Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Sample Preparation and Immobilization on Mica
2.3. Atomic Force Microscopy Imaging
2.4. Image Analysis
3. Results
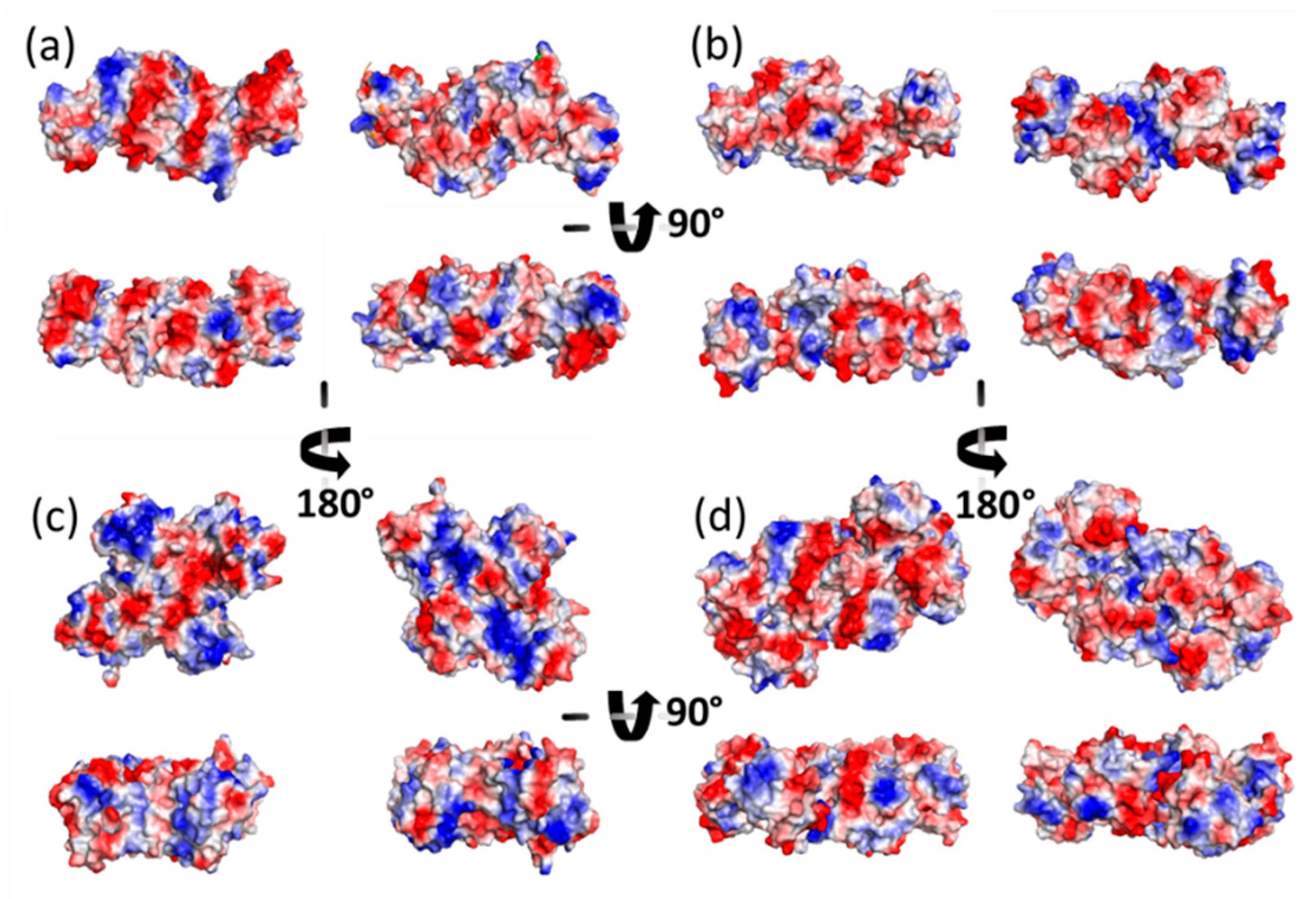
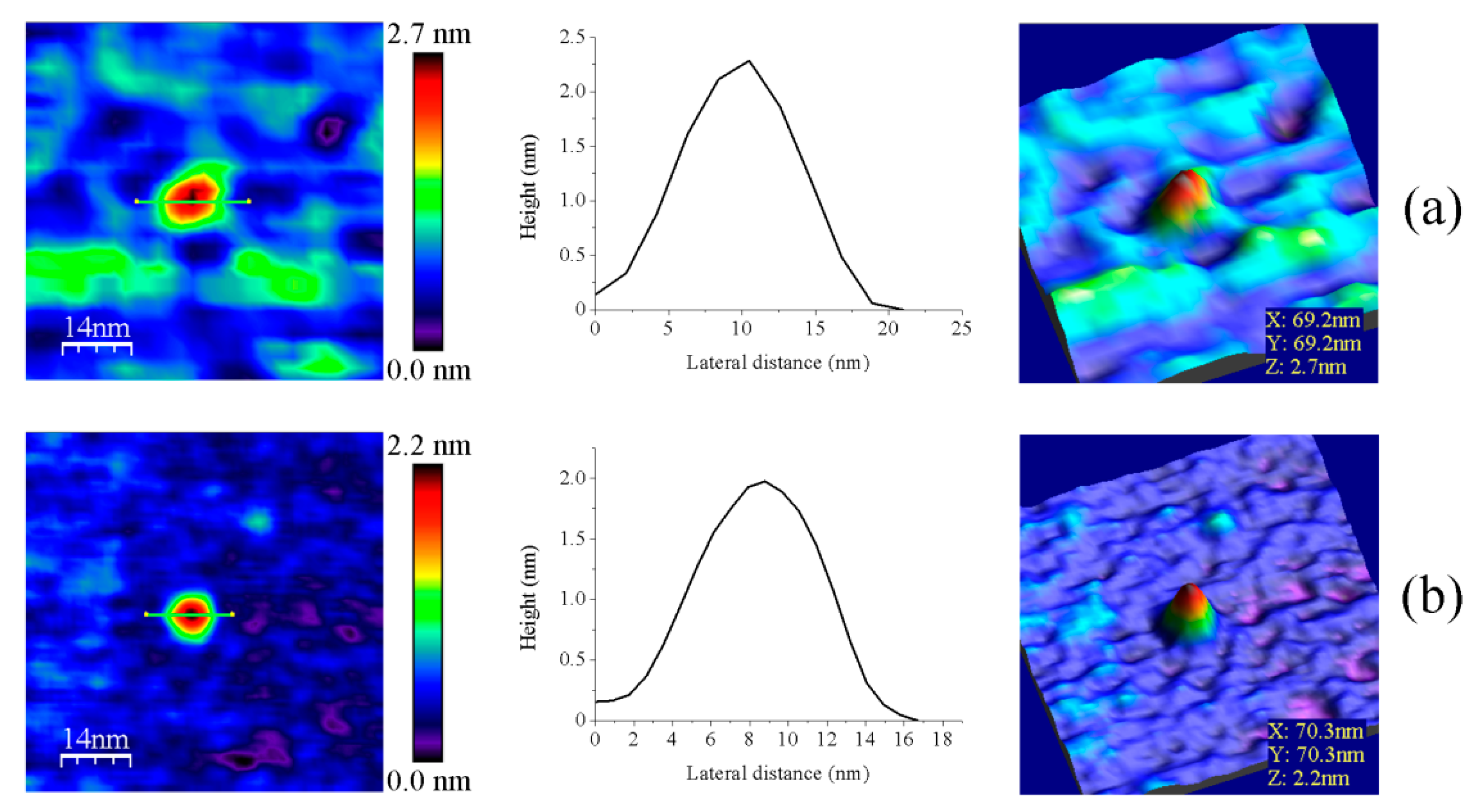
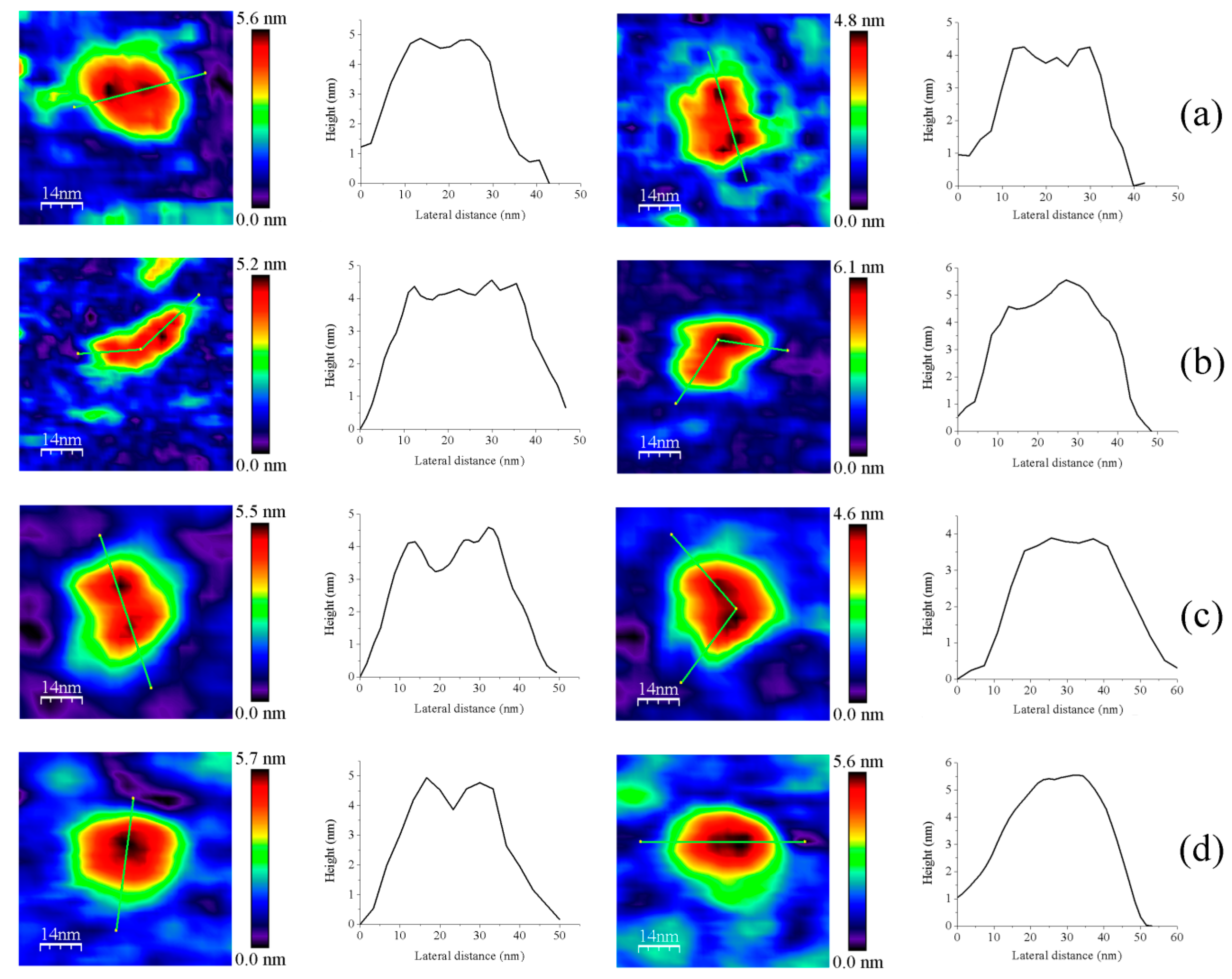
3.1. AFM Allows for Comparison of Topology Patterns of FFTRs
3.2. Redox Protein Partners Modulate the Topology of FFTR AFM Images
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Protein | Abbreviation | Reference | PDB Code | |
|---|---|---|---|---|
| 1 | Fdx-dependent flavin thioredoxin reductase from Gloeobacter violaceus | GvFFTR | [6] | 5J60, 6XTF in complex with Fdx1 |
| 2 | GvFFTR variant where the C-terminal tail, residues Ser307-His317, has been removed | GvFFTR_Δtail | [6] | n.d. |
| 3 | Thioredoxin type-m from Gloeobacter violaceus | GvTrxm | [6] | n.d. |
| 4 | C135S GvFFTR variant | GvFFTRC135S | This work | n.d. |
| 5 | C35S GvTrxm variant | GvTrxC35S | This work | n.d. |
| 6 | Covalent complex of C135S GvFFTR and C35S GvTrx | GvFFTR:GvTrxm | This work | n.d. |
| 7 | Ferredoxin 1 from Gloeobacter violaceus | GvFdx1 | [9] | 6XTF in complex with GvFFTR |
| 8 | Diflavin-linked disulfide oxidoreductase from Gloeobacter violaceus | GvDDOR | [14] | 5ODE |
| 9 | Fdx-dependent flavin thioredoxin reductase 2 from Clostridium acetobutylicum | CaFFTR2 | [7] | 6GNC |
