Human Cystathionine γ-Lyase Is Inhibited by s-Nitrosation: A New Crosstalk Mechanism between NO and H2S
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Expression and Purification
2.3. Differential Scanning Fluorimetry
2.4. Spectrophotometric Measurements
2.5. Enzymatic Activity Assays
2.6. Determination of the Number of Free Exposed Thiols in Human Cystathionine γ-Lyase
2.7. Effect of CSE S-Nitrosation/Denitrosation on PLP Cofactor Load
2.8. Liquid Chromatography-High Resolution Mass Spectrometry (LC-HRMS) Analysis
2.9. Database Searching and Processing
2.10. Statistical Analysis
3. Results
3.1. Effect of GSNO on CSE Enzymatic Activity and Cofactor Load
3.2. Effect of GSNO on Free Exposed Cysteines in WT CSE
3.3. Identification of Cys229 as the Exposed s-Nitrosated Cysteine
3.4. Cys229 s-Nitrosation Does Not Account for GSNO-Inhibition
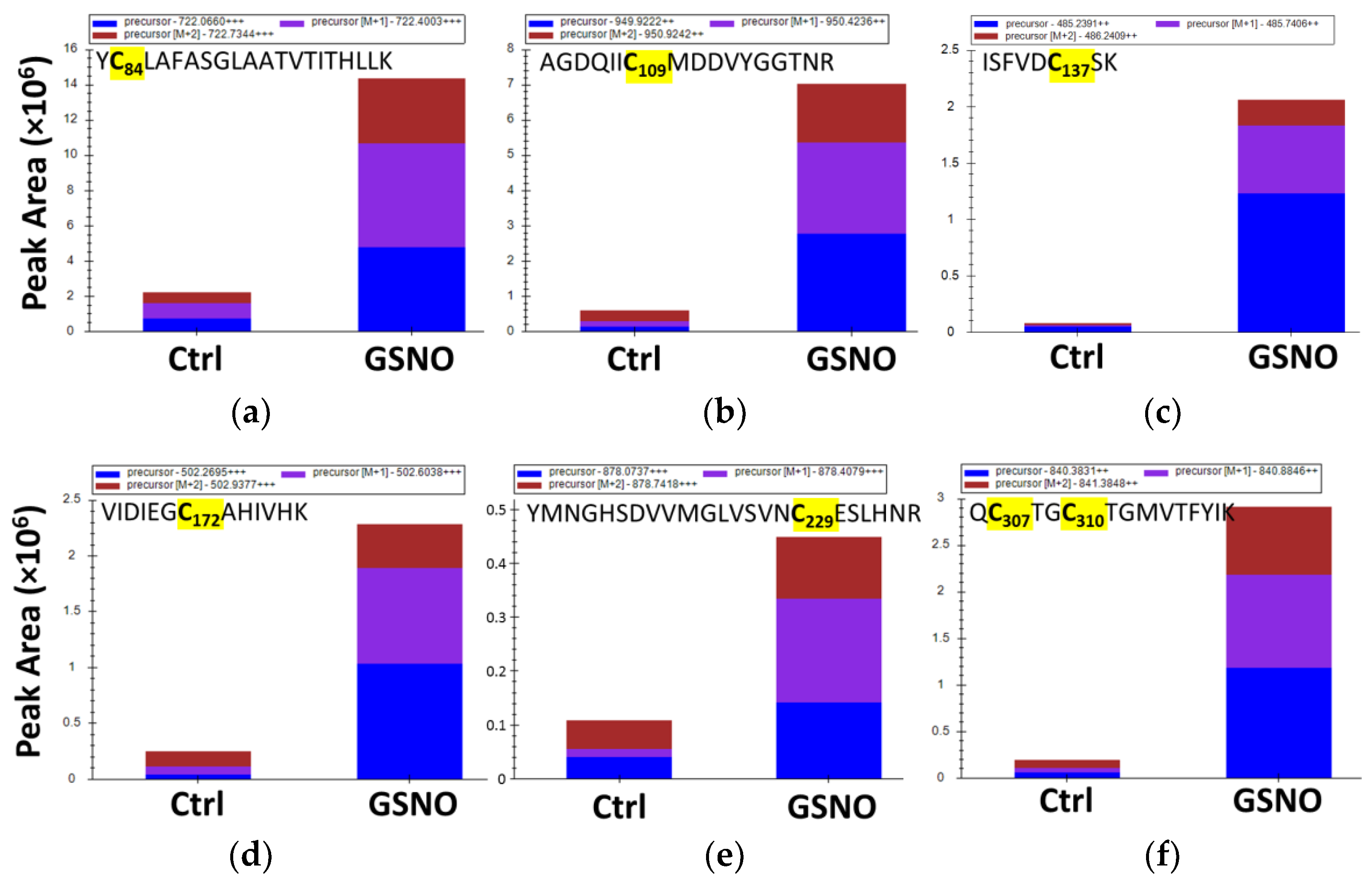
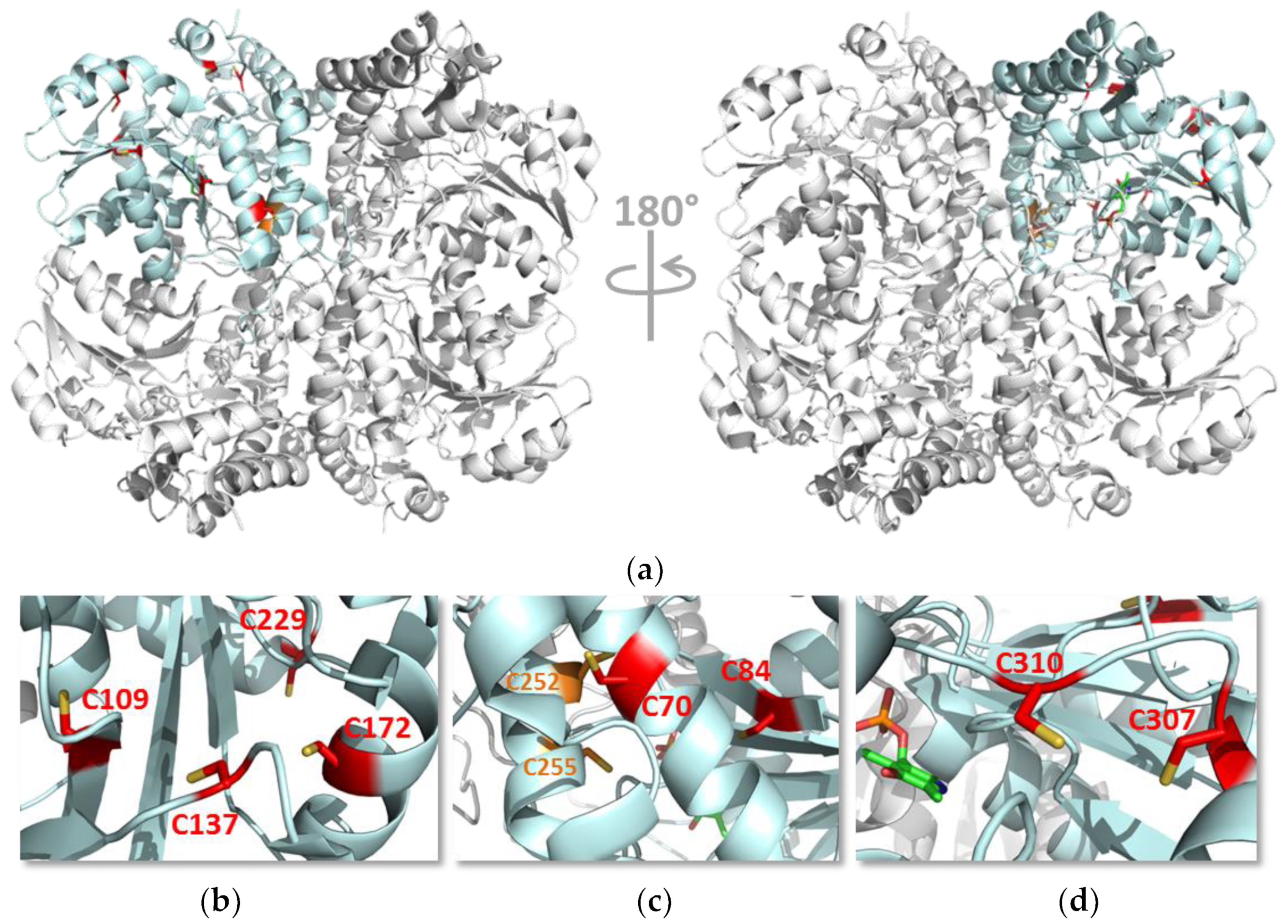
3.5. Identification of Buried s-Nitrosated Cysteines
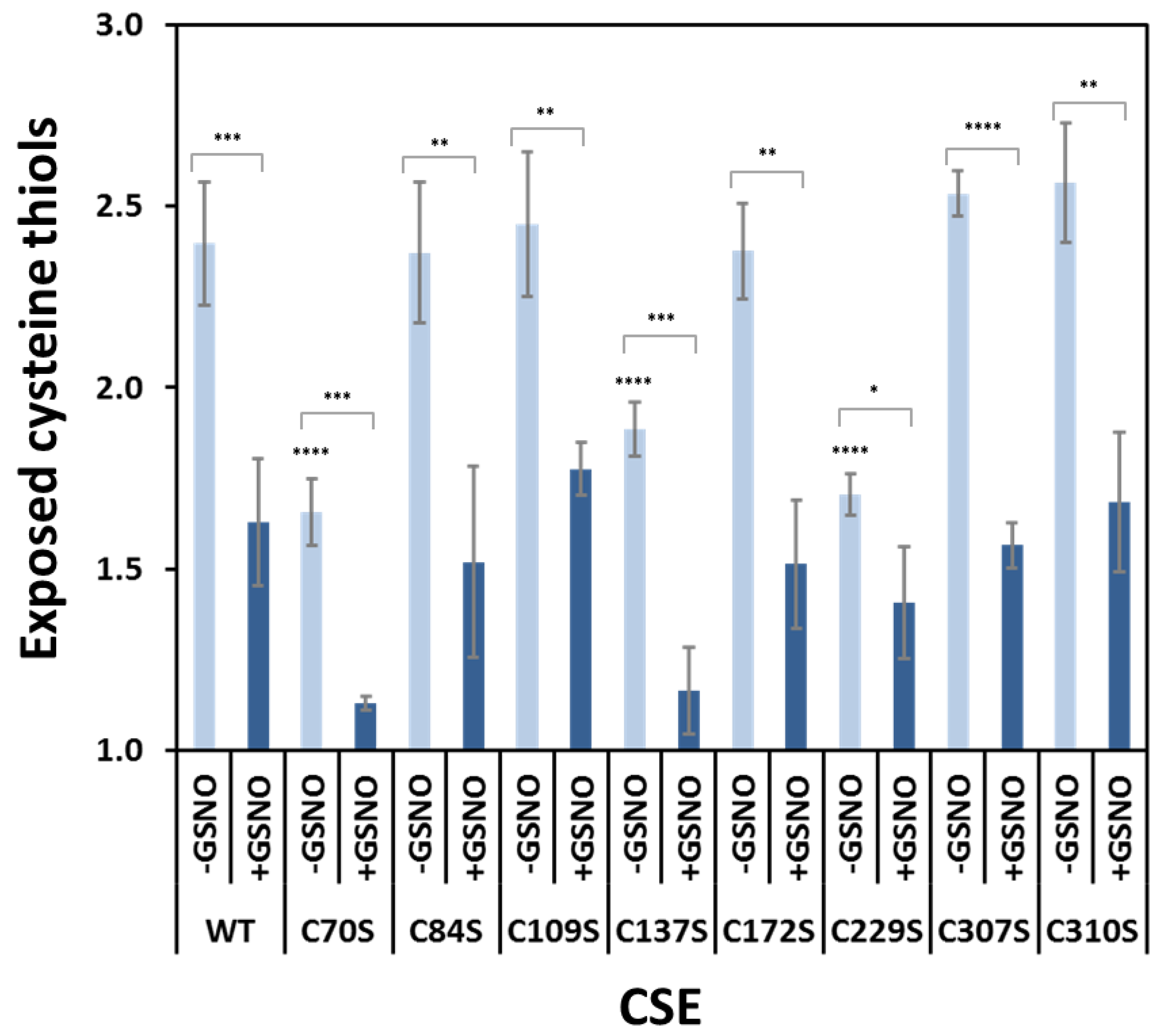
3.6. Functional Analysis of the Serine Variants of Cys70, Cys84, Cys109, Cys137, Cys172, Cys307 and Cys310
3.7. Effect of GSNO on Exposed Cysteines in CSE Cys-to-Ser Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bianco, C.L.; Toscano, J.P.; Fukuto, J.M. Chapter 2—An Integrated View of the Chemical Biology of NO, CO, H2S, and O2 A2. In Nitric Oxide, 3rd ed.; Freeman, B.A., Ignarro, L.J., Eds.; Academic Press: London, UK, 2017; pp. 9–21. [Google Scholar] [CrossRef]
- Fukuto, J.M.; Carrington, S.J.; Tantillo, D.J.; Harrison, J.G.; Ignarro, L.J.; Freeman, B.A.; Chen, A.; Wink, D.A. Small molecule signaling agents: The integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species. Chem. Res. Toxicol. 2012, 25, 769–793. [Google Scholar] [CrossRef]
- Wang, R. Gasotransmitters: Growing pains and joys. Trends Biochem. Sci. 2014, 39, 227–232. [Google Scholar] [CrossRef]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical Biology of H2S Signaling through Persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef]
- Giuffrè, A.; Vicente, J.B. Hydrogen Sulfide Biochemistry and Interplay with Other Gaseous Mediators in Mammalian Physiology. Oxid. Med. Cell Longev. 2018, 2018, 6290931. [Google Scholar] [CrossRef]
