PtrMYB3, a R2R3-MYB Transcription Factor from Poncirus trifoliata, Negatively Regulates Salt Tolerance and Hydrogen Peroxide Scavenging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
2.2. Quantitative Real-Time Polymerase Chain Reaction (qPCR) Analysis
2.3. Cloning and Sequence Analysis of PtrMYB3
2.4. Subcellular Localization
2.5. Transactivation Activity Assay
2.6. Genetic Transformation of Tobacco
2.7. Virus Induced Gene Silencing (VIGS)
2.8. Salt Tolerance Assay
2.9. Dual-Luciferase Reporter Assay
2.10. Hydrogen Peroxide (H2O2) Measurement
2.11. Statistical Analysis
3. Results
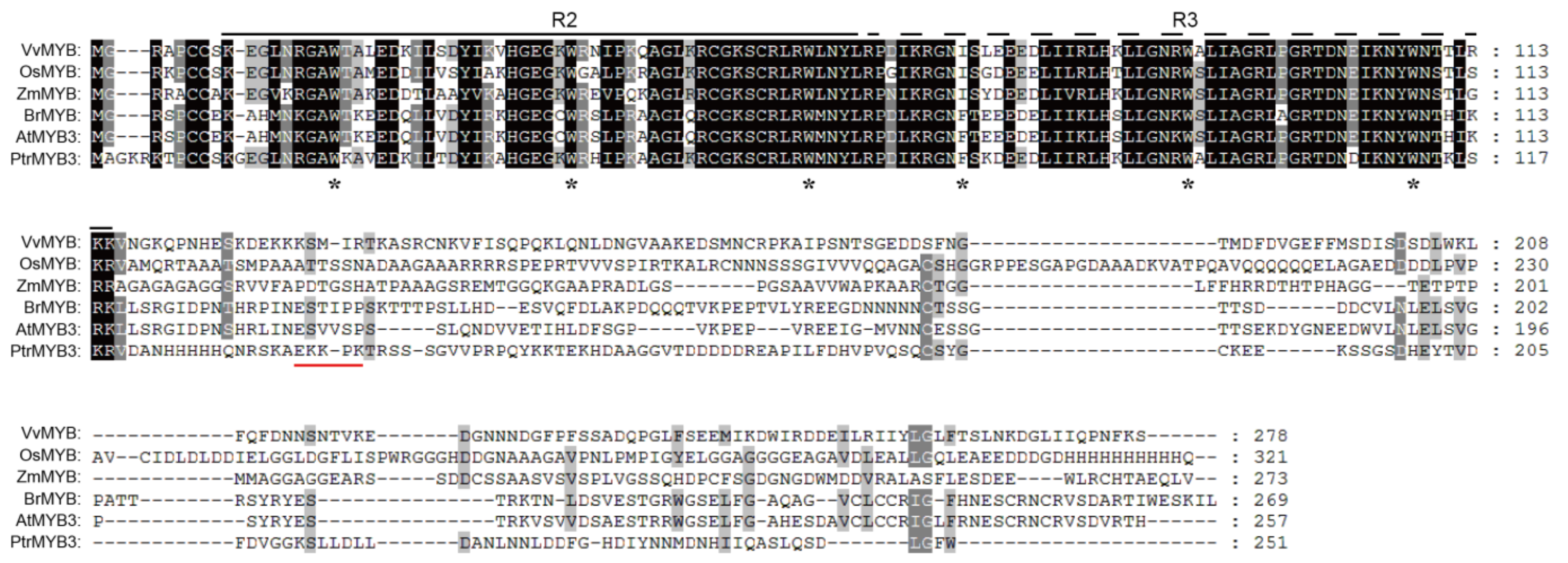
3.1. Cloning and Sequence Analysis of PtrMYB3
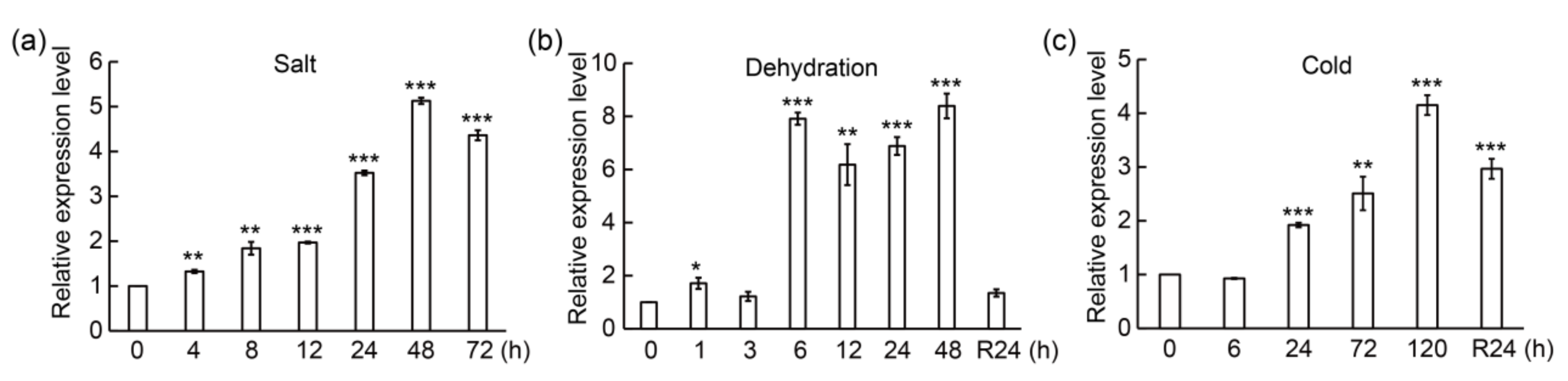
3.2. PtrMYB3 Is Induced by Multiple Abiotic Stresses
3.3. PtrMYB3 Is Localized to Nucleus
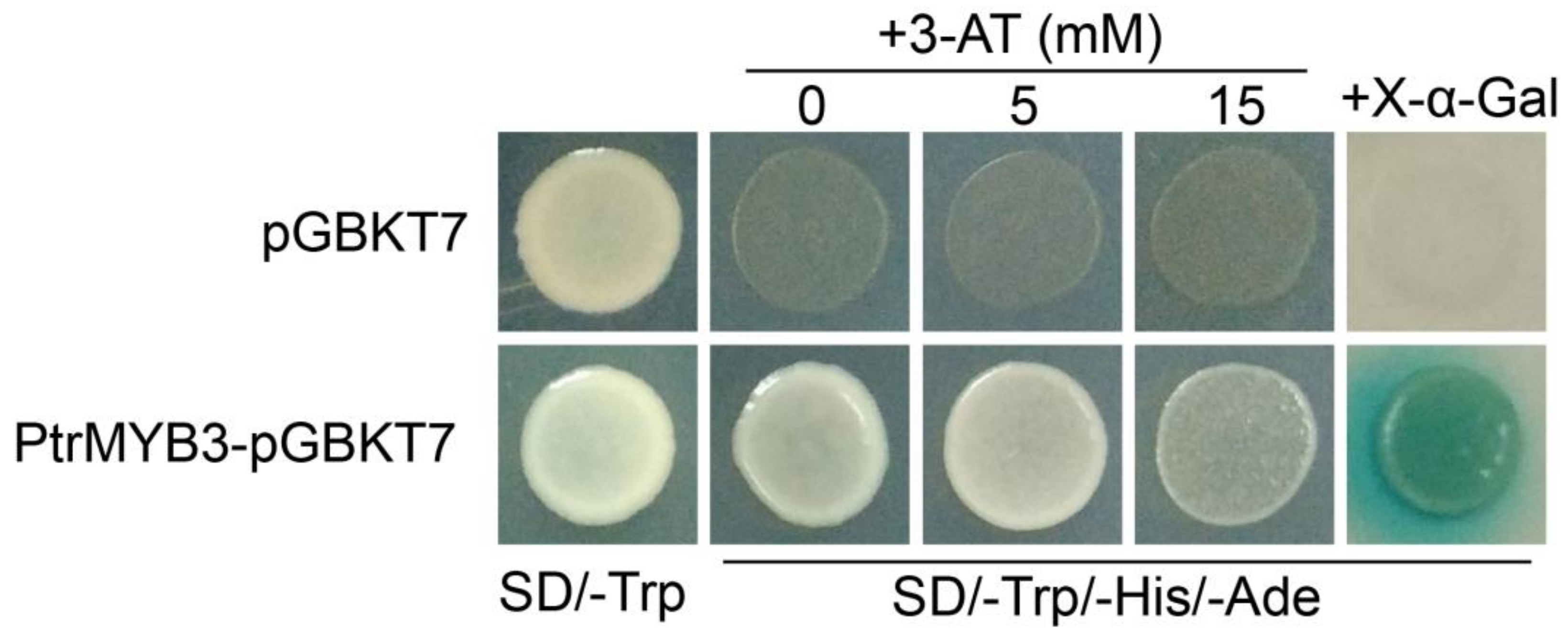
3.4. PtrMYB3 Has Transactivation Activity
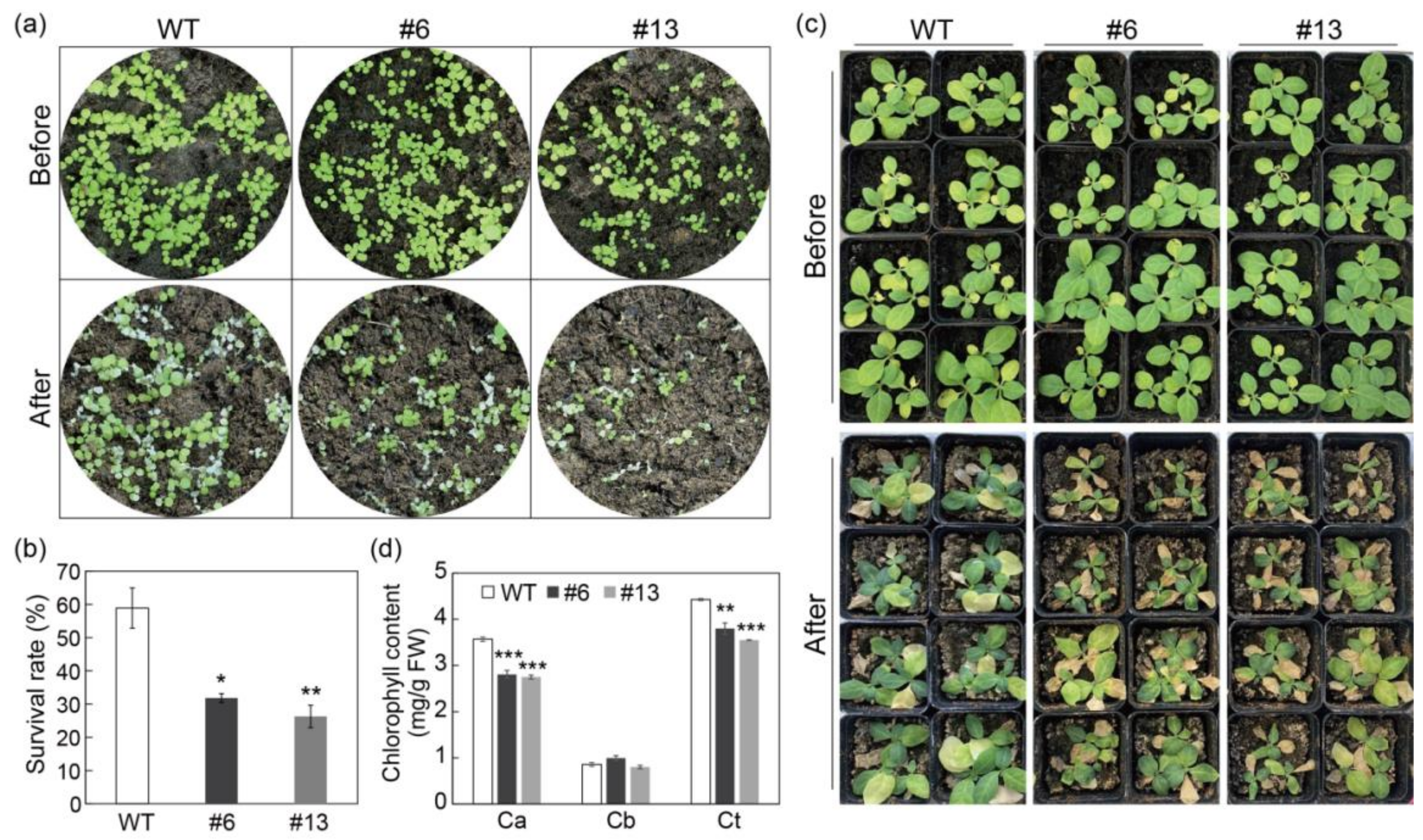
3.5. Overexpression of PtrMYB3 Impairs the Salt Tolerance of Tobacco
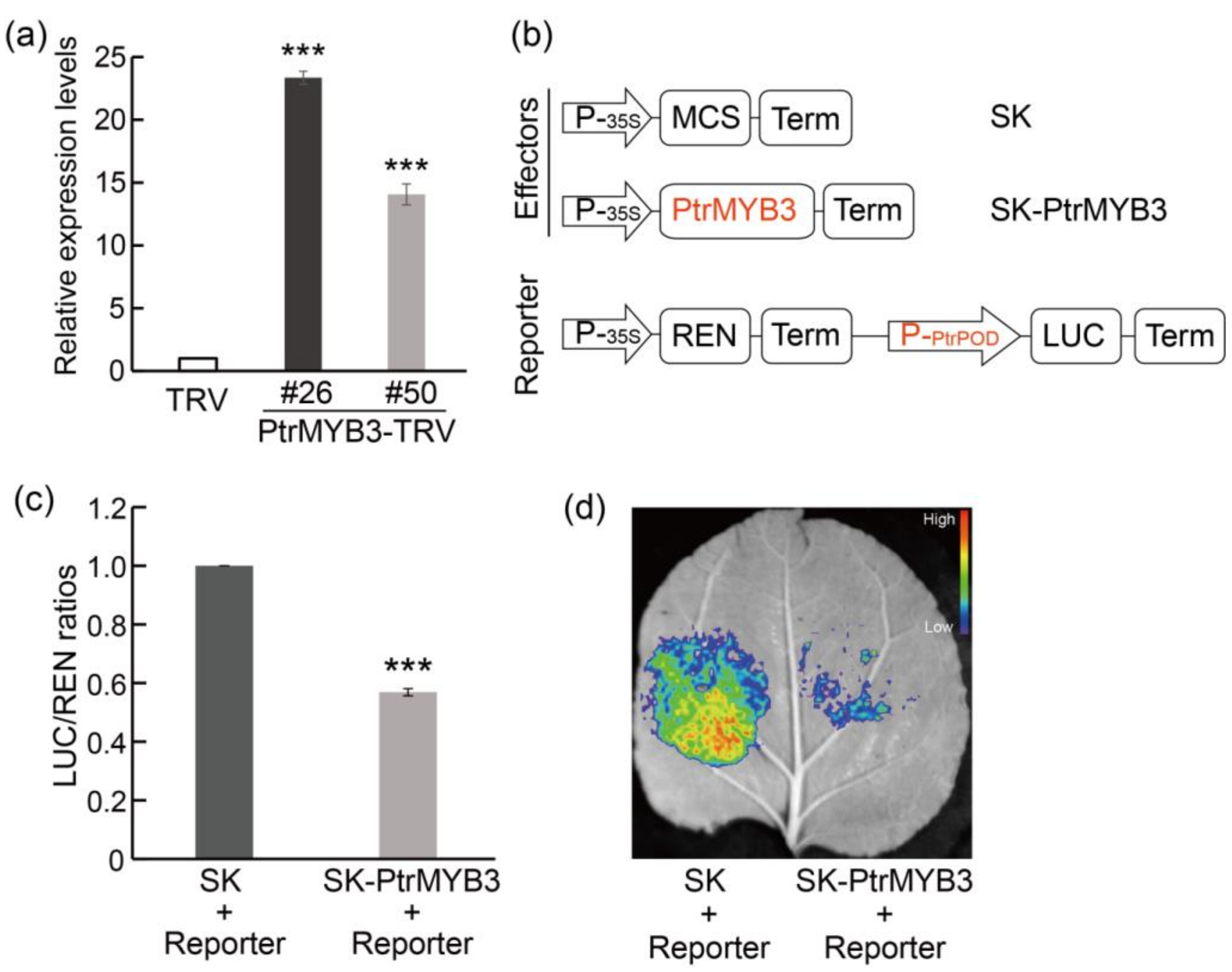
3.6. Silencing of PtrMYB3 in Trifoliate Orange Confers Enhanced Salt Tolerance
3.7. PtrMYB3 Negatively Regulates the Expression of a Peroxidase Gene, PtrPOD