| 10 | Thioredoxin 2 from Clostridium acetobutylicum | CaTrx2 | [7] | 6G9N |
| 11 | C131S CaFFTR2 variant | CaFFTR2C131S | [7] | n.d. |
| 12 | C32S CaTrx2 variant | CaTrx2C32S | [7] | n.d. |
| 13 | Covalent complex of C131SCaFFTR2 and C32SCaTrx2 | CaFFTR2:CaTrx2 | [7] | 6GND |
| Sample | Subpopulation 1 | Subpopulation 2 | Subpopulation 3 |
|---|---|---|---|
| Monomers (nm3) | Homodimers a (nm3) | Homotetramers (nm3) | |
| GvFFTR | 544 ± 25 | 1080 ± 82 | 2133 ± 62 |
| CaFFTR2 | 151 ± 13 | 305 ± 20 | 590 ± 20 |
| GvDDOR | 243 ± 16 | 487 ± 16 | 930 ± 52 |
| GvFFTR_Δtail | 474 ± 22 | 1003 ± 52 | 1942 ± 20 |
| GvFdx1 | 63 ± 3 | ||
| GvFFTR + GvFdx1 | 508 ± 21 | 1120 ± 64 * | 2071 |
| GvFFTR_Δtail + GvFdx1 | 501 ± 20 | 1023 ± 68 * | 2113 |
| GvTrxm | 41 ± 2 | ||
| GvFFTR + GvTrxm | 498 ± 13 | 1069 ± 58 * | |
| GvFFTR_Δtail + GvTrxm | 507 ± 15 | 1048 ± 55 * | |
| GvFFTRC135S | 505 ± 21 | 1020 ± 65 | 2121 ± 114 |
| GvFFTR:GvTrxm | 494 ± 14 | 1053 ± 55 * | 2211 ± 78 |
| CaFFTR2C131S | 157 ± 8 | 313 ± 22 | 643 |
| CaFFTR2:CaTrx2 | 158 ± 8 | 309 ± 19 * |
| Sample | Monomers (%) | Homodimers a (%) | Homotetramers (%) |
|---|---|---|---|
| GvFFTR | 19/53 # | 68/47 # | 13/n.d. # |
| CaFFTR2 | 4/48 # | 88/52 # | 8/n.d. # |
| GvDDOR | 6/57 # | 72/40 # | 22/3 # |
| GvFFTR_Δtail | 12 | 84 | 4 |
| GvFFTR + GvFdx1 | 11 | 86 | 3 |
| GvFFTR_Δtail + GvFdx1 | 15 | 82 | 3 |
| GvFFTR + GvTrxm | 8 | 91 | n.d. |
