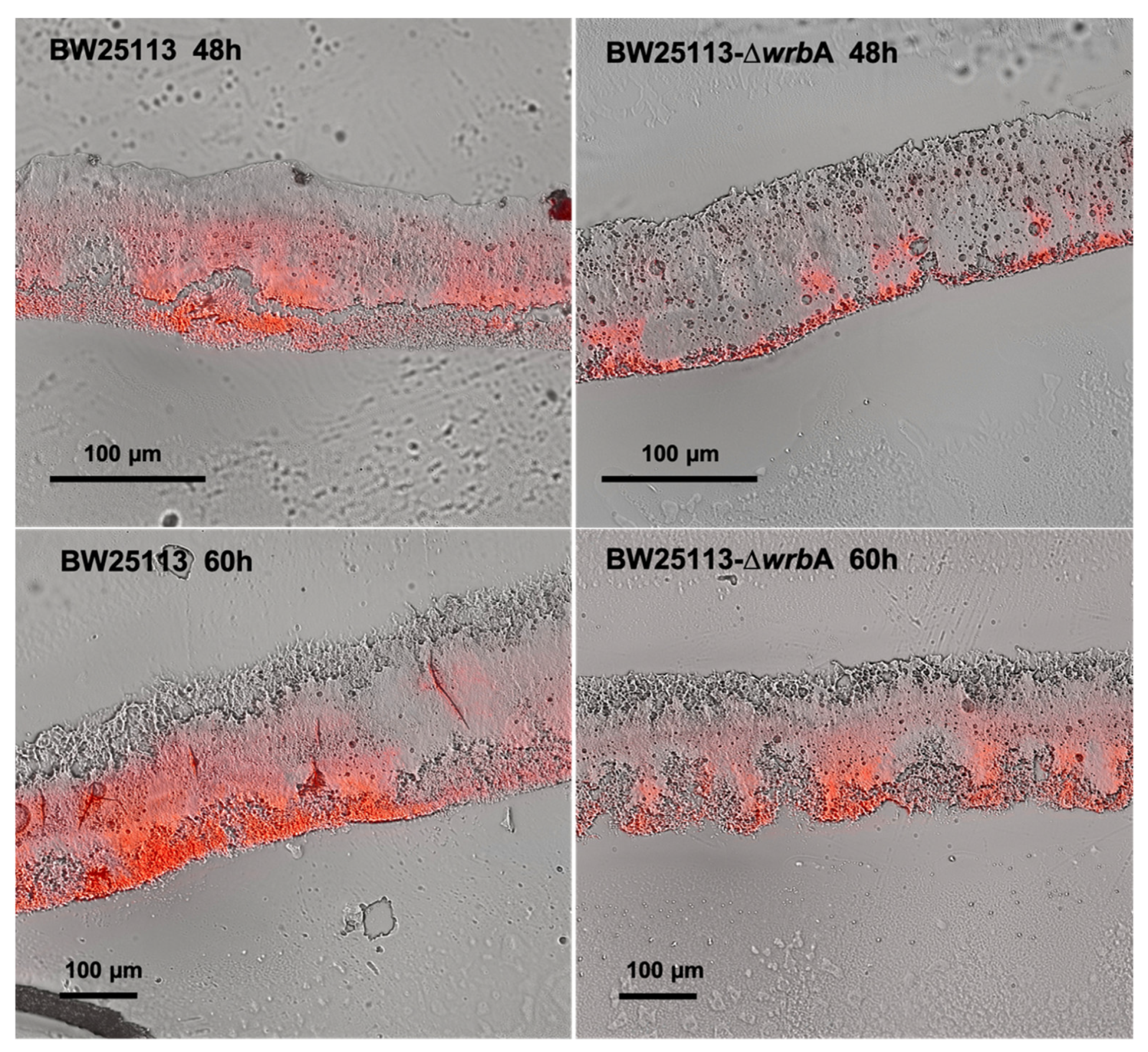
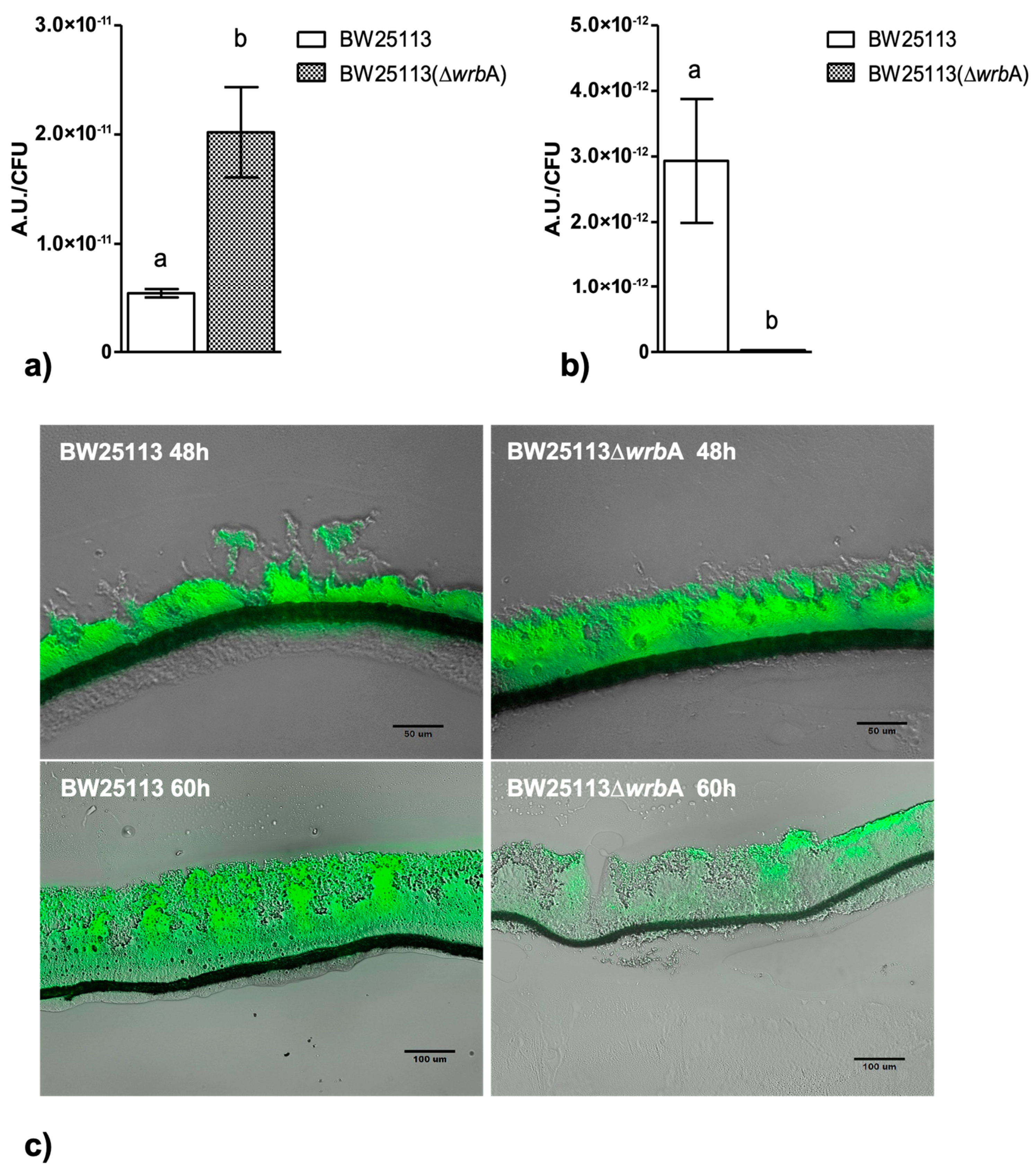
3.2. WrbA Affects the Biofilm Oxidative Stress Profile and Alternative Antioxidant Defenses
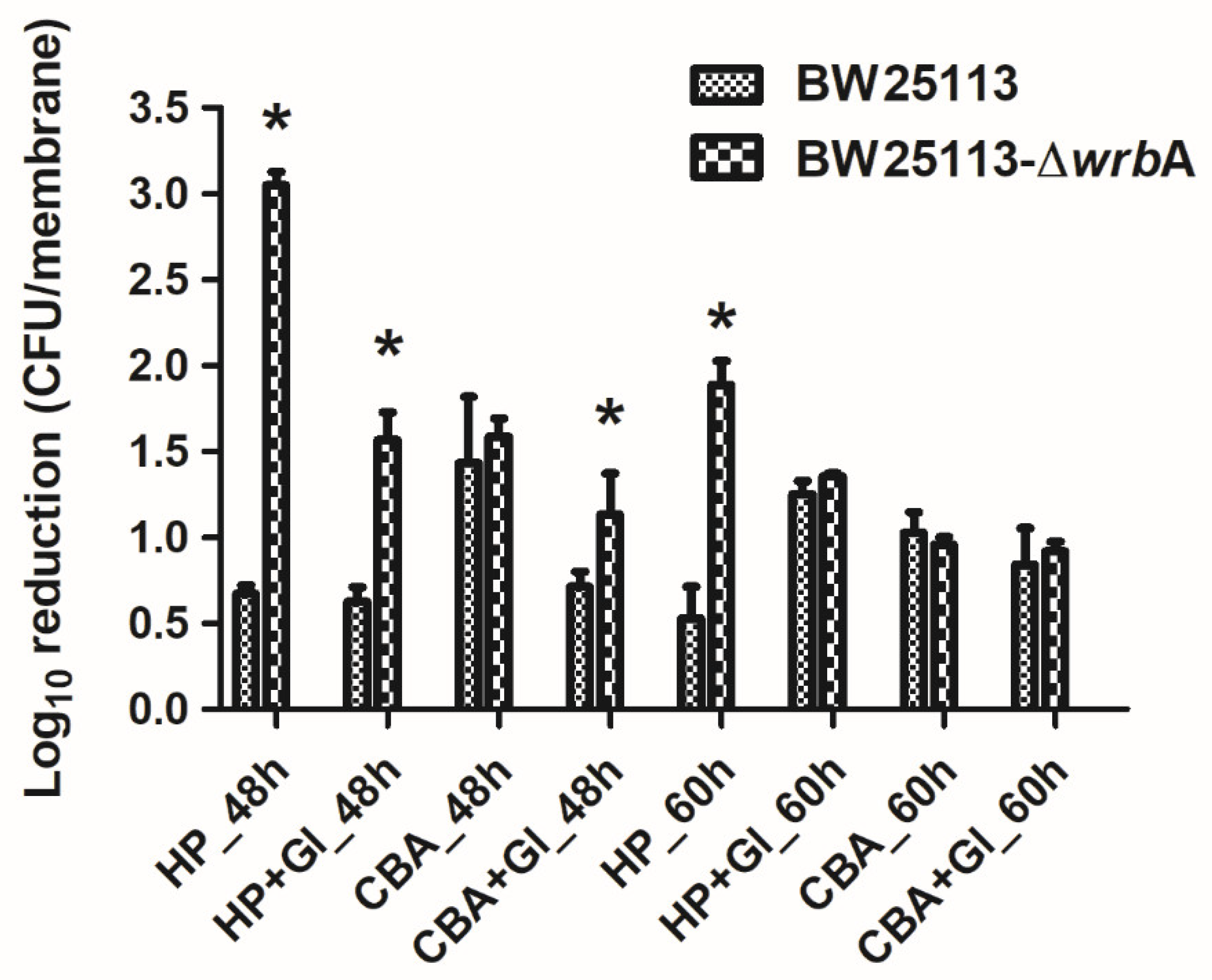
3.3. WrbA Affects the Biofilm Tolerance to H2O2 but Not to 2-Chlorobenzoic Acid
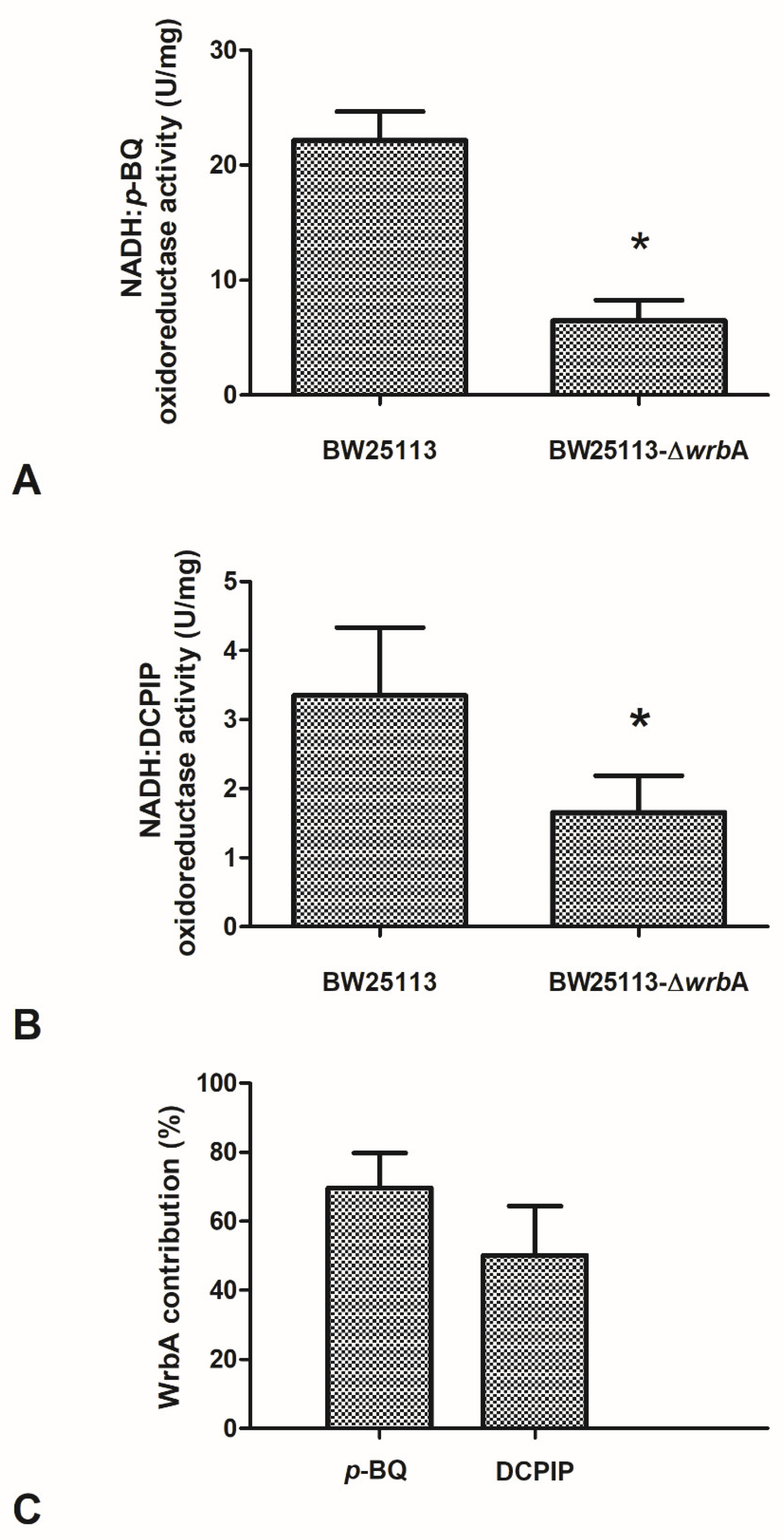
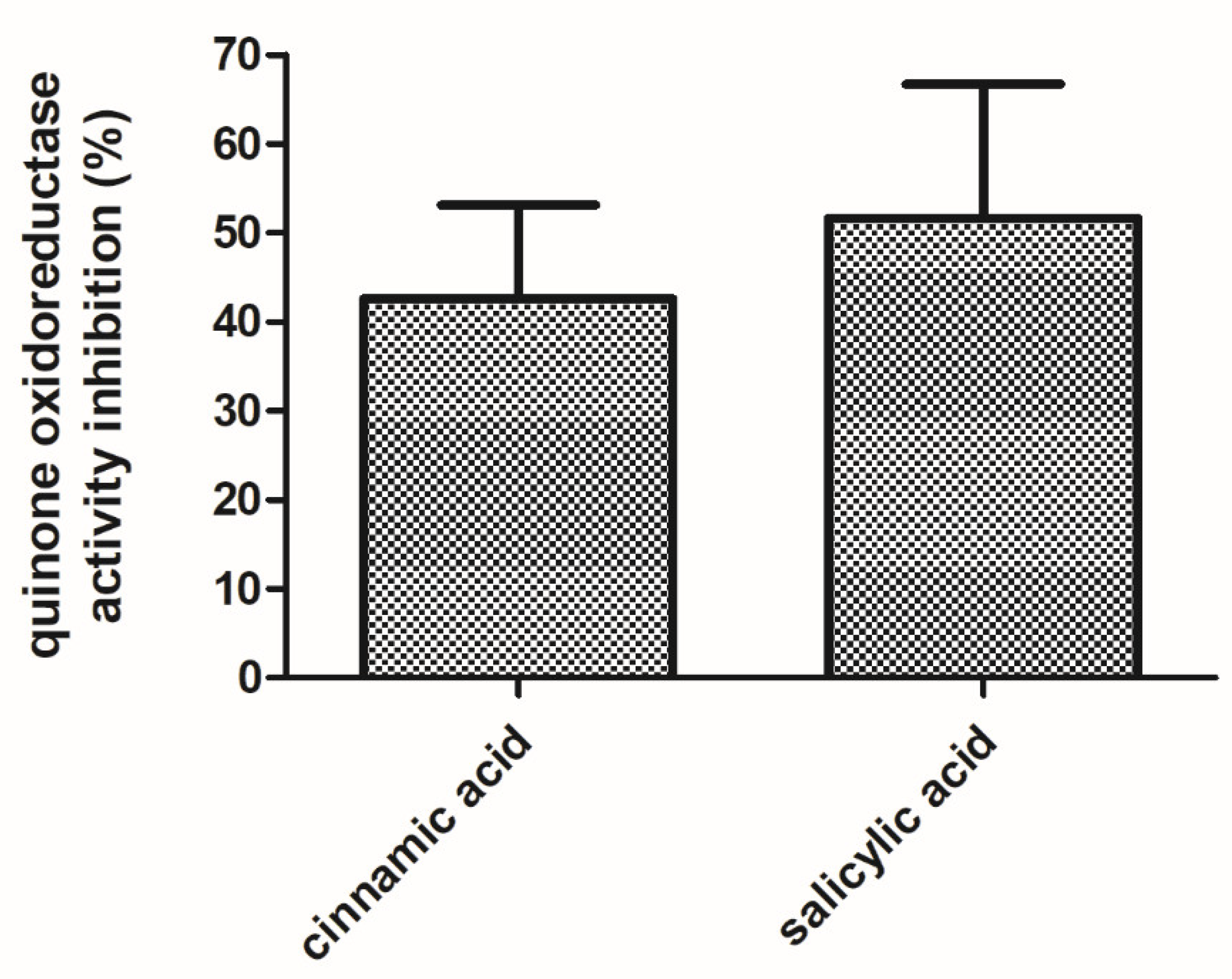
3.4. WrbA Is an Important Functional Component of the E. coli NADH-Dependent Oxidoreductase Activity That Is Inhibited by Cinnamic and Salicylic Acids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flemming, H.-C.; Wuertz, S. Bacteria and Archaea on Earth and Their Abundance in Biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Toyofuku, M.; Inaba, T.; Kiyokawa, T.; Obana, N.; Yawata, Y.; Nomura, N. Environmental Factors That Shape Biofilm Formation. Biosci. Biotechnol. Biochem. 2016, 80, 7–12. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Ponomareva, A.L.; Buzoleva, L.S.; Bogatyrenko, E.A. Abiotic Environmental Factors Affecting the Formation of Microbial Biofilms. Biol. Bull. 2018, 45, 490–496. [Google Scholar] [CrossRef]
- Rossi, E.; Paroni, M.; Landini, P. Biofilm and Motility in Response to Environmental and Host-Related Signals in Gram Negative Opportunistic Pathogens. J. Appl. Microbiol. 2018, 125, 1587–1602. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Bhargava, A. Biofilms and Human Health. Biotechnol. Lett. 2016, 38, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Masters, E.A.; Trombetta, R.P.; de Mesy Bentley, K.L.; Boyce, B.F.; Gill, A.L.; Gill, S.R.; Nishitani, K.; Ishikawa, M.; Morita, Y.; Ito, H.; et al. Evolving Concepts in Bone Infection: Redefining “Biofilm”, “Acute vs. Chronic Osteomyelitis”, “the Immune Proteome” and “Local Antibiotic Therapy. Bone Res. 2019, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Abdullahi, U.F.; Igwenagu, E.; Mu’azu, A.; Aliyu, S.; Umar, M.I. Intrigues of Biofilm: A Perspective in Veterinary Medicine. Vet. World 2016, 9, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Malvi, Y. Animal Infections: The Role of Fungal Biofilms. In Recent Developments in Fungal Diseases of Laboratory Animals; Gupta, A., Singh, N.P., Eds.; Fungal Biology; Springer International Publishing: Cham, Germany, 2019; pp. 149–162. ISBN 978-3-030-18586-2. [Google Scholar]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent Advances in the Understanding of Xanthomonas Citri Ssp. Citri Pathogenesis and Citrus Canker Disease Management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattò, C.; De Vincenti, L.; Cappitelli, F.; D’Attoma, G.; Saponari, M.; Villa, F.; Forlani, F. Non-Lethal Effects of N-Acetylcysteine on Xylella Fastidiosa Strain De Donno Biofilm Formation and Detachment. Microorganisms 2019, 7, 656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, D. Marine Biofilms: A Successful Microbial Strategy With Economic Implications. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef] [Green Version]
