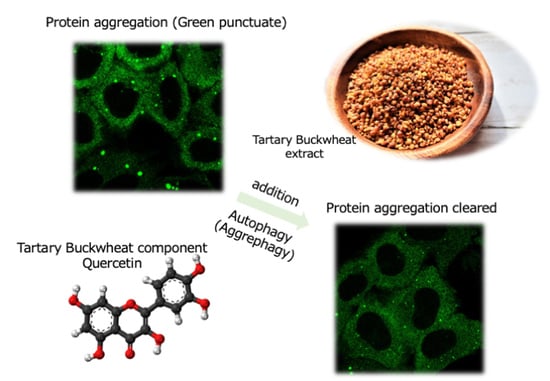
Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Tartary Buckwheat Extract
2.2. Antibodies
2.3. Cell Culture
2.4. MTT (Methylthialazole Tetrazolium) Assay for Cell Death
2.5. Western Blotting
2.6. Immunofluorescence and Confocal Microscope Observation
3. Results
3.1. TBE Induces Autophagy
3.2. TBE Induces Autophagosome Formation
3.3. TBE Enhances Dephosphorylation of mTORC1 Substrates
3.4. TBE Induces Autophagy against Protein Aggregation
3.5. Quercetin Is Responsible for the TBE Effect on Autophagy Induction
3.6. TBE and Quercetin Promote Aggrephagy in Liver Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Nakatogawa, H. Mechanisms Governing Autophagosome Biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A. Autophagy and Disease. J. Biol. Chem. 2018, 293, 5425–5430. [Google Scholar] [CrossRef] [Green Version]
- Fleming, A.; Noda, T.; Yoshimori, T.; Rubinsztein, D.C. Chemical Modulators of Autophagy as Biological Probes and Potential Therapeutics. Nat. Chem. Biol. 2011, 7, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-F.; Wu, M.-Y.; Cai, C.-Z.; Li, M.; Lu, J.-H. Autophagy Modulators from Traditional Chinese Medicine: Mechanisms and Therapeutic Potentials for Cancer and Neurodegenerative Diseases. J. Ethnopharmacol. 2016, 194, 861–876. [Google Scholar] [CrossRef] [PubMed]
- Ikari, S.; Lu, S.-L.; Hao, F.; Imai, K.; Araki, Y.; Yamamoto, Y.; Tsai, C.-Y.; Nishiyama, Y.; Shitan, N.; Yoshimori, T.; et al. Starvation-Induced Autophagy via Calcium-Dependent TFEB Dephosphorylation Is Suppressed by Shigyakusan. PLoS ONE 2020, 15, e0230156. [Google Scholar] [CrossRef]
- Jin, H.-M.; Wei, P. Anti-Fatigue Properties of Tartary Buckwheat Extracts in Mice. Int. J. Mol. Sci. 2011, 12, 4770–4780. [Google Scholar] [CrossRef] [Green Version]
- Sadauskiene, I.; Liekis, A.; Bernotiene, R.; Sulinskiene, J.; Kasauskas, A.; Zekonis, G. The Effects of Buckwheat Leaf and Flower Extracts on Antioxidant Status in Mouse Organs. Oxidative Med. Cell. Longev. 2018, 2018, 1–7. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Qin, P.; Shan, F.; Ren, G. Flavonoid Composition, Antibacterial and Antioxidant Properties of Tartary Buckwheat Bran Extract. Ind. Crop. Prod. 2013, 49, 312–317. [Google Scholar] [CrossRef]
- Kimura, S.; Fujita, N.; Noda, T.; Yoshimori, T. Monitoring Autophagy in Mammalian Cultured Cells through the Dynamics of LC3. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 452, pp. 1–12. ISBN 978-0-12-374547-7. [Google Scholar]
- Kabeya, Y. LC3, a Mammalian Homologue of Yeast Apg8p, Is Localized in Autophagosome Membranes after Processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Kimura, S.; Noda, T.; Yoshimori, T. Dissection of the Autophagosome Maturation Process by a Novel Reporter Protein, Tandem Fluorescent-Tagged LC3. Autophagy 2007, 3, 452–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, T. Regulation of Autophagy through TORC1 and MTORC1. Biomolecules 2017, 7, 52. [Google Scholar] [CrossRef]
- Tanida, I.; Minematsu-Ikeguchi, N.; Ueno, T.; Kominami, E. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 Is a Marker for Autophagy. Autophagy 2005, 1, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Yamamoto, A.; Hatano, M.; Kobayashi, Y.; Kabeya, Y.; Suzuki, K.; Tokuhisa, T.; Ohsumi, Y.; Yoshimori, T. Dissection of Autophagosome Formation Using Apg5-Deficient Mouse Embryonic Stem Cells. J. Cell Biol. 2001, 152, 657–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N. The Role of the Atg1/ULK1 Complex in Autophagy Regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Ruckerbauer, S.; Stierhof, Y.-D.; Berg, C.; Nordheim, A. Human WIPI-1 Puncta-Formation: A Novel Assay to Assess Mammalian Autophagy. FEBS Lett. 2007, 581, 3396–3404. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Arencibia, M.G.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Fingar, D.C.; Richardson, C.J.; Tee, A.R.; Cheatham, L.; Tsou, C.; Blenis, J. MTOR Controls Cell Cycle Progression through Its Cell Growth Effectors S6K1 and 4E-BP1/Eukaryotic Translation Initiation Factor 4E. MCB 2004, 24, 200–216. [Google Scholar] [CrossRef] [Green Version]
- Zaffagnini, G.; Martens, S. Mechanisms of Selective Autophagy. J. Mol. Biol. 2016, 428, 1714–1724. [Google Scholar] [CrossRef] [Green Version]