- Szabo, C. Gasotransmitters in cancer: From pathophysiology to experimental therapy. Nat. Rev. Drug Discov. 2016, 15, 185–203. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Padovani, D.; Leslie, R.A.; Chiku, T.; Banerjee, R. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J. Biol. Chem. 2009, 284, 22457–22466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, R. Catalytic promiscuity and heme-dependent redox regulation of H2S synthesis. Curr. Opin. Chem. Biol. 2017, 37, 115–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabil, O.; Yadav, V.; Banerjee, R. Heme-dependent Metabolite Switching Regulates H2S Synthesis in Response to Endoplasmic Reticulum (ER) Stress. J. Biol. Chem. 2016, 291, 16418–16423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Lin, A.; Banerjee, R. Kinetic properties of polymorphic variants and pathogenic mutants in human cystathionine gamma-lyase. Biochemistry 2008, 47, 6226–6232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hegele, R.A. Genomic basis of cystathioninuria (MIM 219500) revealed by multiple mutations in cystathionine gamma-lyase (CTH). Hum. Genet. 2003, 112, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Tadiboyina, V.T.; Rupar, A.; Atkison, P.; Feigenbaum, A.; Kronick, J.; Wang, J.; Hegele, R.A. Novel mutation in DGUOK in hepatocerebral mitochondrial DNA depletion syndrome associated with cystathioninuria. Am. J. Med. Genet. A 2005, 135, 289–291. [Google Scholar] [CrossRef]
- Kabil, O.; Banerjee, R. Enzymology of H2S biogenesis, decay and signaling. Antioxid. Redox Signal. 2014, 20, 770–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, N.; Paul, B.D.; Gadalla, M.M.; Mustafa, A.K.; Sen, T.; Xu, R.; Kim, S.; Snyder, S.H. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol. Cell 2012, 45, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Badiei, A.; Gieseg, S.; Davies, S.; Izani Othman, M.; Bhatia, M. LPS Up-Regulates Cystathionine gamma -Lyase Gene Expression in Primary Human Macrophages via NF-kappaB/ERK Pathway. Inflamm. Allergy Drug Targets 2015, 14, 99–104. [Google Scholar] [CrossRef]