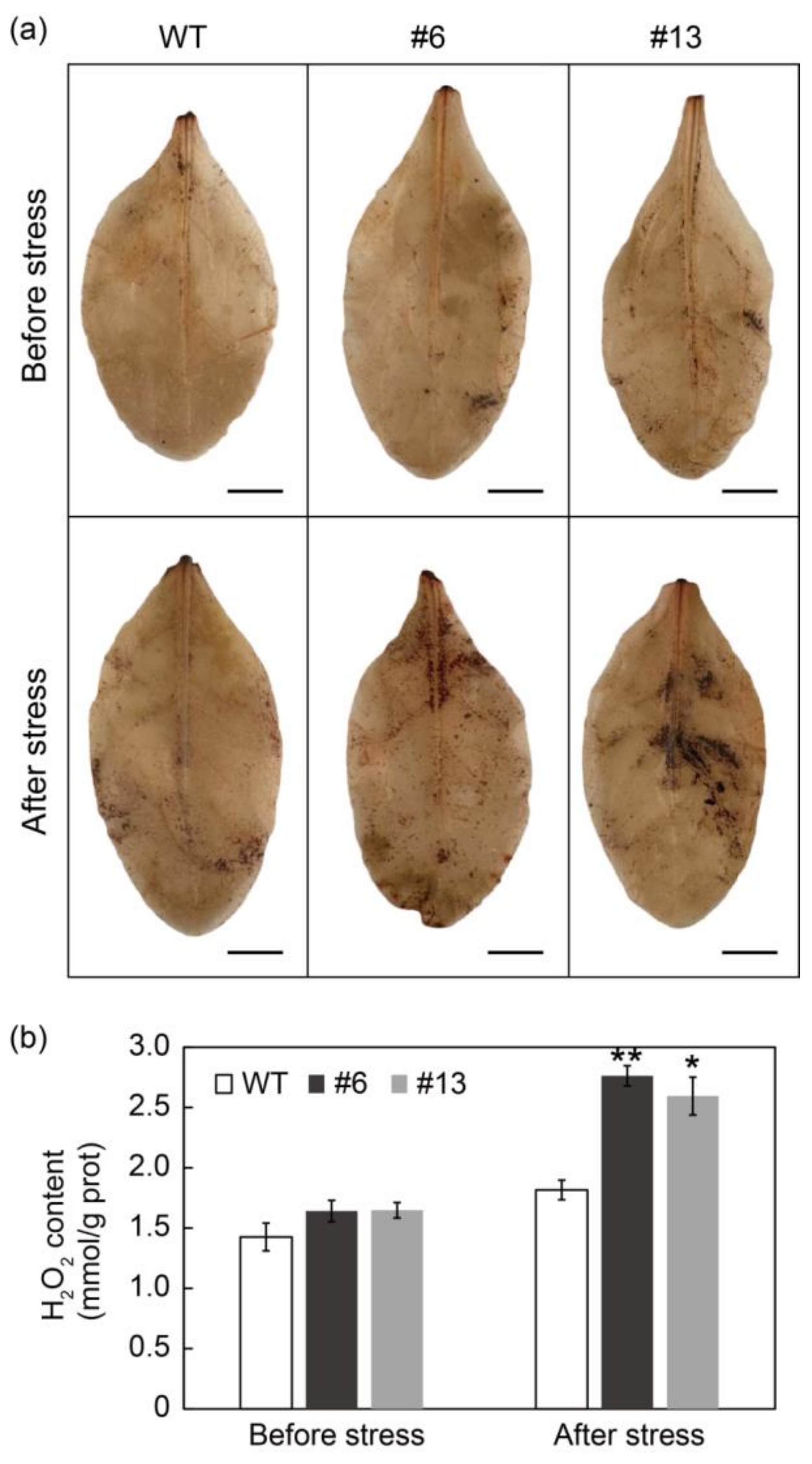
3.8. PtrMYB3 Transgenic Tobacco Plants Accumulated Excess H2O2 after Salt Stress
4. Discussion
4.1. PtrMYB3 Is Induced by Various Stresses
4.2. PtrMYB3 Negatively Regulates Salt Tolerance
4.3. PtrMYB3 Regulates PtrPOD-Mediated H2O2 Accumulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-H.; Peng, T.; Dai, W. Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Li, C.; Ng, C.K.Y.; Fan, L.-M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 2012, 1819, 67–77. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Yang, S.-S.; Liang, Z.; Feng, B.-R.; Liu, L.; Huang, Y.-B.; Tang, Y.-X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wang, X.; Xu, Y.; Deng, X.; Xu, Q. Genome-wide analysis of the R2R3-MYB transcription factor gene family in sweet orange (Citrus sinensis). Mol. Biol. Rep. 2014, 41, 6769–6785. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Zhang, S.; Feng, M.; Chen, W.; Zhou, X.; Lu, J.; Wang, Y.; Li, Y.; Jiang, C.-Z.; Gan, S.-S.; Ma, N.; et al. In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat. Plants 2019, 5, 290–299. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Y.; Liang, Y.; Chen, L.; Chen, W.; Cheng, B. Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 2019, 137, 179–188. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, C.; Liu, Y.; Zhao, L.; Zhang, S.; Huang, X. A novel MYB transcription factor regulates ascorbic acid synthesis and affects cold tolerance. Plant Cell Environ. 2019, 42, 832–845. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Zheng, Y.; Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Song, M.; Guo, Y.; Liu, L.; Xue, H.; Dai, H.; Zhang, Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Li, S.; Ding, A.; Zhang, Z.; Hao, Q.; Wang, K.; Liu, Q.; Liu, Q. The novel rose MYB transcription factor RhMYB96 enhances salt tolerance in transgenic Arabidopsis. Plant Mol. Biol. Rep. 2018, 36, 406–417. [Google Scholar] [CrossRef]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.-W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef] [Green Version]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-K.; Cao, Z.-H.; Hao, Y.-J. Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 2014, 150, 76–87. [Google Scholar] [CrossRef]
- Shan, H.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H.; et al. Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Cai, H.; Tian, S.; Liu, C.; Dong, H. Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene 2011, 485, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Rahaie, M.; Xue, G.-P.; Naghavi, M.R.; Alizadeh, H.; Schenk, P.M. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010, 29, 835–844. [Google Scholar] [CrossRef]
- Ahmad, P. Water Stress and Crop Plants: A Sustainable Approach; John Wiley & Sons: Hoboken, NJ, USA, 2016; Volume 1, pp. 1–756. [Google Scholar]
- De Rossi, S.; Di Marco, G.; Bruno, L.; Gismondi, A.; Canini, A. Investigating the drought and salinity effect on the redox components of Sulla Coronaria (L.) Medik. Antioxidants 2021, 10, 1048. [Google Scholar] [CrossRef]
- Hossain, M.S.; ElSayed, A.I.; Moore, M.; Dietz, K.J. Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. J. Exp. Bot. 2017, 68, 1283–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Dai, W.; Du, J.; Ming, R.; Dahro, B.; Liu, J.-H. ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 2019, 17, 1316–1332. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.; Liu, J.-H. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 2018, 69, 2677–2692. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Wang, M.; Hu, J.; Wang, W.; Fu, X.; Liu, J.-H. PtrABF of Poncirus trifoliata functions in dehydration tolerance by reducing stomatal density and maintaining reactive oxygen species homeostasis. J. Exp. Bot. 2015, 66, 5911–5927. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Xu, Y.; Jiang, X.; Yu, H.; Jia, H.; Tan, C.; Hu, G.; Hu, Y.; Rao, M.J.; Deng, X.; et al. Genome of a citrus rootstock and global DNA demethylation caused by heterografting. Hortic. Res. 2021, 8, 69. [Google Scholar] [CrossRef]
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.; Rawat, N.; Du, D.; Parajuli, S.; Yu, Q.; You, Q.; Rokhsar, D.S.; et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Liu, J.-H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 88. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar] [CrossRef]
- Dai, W.; Wang, M.; Gong, X.; Liu, J.-H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Wang, Y.; Xie, Z.; Guo, D.; Chen, C.; Fan, Q.; Deng, X.; Liu, J.-H. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata. Plant Biotechnol. J. 2019, 17, 1394–1407. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Albert, N.W.; Thrimawithana, A.H.; McGhie, T.K.; Clayton, W.A.; Deroles, S.C.; Schwinn, K.E.; Bowman, J.L.; Jordan, B.R.; Davies, K.M. Genetic analysis of the liverwort Marchantia polymorpha reveals that R2R3MYB activation of flavonoid production in response to abiotic stress is an ancient character in land plants. New Phytol. 2018, 218, 554–566. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Sun, T.; Pan, Q.; Anupol, N.; Chen, H.; Shi, J.; Liu, F.; Deqiang, D.; Wang, C.; Zhao, J.; et al. RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 2019, 17, 2078–2095. [Google Scholar] [CrossRef] [Green Version]
- Navrot, N.; Collin, V.r.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.-P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Xing, C.; Yao, Z.; Huang, X. PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 2017, 15, 1186–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.-Q.; Ba, L.-J.; Shan, W.; Xiao, Y.-Y.; Lu, W.-J.; Kuang, J.-F.; Chen, J.-Y. A banana R2R3-MYB transcription factor MaMYB3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and MabHLH6. Plant J. 2018, 96, 1191–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.