- Kirkin, V.; Lamark, T.; Sou, Y.-S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.-A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic Levels of P62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, T.; Masaki, K.; Kashiwagi, T. An Enzyme Degrading Rutin in Tartary Buckwheat Seeds. Nippon Shokuhin Kogyo Gakkaishi 1992, 39, 994–1000. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Yuan, Y.; Gu, J.; Chen, P. Purification, characterization and partial primary structure analysis of rutin-degrading enzyme in tartary buckwheat seeds. Sheng Wu Gong Cheng Xue Bao 2017, 33, 796–807. [Google Scholar] [CrossRef]
- French, S.; Masouminia, M.; Samadzadeh, S.; Tillman, B.; Mendoza, A.; French, B. Role of Protein Quality Control Failure in Alcoholic Hepatitis Pathogenesis. Biomolecules 2017, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of Starvation-Induced and Constitutive Autophagy in Atg7-Deficient Mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, R.; Li, J.; Mao, J.; Lei, Y.; Wu, J.; Zeng, J.; Zhang, T.; Wu, H.; Chen, L.; et al. Quercetin Induces Protective Autophagy in Gastric Cancer Cells: Involvement of Akt-MTOR- and Hypoxia-Induced Factor 1α-Mediated Signaling. Autophagy 2011, 7, 966–978. [Google Scholar] [CrossRef] [Green Version]
- Klappan, A.K.; Hones, S.; Mylonas, I.; Brüning, A. Proteasome Inhibition by Quercetin Triggers Macroautophagy and Blocks MTOR Activity. Histochem. Cell Biol. 2012, 137, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ji, X.-J.; Zhou, Y.-X.; Yao, X.-Q.; Liu, Y.-Q.; Zhang, F.; Yin, X.-X. Quercetin Inhibits the MTORC1/P70S6K Signaling-Mediated Renal Tubular Epithelial–Mesenchymal Transition and Renal Fibrosis in Diabetic Nephropathy. Pharmacol. Res. 2015, 99, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhao, H.; Wang, Z.; Zhang, C.; Bian, Y.; Liu, X.; Zhang, C.; Zhang, X.; Zhao, Y. Quercetin Promotes in Vitro Maturation of Oocytes from Humans and Aged Mice. Cell Death Dis. 2020, 11, 965. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, X.; Wang, D.; Peng, P.; Xu, X.; Gao, B.; Zheng, C.; Wang, H.; Jia, H.; Shang, Q.; et al. Quercetin Suppresses Apoptosis and Attenuates Intervertebral Disc Degeneration via the SIRT1-Autophagy Pathway. Front. Cell Dev. Biol. 2020, 8, 613006. [Google Scholar] [CrossRef]
- Daw, S.; Law, S. Quercetin Induces Autophagy in Myelodysplastic Bone Marrow Including Hematopoietic Stem/Progenitor Compartment. Environ. Toxicol. 2021, 36, 149–167. [Google Scholar] [CrossRef]
- Li, D.L.; Mao, L.; Gu, Q.; Wei, F.; Gong, Y.-Y. Quercetin Protects Retina External Barrier from Oxidative Stress Injury by Promoting Autophagy. Cutan. Ocul. Toxicol. 2021, 40, 7–13. [Google Scholar] [CrossRef]
- Du, G.; Zhao, Z.; Chen, Y.; Li, Z.; Tian, Y.; Liu, Z.; Liu, B.; Song, J. Quercetin Attenuates Neuronal Autophagy and Apoptosis in Rat Traumatic Brain Injury Model via Activation of PI3K/Akt Signaling Pathway. Neurol. Res. 2016, 38, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, Y.; Shaw, A.M.; Goldfine, H.; Tian, J.; Cai, J. Enhancing TFEB-Mediated Cellular Degradation Pathways by the MTORC1 Inhibitor Quercetin. Oxidative Med. Cell. Longev. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-Deficient Mice Develop Multiple Liver Tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Xu, Y.; Zhang, S.; Sun, J.; Liu, P.; Xiao, L.; Tang, Y.; Liu, L.; Yao, P. Quercetin Attenuates Chronic Ethanol-Induced Hepatic Mitochondrial Damage through Enhanced Mitophagy. Nutrients 2016, 8, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Guo, X.; Zhou, F.; Xiao, L.; Liu, J.; Jiang, C.; Xing, M.; Yao, P. Quercetin Alleviates Ethanol-Induced Liver Steatosis Associated with Improvement of Lipophagy. Food Chem. Toxicol. 2019, 125, 21–28. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikari, S.; Yang, Q.; Lu, S.-L.; Liu, Y.; Hao, F.; Tong, G.; Lu, S.; Noda, T. Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations. Antioxidants 2021, 10, 1217. https://doi.org/10.3390/antiox10081217
Ikari S, Yang Q, Lu S-L, Liu Y, Hao F, Tong G, Lu S, Noda T. Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations. Antioxidants. 2021; 10(8):1217. https://doi.org/10.3390/antiox10081217
Chicago/Turabian StyleIkari, Sumiko, Qiang Yang, Shiou-Ling Lu, Yuancai Liu, Feike Hao, Guoqiang Tong, Shiguang Lu, and Takeshi Noda. 2021. "Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations" Antioxidants 10, no. 8: 1217. https://doi.org/10.3390/antiox10081217
APA StyleIkari, S., Yang, Q., Lu, S.-L., Liu, Y., Hao, F., Tong, G., Lu, S., & Noda, T. (2021). Quercetin in Tartary Buckwheat Induces Autophagy against Protein Aggregations. Antioxidants, 10(8), 1217. https://doi.org/10.3390/antiox10081217