- Ozaki, T.; Tsubota, M.; Sekiguchi, F.; Kawabata, A. Involvement of NF-kappaB in the upregulation of cystathionine-gamma-lyase, a hydrogen sulfide-forming enzyme, and bladder pain accompanying cystitis in mice. Clin. Exp. Pharmacol. Physiol. 2018, 45, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Dickhout, J.G.; Carlisle, R.E.; Jerome, D.E.; Mohammed-Ali, Z.; Jiang, H.; Yang, G.; Mani, S.; Garg, S.K.; Banerjee, R.; Kaufman, R.J.; et al. Integrated stress response modulates cellular redox state via induction of cystathionine gamma-lyase: Cross-talk between integrated stress response and thiol metabolism. J. Biol. Chem. 2012, 287, 7603–7614. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, J.; Sato, W.; Kosugi, T.; Yamamoto, T.; Kimura, T.; Taniguchi, S.; Kojima, H.; Maruyama, S.; Imai, E.; Matsuo, S.; et al. Distribution of hydrogen sulfide (H(2)S)-producing enzymes and the roles of the H(2)S donor sodium hydrosulfide in diabetic nephropathy. Clin. Exp. Nephrol. 2013, 17, 32–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusuf, M.; Kwong Huat, B.T.; Hsu, A.; Whiteman, M.; Bhatia, M.; Moore, P.K. Streptozotocin-induced diabetes in the rat is associated with enhanced tissue hydrogen sulfide biosynthesis. Biochem. Biophys. Res. Commun. 2005, 333, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Guo, Z.; Wang, S. Regulation of cystathionine gamma-lyase in mammalian cells by hypoxia. Biochem. Genet. 2014, 52, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Guo, Z.; Wang, S. The effect of certain conditions in the regulation of cystathionine gamma-lyase by exogenous hydrogen sulfide in mammalian cells. Biochem. Genet. 2013, 51, 503–513. [Google Scholar] [CrossRef]
- Agrawal, N.; Banerjee, R. Human polycomb 2 protein is a SUMO E3 ligase and alleviates substrate-induced inhibition of cystathionine beta-synthase sumoylation. PLoS ONE 2008, 3, e4032. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Ji, D.; Li, Y.; Cao, Y.; Zhang, S.; Yan, W.; Xue, K.; Chai, J.; Wu, Y.; Liu, H.; et al. Abnormal nitration and S-sulfhydration modification of Sp1-CSE-H2S pathway trap the progress of hyperhomocysteinemia into a vicious cycle. Free Radic. Biol. Med. 2021, 164, 20–33. [Google Scholar] [CrossRef]