- Meesilp, N.; Mesil, N. Effect of Microbial Sanitizers for Reducing Biofilm Formation of Staphylococcus Aureus and Pseudomonas Aeruginosa on Stainless Steel by Cultivation with UHT Milk. Food Sci. Biotechnol. 2019, 28, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.; Pullerits, K.; Keucken, A.; Persson, K.M.; Paul, C.J.; Rådström, P. Bacterial Release from Pipe Biofilm in a Full-Scale Drinking Water Distribution System. Npj Biofilms Microbiomes 2019, 5, 1–8. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas Aeruginosa Biofilms to Antimicrobial Agents-How P. Aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [Green Version]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of Intervention, Treatment, and Antibiotic Resistance of Biofilm-Forming Microorganisms. Heliyon 2019, 5. [Google Scholar] [CrossRef] [Green Version]
- Jang, Y.S.; Mosolygó, T. Inhibition of Bacterial Biofilm Formation by Phytotherapeutics with Focus on Overcoming Antimicrobial Resistance. Curr. Pharm. Des. 2020, 26, 2807–2816. [Google Scholar] [CrossRef]
- Li, J.; Li, S.; Li, H.; Guo, X.; Guo, D.; Yang, Y.; Wang, X.; Zhang, C.; Shan, Z.; Xia, X.; et al. Antibiofilm Activity of Shikonin against Listeria Monocytogenes and Inhibition of Key Virulence Factors. Food Control 2021, 120, 107558. [Google Scholar] [CrossRef]
- Santos, C.A.; Almeida, F.A.; Quecán, B.X.V.; Pereira, P.A.P.; Gandra, K.M.B.; Cunha, L.R.; Pinto, U.M. Bioactive Properties of Syzygium Cumini (L.) Skeels Pulp and Seed Phenolic Extracts. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Jafri, H.; Ahmad, I. Thymus Vulgaris Essential Oil and Thymol Inhibit Biofilms and Interact Synergistically with Antifungal Drugs against Drug Resistant Strains of Candida Albicans and Candida Tropicalis. J. Mycol. Medicale 2020, 30, 100911. [Google Scholar] [CrossRef]
- Villa, F.; Cappitelli, F. Plant-Derived Bioactive Compounds at Sub-Lethal Concentrations: Towards Smart Biocide-Free Antibiofilm Strategies. Phytochem. Rev. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Szczepanski, S.; Lipski, A. Essential Oils Show Specific Inhibiting Effects on Bacterial Biofilm Formation. Food Control 2014, 36, 224–229. [Google Scholar] [CrossRef]
- Čabarkapa, I.; Čolović, R.; Đuragić, O.; Popović, S.; Kokić, B.; Milanov, D.; Pezo, L. Anti-Biofilm Activities of Essential Oils Rich in Carvacrol and Thymol against Salmonella Enteritidis. Biofouling 2019, 35, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Pitts, B.; Stewart, P.S.; Giussani, B.; Roncoroni, S.; Albanese, D.; Giordano, C.; Tunesi, M.; Cappitelli, F. Efficacy of Zosteric Acid Sodium Salt on the Yeast Biofilm Model Candida Albicans. Microb. Ecol. 2011, 62, 584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattò, C.; Dell’Orto, S.; Villa, F.; Villa, S.; Gelain, A.; Vitali, A.; Marzano, V.; Baroni, S.; Forlani, F.; Cappitelli, F. Unravelling the Structural and Molecular Basis Responsible for the Anti-Biofilm Activity of Zosteric Acid. PLoS ONE 2015, 10, e0131519. [Google Scholar] [CrossRef] [Green Version]