| GvFFTR_Δtail + GvTrxm | 12 | 88 | n.d. |
| GvFFTRC135S | 21 | 75 | 4 |
| GvFFTR:GvTrxm | 10 | 84 | 6 |
| CaFFTR2C131S | 10 | 88 | 2 |
| CaFFTR2:CaTrx2 | 9 | 91 | n.d. |


Appendix B

References
- Balsera, M.; Buchanan, B.B. Evolution of the thioredoxin system as a step enabling adaptation to oxidative stress. Free Radic. Biol. Med. 2019, 140, 28–35. [Google Scholar] [CrossRef]
- Eklund, H.; Gleason, F.K.; Holmgren, A. Structural and functional relations among thioredoxins of different species. Proteins 1991, 11, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, J.; Ballou, D.P.; Williams, C.H., Jr. Reactivity of Thioredoxin as a Protein Thiol-Disulfide Oxidoreductase. Chem. Rev. 2011, 111, 5768–5783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Buey, R.M.; Galindo-Trigo, S.; Lopez-Maury, L.; Velazquez-Campoy, A.; Luis Revuelta, J.; Javier Florencio, F.; de Pereda, J.M.; Schurmann, P.; Buchanan, B.B.; Balsera, M. A New Member of the Thioredoxin Reductase Family from Early Oxygenic Photosynthetic Organisms. Mol. Plant 2017, 10, 212–215. [Google Scholar] [CrossRef] [Green Version]
- Buey, R.M.; Fernandez-Justel, D.; de Pereda, J.M.; Revuelta, J.L.; Schurmann, P.; Buchanan, B.B.; Balsera, M. Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases. Proc. Natl. Acad. Sci. USA 2018, 115, 12967–12972. [Google Scholar] [CrossRef] [Green Version]