-S.; Wang, W.; Zhang, Q.; Liu, J.-H. A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 2013, 162, 1178–1194. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, T.; Guo, D.; Liu, J. PtrMYB3, a R2R3-MYB Transcription Factor from Poncirus trifoliata, Negatively Regulates Salt Tolerance and Hydrogen Peroxide Scavenging. Antioxidants 2021, 10, 1388. https://doi.org/10.3390/antiox10091388
Wei T, Guo D, Liu J. PtrMYB3, a R2R3-MYB Transcription Factor from Poncirus trifoliata, Negatively Regulates Salt Tolerance and Hydrogen Peroxide Scavenging. Antioxidants. 2021; 10(9):1388. https://doi.org/10.3390/antiox10091388
Chicago/Turabian StyleWei, Tonglu, Dalong Guo, and Jihong Liu. 2021. "PtrMYB3, a R2R3-MYB Transcription Factor from Poncirus trifoliata, Negatively Regulates Salt Tolerance and Hydrogen Peroxide Scavenging" Antioxidants 10, no. 9: 1388. https://doi.org/10.3390/antiox10091388
APA StyleWei, T., Guo, D., & Liu, J. (2021). PtrMYB3, a R2R3-MYB Transcription Factor from Poncirus trifoliata, Negatively Regulates Salt Tolerance and Hydrogen Peroxide Scavenging. Antioxidants, 10(9), 1388. https://doi.org/10.3390/antiox10091388