- Cattò, C.; Grazioso, G.; Dell’Orto, S.; Gelain, A.; Villa, S.; Marzano, V.; Vitali, A.; Villa, F.; Cappitelli, F.; Forlani, F. The Response of Escherichia Coli Biofilm to Salicylic Acid. Biofouling 2017, 33, 235–251. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.A.K.S.; Rudden, M.; Smyth, T.J.; Dooley, J.S.G.; Marchant, R.; Banat, I.M. Natural Quorum Sensing Inhibitors Effectively Downregulate Gene Expression of Pseudomonas Aeruginosa Virulence Factors. Appl. Microbiol. Biotechnol. 2019, 103, 3521–3535. [Google Scholar] [CrossRef] [Green Version]
- Rajkumari, J.; Borkotoky, S.; Murali, A.; Suchiang, K.; Mohanty, S.K.; Busi, S. Cinnamic Acid Attenuates Quorum Sensing Associated Virulence Factors and Biofilm Formation in Pseudomonas Aeruginosa PAO1. Biotechnol. Lett. 2018, 40, 1087–1100. [Google Scholar] [CrossRef]
- Villa, F.; Remelli, W.; Forlani, F.; Vitali, A.; Cappitelli, F. Altered Expression Level of Escherichia Coli Proteins in Response to Treatment with the Antifouling Agent Zosteric Acid Sodium Salt. Environ. Microbiol. 2012, 14, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Villa, F.; Secundo, F.; Forlani, F.; Cattò, C.; Cappitelli, F. Biochemical and Molecular Changes of the Zosteric Acid-Treated Escherichia Coli Biofilm on a Mineral Surface. Ann. Microbiol. 2021, 71, 3. [Google Scholar] [CrossRef]
- Sauer, H.; Wartenberg, M.; Hescheler, J. Reactive Oxygen Species as Intracellular Messengers during Cell Growth and Differentiation. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2001, 11, 173–186. [Google Scholar] [CrossRef]
- Čáp, M.; Váchová, L.; Palková, Z. Reactive Oxygen Species in the Signaling and Adaptation of Multicellular Microbial Communities. Oxid. Med. Cell. Longev. 2012, 2012, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natalello, A.; Doglia, S.M.; Carey, J.; Grandori, R. Role of Flavin Mononucleotide in the Thermostability and Oligomerization of Escherichia Coli Stress-Defense Protein WrbA. Biochemistry 2007, 46, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Patridge, E.V.; Ferry, J.G. WrbA from Escherichia Coli and Archaeoglobus Fulgidus Is an NAD(P)H:Quinone Oxidoreductase. J. Bacteriol. 2006, 188, 3498–3506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moshiri, J.; Kaur, D.; Hambira, C.M.; Sandala, J.L.; Koopman, J.A.; Fuchs, J.R.; Gunn, J.S. Identification of a Small Molecule Anti-Biofilm Agent Against Salmonella Enterica. Front. Microbiol. 2018, 9, 2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattò, C.; Cappitelli, F. Testing Anti-Biofilm Polymeric Surfaces: Where to Start? Int. J. Mol. Sci. 2019, 20, 3794. [Google Scholar] [CrossRef] [Green Version]
- Villa, F.; Albanese, D.; Giussani, B.; Stewart, P.S.; Daffonchio, D.; Cappitelli, F. Hindering Biofilm Formation with Zosteric Acid. Biofouling 2010, 26, 739–752. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia Coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [Green Version]