- Kevil, C.G.; Cortese-Krott, M.M.; Nagy, P.; Papapetropoulos, A.; Feelisch, M.; Szabo, C. Cooperative Interactions Between NO and H2S: Chemistry, Biology, Physiology, Pathophysiology. In Nitric Oxide, Biology and Pathobiology, 3rd ed.; Ignarro, L.J., Freeman, B.A., Eds.; Academic Press: London, UK, 2017. [Google Scholar]
- Cortese-Krott, M.M.; Koning, A.; Kuhnle, G.G.C.; Nagy, P.; Bianco, C.L.; Pasch, A.; Wink, D.A.; Fukuto, J.M.; Jackson, A.A.; van Goor, H.; et al. The Reactive Species Interactome: Evolutionary Emergence, Biological Significance, and Opportunities for Redox Metabolomics and Personalized Medicine. Antioxid. Redox Signal. 2017, 27, 684–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortese-Krott, M.M.; Kuhnle, G.G.; Dyson, A.; Fernandez, B.O.; Grman, M.; DuMond, J.F.; Barrow, M.P.; McLeod, G.; Nakagawa, H.; Ondrias, K.; et al. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc. Natl. Acad. Sci. USA 2015, 112, E4651–E4660. [Google Scholar] [CrossRef] [Green Version]
- Filipovic, M.R.; Miljkovic, J.; Nauser, T.; Royzen, M.; Klos, K.; Shubina, T.; Koppenol, W.H.; Lippard, S.J.; Ivanovic-Burmazovic, I. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J. Am. Chem. Soc. 2012, 134, 12016–12027. [Google Scholar] [CrossRef] [PubMed]
- Carballal, S.; Cuevasanta, E.; Marmisolle, I.; Kabil, O.; Gherasim, C.; Ballou, D.P.; Banerjee, R.; Alvarez, B. Kinetics of reversible reductive carbonylation of heme in human cystathionine beta-synthase. Biochemistry 2013, 52, 4553–4562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballal, S.; Cuevasanta, E.; Yadav, P.K.; Gherasim, C.; Ballou, D.P.; Alvarez, B.; Banerjee, R. Kinetics of Nitrite Reduction and Peroxynitrite Formation by Ferrous Heme in Human Cystathionine beta-Synthase. J. Biol. Chem. 2016, 291, 8004–8013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puranik, M.; Weeks, C.L.; Lahaye, D.; Kabil, O.; Taoka, S.; Nielsen, S.B.; Groves, J.T.; Banerjee, R.; Spiro, T.G. Dynamics of carbon monoxide binding to cystathionine beta-synthase. J. Biol. Chem. 2006, 281, 13433–13438. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.B.; Colaco, H.G.; Malagrino, F.; Santo, P.E.; Gutierres, A.; Bandeiras, T.M.; Leandro, P.; Brito, J.A.; Giuffrè, A. A Clinically Relevant Variant of the Human Hydrogen Sulfide-Synthesizing Enzyme Cystathionine beta-Synthase: Increased CO Reactivity as a Novel Molecular Mechanism of Pathogenicity? Oxid. Med. Cell Longev. 2017, 2017, 8940321. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.B.; Colaco, H.G.; Mendes, M.I.; Sarti, P.; Leandro, P.; Giuffrè, A. NO* binds human cystathionine beta-synthase quickly and tightly. J. Biol. Chem. 2014, 289, 8579–8587. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.B.; Colaco, H.G.; Sarti, P.; Leandro, P.; Giuffrè, A. S-Adenosyl-l-methionine Modulates CO and NO* Binding to the Human H2S-generating Enzyme Cystathionine beta-Synthase. J. Biol. Chem. 2016, 291, 572–581. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.B.; Malagrino, F.; Arese, M.; Forte, E.; Sarti, P.; Giuffrè, A. Bioenergetic relevance of hydrogen sulfide and the interplay between gasotransmitters at human cystathionine beta-synthase. Biochim. Biophys. Acta 2016, 1857, 1127–1138. [Google Scholar] [CrossRef]
- Cooper, C.E.; Brown, G.C. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 2008, 40, 533–539. [Google Scholar] [CrossRef]
- Kolluru, G.K.; Yuan, S.; Shen, X.; Kevil, C.G. H2S regulation of nitric oxide metabolism. Methods Enzymol. 2015, 554, 271–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szijarto, I.A.; Marko, L.; Filipovic, M.R.; Miljkovic, J.L.; Tabeling, C.; Tsvetkov, D.; Wang, N.; Rabelo, L.A.; Witzenrath, M.; Diedrich, A.; et al. Cystathionine gamma-Lyase-Produced Hydrogen Sulfide Controls Endothelial NO Bioavailability and Blood Pressure. Hypertension 2018, 71, 1210–1217. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ju, Y.; Fu, M.; Zhang, Y.; Pei, Y.; Racine, M.; Baath, S.; Merritt, T.J.S.; Wang, R.; Wu, L. Cystathionine gamma-lyase/hydrogen sulfide system is essential for adipogenesis and fat mass accumulation in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Asimakopoulou, A.; Panopoulos, P.; Chasapis, C.T.; Coletta, C.; Zhou, Z.; Cirino, G.; Giannis, A.; Szabo, C.; Spyroulias, G.A.; Papapetropoulos, A. Selectivity of commonly used pharmacological inhibitors for cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE). Br. J. Pharmacol. 2013, 169, 922–932. [Google Scholar] [CrossRef] [Green Version]