- Cherepanov, P.P.; Wackernagel, W. Gene Disruption in Escherichia Coli: TcR and KmR Cassettes with the Option of Flp-Catalyzed Excision of the Antibiotic-Resistance Determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Morooka, N.; Yamamoto, Y.; Fujita, K.; Isono, K.; Choi, S.; Ohtsubo, E.; Baba, T.; Wanner, B.L.; Mori, H.; et al. Highly Accurate Genome Sequences of Escherichia Coli K-12 Strains MG1655 and W3110. Mol. Syst. Biol. 2006, 2, 2006.0007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of Antibiotic Penetration Limitation in Klebsiella Pneumoniae Biofilm Resistance to Ampicillin and Ciprofloxacin. Antimicrob. Agents Chemother. 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J. Determination of Viable Cell Counts: Bacterial Growth Curves. Experiments in Molecular Genetics; Miller, J.H., Ed.; Cold Spring Harbor: New York, NY, USA, 1972. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Jakubowski, W.; Biliński, T.; Bartosz, G. Oxidative Stress during Aging of Stationary Cultures of the Yeast Saccharomyces Cerevisiae. Free Radic. Biol. Med. 2000, 28, 659–664. [Google Scholar] [CrossRef]
- Iwase, T.; Tajima, A.; Sugimoto, S.; Okuda, K.; Hironaka, I.; Kamata, Y.; Takada, K.; Mizunoe, Y. A Simple Assay for Measuring Catalase Activity: A Visual Approach. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Villa, F.; Remelli, W.; Forlani, F.; Gambino, M.; Landini, P.; Cappitelli, F. Effects of Chronic Sub-Lethal Oxidative Stress on Biofilm Formation by Azotobacter Vinelandii. Biofouling 2012, 28, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete Set of ORF Clones of Escherichia Coli ASKA Library (A Complete S et of E. Coli K -12 ORF A Rchive): Unique Resources for Biological Research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idalia, V.-M.N.; Bernardo, F. Escherichia Coli as a Model Organism and Its Application in Biotechnology. IEscherichia Colii Recent Adv. Physiol. Pathog. Biotechnol. Appl. 2017. [Google Scholar] [CrossRef] [Green Version]
- Adamczyk, P.A.; Reed, J.L. Escherichia Coli as a Model Organism for Systems Metabolic Engineering. Curr. Opin. Syst. Biol. 2017, 6, 80–88. [Google Scholar] [CrossRef]
- Blount, Z.D. The Unexhausted Potential of E. Coli. eLife 2015, 4, e05826. [Google Scholar] [CrossRef]
- Beloin, C.; Valle, J.; Latour-Lambert, P.; Faure, P.; Kzreminski, M.; Balestrino, D.; Haagensen, J.A.J.; Molin, S.; Prensier, G.; Arbeille, B.; et al. Global Impact of Mature Biofilm Lifestyle on Escherichia Coli K-12 Gene Expression. Mol. Microbiol. 2004, 51, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Besharova, O.; Suchanek, V.M.; Hartmann, R.; Drescher, K.; Sourjik, V. Diversification of Gene Expression during Formation of Static Submerged Biofilms by Escherichia Coli. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Hufnagel, D.A.; Depas, W.H.; Chapman, M.R. The Biology of the Escherichia Coli Extracellular Matrix. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehman, A.P.; Long, S.R. Exopolysaccharides from Sinorhizobium Meliloti Can Protect against H2O2-Dependent Damage. J. Bacteriol. 2013, 195, 5362–5369. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Lee, S.M.; Mao, Y. Protective Effect of Exopolysaccharide Colanic Acid of Escherichia Coli O157:H7 to Osmotic and Oxidative Stress. Int. J. Food Microbiol. 2004, 93, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Arce Miranda, J.E.; Sotomayor, C.E.; Albesa, I.; Paraje, M.G. Oxidative and Nitrosative Stress in Staphylococcus Aureus Biofilm. FEMS Microbiol. Lett. 2011, 315, 23–29. [Google Scholar] [CrossRef]
- Arita-Morioka, K.I.; Yamanaka, K.; Mizunoe, Y.; Tanaka, Y.; Ogura, T.; Sugimoto, S. Inhibitory effects of Myricetin derivatives on curli-dependent biofilm formation in Escherichia coli. Sci. Rep. 2018, 8, 8452. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.L.; Chapman, M.R. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Soo, V.W.; Wood, T.K. Antitoxin MqsA represses curli formation through the master biofilm regulator CsgD. Sci. Rep. 2013, 3, 3186. [Google Scholar] [CrossRef] [Green Version]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Kirisits, M.J.; Prost, L.; Starkey, M.; Parsek, M.R. Characterization of Colony Morphology Variants Isolated from Pseudomonas Aeruginosa Biofilms. Appl. Environ. Microbiol. 2005, 71, 4809–4821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Fei, C.; Mao, S.; Moreau, A.; Wingreen, N.S.; Košmrlj, A.; Stone, H.A.; Bassler, B.L. Mechanical Instability and Interfacial Energy Drive Biofilm Morphogenesis. eLife 2019, 8, e43920. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Mao, S.; Yan, J.; Alert, R.; Stone, H.A.; Bassler, B.L.; Wingreen, N.S.; Košmrlj, A. Nonuniform Growth and Surface Friction Determine Bacterial Biofilm Morphology on Soft Substrates. Proc. Natl. Acad. Sci. USA 2020, 117, 7622–7632. [Google Scholar] [CrossRef] [Green Version]
- Carey, J.; Brynda, J.; Wolfová, J.; Grandori, R.; Gustavsson, T.; Ettrich, R.; Smatanová, I.K. WrbA Bridges Bacterial Flavodoxins and Eukaryotic NAD(P)H:Quinone Oxidoreductases. Protein Sci. 2007, 16, 2301–2305. [Google Scholar] [CrossRef]
- Green, L.K.; La Flamme, A.C.; Ackerley, D.F. Pseudomonas Aeruginosa MdaB and WrbA Are Water-Soluble Two-Electron Quinone Oxidoreductases with the Potential to Defend against Oxidative Stress. J. Microbiol. 2014, 52, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Okegbe, C.; Price-Whelan, A.; Sakhtah, H.; Hunter, R.C.; Newman, D.K. Bacterial Community Morphogenesis Is Intimately Linked to the Intracellular Redox State. J. Bacteriol. 2013, 195, 1371–1380. [Google Scholar] [CrossRef] [Green Version]
- DePas, W.H.; Hufnagel, D.A.; Lee, J.S.; Blanco, L.P.; Bernstein, H.C.; Fisher, S.T.; James, G.A.; Stewart, P.S.; Chapman, M.R. Iron Induces Bimodal Population Development by Escherichia Coli. Proc. Natl. Acad. Sci. USA 2013, 110, 2629–2634. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. The Molecular Mechanisms and Physiological Consequences of Oxidative Stress: Lessons from a Model Bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.L.; Ding, Y.; Liu, Y.; Cai, Z.; Zhou, J.; Swarup, S.; Drautz-Moses, D.I.; Schuster, S.C.; Kjelleberg, S.; Givskov, M.; et al. Reactive Oxygen Species Drive Evolution of Pro-Biofilm Variants in Pathogens by Modulating Cyclic-Di-GMP Levels. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Morvay, A.A.; Shi, X.; Suo, Y.; Shi, C.; Knøchel, S. Comparison of Oxidative Stress Response and Biofilm Formation of Listeria Monocytogenes Serotypes 4b and 1/2a. Food Control 2018, 85, 416–422. [Google Scholar] [CrossRef]
- Jalal, N.; Lee, S.F. The MsrAB Reducing Pathway of Streptococcus Gordonii Is Needed for Oxidative Stress Tolerance, Biofilm Formation, and Oral Colonization in Mice. PLoS ONE 2020, 15, e0229375. [Google Scholar] [CrossRef]
- Gambino, M.; Cappitelli, F. Mini-Review: Biofilm Responses to Oxidative Stress. Biofouling 2016, 32, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, A.; Mavi, P.S.; Bhatt, D.; Kumar, A. Thiol Reductive Stress Induces Cellulose-Anchored Biofilm Formation in Mycobacterium Tuberculosis. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Jeon, B. Role of Alkyl Hydroperoxide Reductase (AhpC) in the Biofilm Formation of Campylobacter Jejuni. PLoS ONE 2014, 9, e87312. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Zhang, C.; Chen, G.; Wu, M.; Liu, W.; Ding, C.; Dong, Q.; Fan, E.; Liu, Q. Reactive Oxygen Species Inhibit Biofilm Formation of Listeria Monocytogenes. Microb. Pathog. 2019, 127, 183–189. [Google Scholar] [CrossRef]
- Loprasert, S.; Sallabhan, R.; Whangsuk, W.; Mongkolsuk, S. Characterization and Mutagenesis of Fur Gene from Burkholderia Pseudomallei. Gene 2000, 254, 129–137. [Google Scholar] [CrossRef]
- Xie, K.; Peng, H.; Hu, H.; Wang, W.; Zhang, X. OxyR, an Important Oxidative Stress Regulator to Phenazines Production and Hydrogen Peroxide Resistance in Pseudomonas Chlororaphis GP72. Microbiol. Res. 2013, 168, 646–653. [Google Scholar] [CrossRef]
- Wu, J.; Lin, X.; Xie, H.; Burne, R. OxyR Is Involved in Coordinate Regulation of Expression of FimA and Sod Genes in Porphyromonas Gingivalis. FEMS Microbiol. Lett. 2008, 282, 188–195. [Google Scholar] [CrossRef] [Green Version]
- Collet, A.; Cosette, P.; Beloin, C.; Ghigo, J.-M.; Rihouey, C.; Lerouge, P.; Junter, G.-A.; Jouenne, T. Impact of RpoS Deletion on the Proteome of Escherichia Coli Grown Planktonically and as Biofilm. J. Proteome Res. 2008, 7, 4659–4669. [Google Scholar] [CrossRef] [PubMed]
- Benov, L.; Sequeira, F. Escherichia Coli Deltafur Mutant Displays Low HPII Catalase Activity in Stationary Phase. Redox Rep. Commun. Free Radic. Res. 2003, 8, 379–383. [Google Scholar] [CrossRef]