- Foster, M.W.; Hess, D.T.; Stamler, J.S. Protein S-nitrosylation in health and disease: A current perspective. Trends Mol. Med. 2009, 15, 391–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, N.; Doulias, P.T.; Tenopoulou, M.; Raju, K.; Ischiropoulos, H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J. Biol. Chem. 2013, 288, 26473–26479. [Google Scholar] [CrossRef] [Green Version]
- Smith, B.C.; Marletta, M.A. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Curr. Opin. Chem. Biol. 2012, 16, 498–506. [Google Scholar] [CrossRef] [Green Version]
- Hess, D.T.; Matsumoto, A.; Kim, S.O.; Marshall, H.E.; Stamler, J.S. Protein S-nitrosylation: Purview and parameters. Nat. Rev. Mol. Cell Biol. 2005, 6, 150–166. [Google Scholar] [CrossRef]
- Sun, Q.; Collins, R.; Huang, S.; Holmberg-Schiavone, L.; Anand, G.S.; Tan, C.H.; van-den-Berg, S.; Deng, L.W.; Moore, P.K.; Karlberg, T.; et al. Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S. J. Biol. Chem. 2009, 284, 3076–3085. [Google Scholar] [CrossRef] [Green Version]
- Thorson, M.K.; Majtan, T.; Kraus, J.P.; Barrios, A.M. Identification of cystathionine beta-synthase inhibitors using a hydrogen sulfide selective probe. Angew. Chem. Int. Ed. Engl. 2013, 52, 4641–4644. [Google Scholar] [CrossRef]
- Zuhra, K.; Sousa, P.M.F.; Paulini, G.; Lemos, A.R.; Kalme, Z.; Bisenieks, I.; Bisenieks, E.; Vigante, B.; Duburs, G.; Bandeiras, T.M.; et al. Screening Pyridine Derivatives against Human Hydrogen Sulfide-synthesizing Enzymes by Orthogonal Methods. Sci. Rep. 2019, 9, 684. [Google Scholar] [CrossRef]
- Noble, D.R.; Williams, D.L. Structure-reactivity studies of the Cu(2+)-catalyzed decomposition of four S-nitrosothiols based around the S-Nitrosocysteine/S-nitrosoglutathione structures. Nitric Oxide 2000, 4, 392–398. [Google Scholar] [CrossRef]