- Tanaka, K.; Handel, K.; Loewen, P.C.; Takahashi, H. Identification and Analysis of the RpoS-Dependent Promoter of KatE, Encoding Catalase HPII in Escherichia Coli. Biochim. Biophys. Acta 1997, 1352, 161–166. [Google Scholar] [CrossRef]
- Gottesman, S. Trouble Is Coming: Signaling Pathways That Regulate General Stress Responses in Bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schellhorn, H.E. Function, Evolution, and Composition of the RpoS Regulon in Escherichia Coli. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Horita, M.; Wang, D.-H.; Tsutsui, K.; Sano, K.; Masuoka, N.; Kira, S. Involvement of Oxidative Stress in Hydroquinone-Induced Cytotoxicity in Catalase-Deficient Escherichia Coli Mutants. Free Radic. Res. 2005, 39, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Benoit, S.L.; Maier, R.J. Helicobacter Catalase Devoid of Catalytic Activity Protects the Bacterium against Oxidative Stress. J. Biol. Chem. 2016, 291, 23366–23373. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.; Chiang, S.M.; Joyce, C.; Yu, R.; Schellhorn, H.E. Polymorphism and Selection of RpoS in Pathogenic Escherichia Coli. BMC Microbiol. 2009, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Corona-Izquierdo, F.P.; Membrillo-Hernández, J. A Mutation in RpoS Enhances Biofilm Formation in Escherichia Coli during Exponential Phase of Growth. FEMS Microbiol. Lett. 2002, 211, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, J.R.; Yim, M.-S.; Saliba, J.H.; Chung, W.-H.; Wong, K.-Y.; Leung, K.T. Role of RpoS in Escherichia Coli O157:H7 Strain H32 Biofilm Development and Survival. Appl. Environ. Microbiol. 2012, 78, 8331–8339. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Wu, Y.; Chen, Y.; Xu, F.; Halliday, N.; Gao, K.; Chan, K.G.; Cámara, M. RpoS Differentially Affects the General Stress Response and Biofilm Formation in the Endophytic Serratia Plymuthica G3. Res. Microbiol. 2016, 167, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Zlatkov, N.; Uhlin, B.E. Absence of Global Stress Regulation in Escherichia Coli Promotes Pathoadaptation and Novel C-Di-GMP-Dependent Metabolic Capability. Sci. Rep. 2019, 9, 2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, A.; Taniuchi, A.; May, T.; Kawata, K.; Okabe, S. Increased Antibiotic Resistance of Escherichia Coli in Mature Biofilms. Appl. Environ. Microbiol. 2009, 75, 4093–4100. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus Biofilm: An Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Warth, A.D. Mechanism of Action of Benzoic Acid on Zygosaccharomyces Bailii: Effects on Glycolytic Metabolite Levels, Energy Production, and Intracellular PH. Appl. Environ. Microbiol. 1991, 57, 3410–3414. [Google Scholar] [CrossRef] [Green Version]