- Singh, R.J.; Hogg, N.; Joseph, J.; Kalyanaraman, B. Mechanism of nitric oxide release from S-nitrosothiols. J. Biol. Chem. 1996, 271, 18596–18603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stubauer, G.; Giuffrè, A.; Sarti, P. Mechanism of S-nitrosothiol formation and degradation mediated by copper ions. J. Biol. Chem. 1999, 274, 28128–28133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortese-Krott, M.M.; Fernandez, B.O.; Santos, J.L.; Mergia, E.; Grman, M.; Nagy, P.; Kelm, M.; Butler, A.; Feelisch, M. Nitrosopersulfide (SSNO(-)) accounts for sustained NO bioactivity of S-nitrosothiols following reaction with sulfide. Redox Biol. 2014, 2, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.R.; Clover, T.; Olaitan, A.D.; Becker, C.; Solouki, T.; Farmer, P.J. The reaction between GSNO and H2S: On the generation of NO, HNO and N2O. Nitric Oxide 2018, 77, 96–105. [Google Scholar] [CrossRef]
- Kumar, M.R.; Farmer, P.J. Characterization of Polysulfides, Polysulfanes, and Other Unique Species in the Reaction between GSNO and H2S. Molecules 2019, 24, 3090. [Google Scholar] [CrossRef] [Green Version]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009, 37, D169–D174. [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcock, L.J.; Perkins, M.V.; Chalker, J.M. Chemical methods for mapping cysteine oxidation. Chem. Soc. Rev. 2018, 47, 231–268. [Google Scholar] [CrossRef] [Green Version]
- Bonzon-Kulichenko, E.; Camafeita, E.; Lopez, J.A.; Gomez-Serrano, M.; Jorge, I.; Calvo, E.; Nunez, E.; Trevisan-Herraz, M.; Bagwan, N.; Barcena, J.A.; et al. Improved integrative analysis of the thiol redox proteome using filter-aided sample preparation. J. Proteomics. 2020, 214, 103624. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Iwabuchi, T.; Soga, T.; Kato, Y.; Yamamoto, T.; Takano, N.; Hishiki, T.; Ueno, Y.; Ikeda, S.; Sakuragawa, T.; et al. Cystathionine beta-synthase as a carbon monoxide-sensitive regulator of bile excretion. Hepatology 2009, 49, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Suematsu, M.; Nakamura, T.; Tokumoto, Y.; Yamamoto, T.; Kajimura, M.; Kabe, Y. CO-CBS-H2S Axis: From Vascular Mediator to Cancer Regulator. Microcirculation 2016, 23, 183–190. [Google Scholar] [CrossRef]
- Morikawa, T.; Kajimura, M.; Nakamura, T.; Hishiki, T.; Nakanishi, T.; Yukutake, Y.; Nagahata, Y.; Ishikawa, M.; Hattori, K.; Takenouchi, T.; et al. Hypoxic regulation of the cerebral microcirculation is mediated by a carbon monoxide-sensitive hydrogen sulfide pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 1293–1298. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Chen, Y.; Zhu, N.; Zhao, S.; Fan, J.; Liu, E. Hydrogen sulfide inhibits development of atherosclerosis through up-regulating protein S-nitrosylation. Biomed. Pharmacother. 2016, 83, 466–476. [Google Scholar] [CrossRef]
- Cao, X.; Ding, L.; Xie, Z.Z.; Yang, Y.; Whiteman, M.; Moore, P.K.; Bian, J.S. A Review of Hydrogen Sulfide Synthesis, Metabolism, and Measurement: Is Modulation of Hydrogen Sulfide a Novel Therapeutic for Cancer? Antioxid. Redox Signal. 2019, 31, 1–38. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, D.G.F.; Nunes, J.; Tomé, C.S.; Zuhra, K.; Costa, J.M.F.; Antunes, A.M.M.; Giuffrè, A.; Vicente, J.B. Human Cystathionine γ-Lyase Is Inhibited by s-Nitrosation: A New Crosstalk Mechanism between NO and H2S. Antioxidants 2021, 10, 1391. https://doi.org/10.3390/antiox10091391
Fernandes DGF, Nunes J, Tomé CS, Zuhra K, Costa JMF, Antunes AMM, Giuffrè A, Vicente JB. Human Cystathionine γ-Lyase Is Inhibited by s-Nitrosation: A New Crosstalk Mechanism between NO and H2S. Antioxidants. 2021; 10(9):1391. https://doi.org/10.3390/antiox10091391
Chicago/Turabian StyleFernandes, Dalila G. F., João Nunes, Catarina S. Tomé, Karim Zuhra, João M. F. Costa, Alexandra M. M. Antunes, Alessandro Giuffrè, and João B. Vicente. 2021. "Human Cystathionine γ-Lyase Is Inhibited by s-Nitrosation: A New Crosstalk Mechanism between NO and H2S" Antioxidants 10, no. 9: 1391. https://doi.org/10.3390/antiox10091391
APA StyleFernandes, D. G. F., Nunes, J., Tomé, C. S., Zuhra, K., Costa, J. M. F., Antunes, A. M. M., Giuffrè, A., & Vicente, J. B. (2021). Human Cystathionine γ-Lyase Is Inhibited by s-Nitrosation: A New Crosstalk Mechanism between NO and H2S. Antioxidants, 10(9), 1391. https://doi.org/10.3390/antiox10091391