- Bouarab-Chibane, L.; Forquet, V.; Lantéri, P.; Clément, Y.; Léonard-Akkari, L.; Oulahal, N.; Degraeve, P.; Bordes, C. Antibacterial Properties of Polyphenols: Characterization and QSAR (Quantitative Structure-Activity Relationship) Models. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Mangalappalli-Illathu, A.K.; Korber, D.R. Adaptive Resistance and Differential Protein Expression of Salmonella Enterica Serovar Enteritidis Biofilms Exposed to Benzalkonium Chloride. Antimicrob. Agents Chemother. 2006, 50, 3588–3596. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Wang, S.; Huang, L.; Sa, W.; Li, J.; Huang, J.; Dai, M.; Cheng, G. Development of Resistance in Escherichia Coli ATCC25922 under Exposure of Sub-Inhibitory Concentration of Olaquindox. Antibiotics 2020, 9, 791. [Google Scholar] [CrossRef]
- Shah, D.; Zhang, Z.; Khodursky, A.; Kaldalu, N.; Kurg, K.; Lewis, K. Persisters: A Distinct Physiological State of E. Coli. BMC Microbiol. 2006, 6, 53. [Google Scholar] [CrossRef] [Green Version]
- Amato, S.M.; Orman, M.A.; Brynildsen, M.P. Metabolic Control of Persister Formation in Escherichia Coli. Mol. Cell 2013, 50, 475–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avican, K.; Fahlgren, A.; Huss, M.; Heroven, A.K.; Beckstette, M.; Dersch, P.; Fällman, M. Reprogramming of Yersinia from Virulent to Persistent Mode Revealed by Complex In Vivo RNA-Seq Analysis. PLoS Pathog. 2015, 11, e1004600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, D.-E.; Smalley, D.J.; Conway, T. Gene Expression Profiling of Escherichia Coli Growth Transitions: An Expanded Stringent Response Model. Mol. Microbiol. 2002, 45, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zetterström, C.E.; Gabrielsen, M.; Beckham, K.S.H.; Tree, J.J.; Macdonald, S.E.; Byron, O.; Mitchell, T.J.; Gally, D.L.; Herzyk, P.; et al. Identification of Bacterial Target Proteins for the Salicylidene Acylhydrazide Class of Virulence-Blocking Compounds. J. Biol. Chem. 2011, 286, 29922–29931. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Sequence a | Description |
|---|---|---|
| U-wrbA | TGCGACAAAATTACGTGCTTG | wrbA-locus-specific primer (5′ end aligns at the position 144 nt upstream of the start codon of the wrbA gene) |
| D-wrbA | GCGACTTCGAAAATGGCCTC | wrbA-locus-specific primer (5′ end aligns at the position 115 nt downstream of the stop codon of the wrbA gene) |
| k1 | CAGTCATAGCCGAATAGCCT | Kanamycin-resistance-specific primer [42] |
| k2 | CGGTGCCCTGAATGAACTGC | Kanamycin-resistance-specific primer [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, F.; Cattò, C.; Mugnai, G.; Villa, F.; Forlani, F. Effects of the Quinone Oxidoreductase WrbA on Escherichia coli Biofilm Formation and Oxidative Stress. Antioxidants 2021, 10, 919. https://doi.org/10.3390/antiox10060919
Rossi F, Cattò C, Mugnai G, Villa F, Forlani F. Effects of the Quinone Oxidoreductase WrbA on Escherichia coli Biofilm Formation and Oxidative Stress. Antioxidants. 2021; 10(6):919. https://doi.org/10.3390/antiox10060919
Chicago/Turabian StyleRossi, Federico, Cristina Cattò, Gianmarco Mugnai, Federica Villa, and Fabio Forlani. 2021. "Effects of the Quinone Oxidoreductase WrbA on Escherichia coli Biofilm Formation and Oxidative Stress" Antioxidants 10, no. 6: 919. https://doi.org/10.3390/antiox10060919