- Hammel, K.E.; Cornwell, K.L.; Buchanan, B.B. Ferredoxin/flavoprotein-linked pathway for the reduction of thioredoxin. Proc. Natl. Acad. Sci. USA 1983, 80, 3681–3685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buey, R.M.; Fernandez-Justel, D.; Gonzalez-Holgado, G.; Martinez-Julvez, M.; Gonzalez-Lopez, A.; Velazquez-Campoy, A.; Medina, M.; Buchanan, B.B.; Balsera, M. Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria. Plant Physiol. 2021, 186, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Lennon, B.W.; Williams, C.H.; Ludwig, M.L. Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000, 289, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Kuriyan, J.; Krishna, T.S.R.; Wong, L.; Guenther, B.; Pahler, A.; Williams, C.H.; Model, P. Convergent evolution of similar function in two structurally divergent enzymes. Nature. 1991, 352, 172–174. [Google Scholar] [CrossRef]
- Cornish, P.V.; Ha, T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007, 2, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buey, R.M.; Arellano, J.B.; Lopez-Maury, L.; Galindo-Trigo, S.; Velazquez-Campoy, A.; Revuelta, J.L.; de Pereda, J.M.; Florencio, F.J.; Schurmann, P.; Buchanan, B.B.; et al. Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase. Proc. Natl. Acad. Sci. USA 2017, 114, 12725–12730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcuello, C.; de Miguel, R.; Gomez-Moreno, C.; Martinez-Julvez, M.; Lostao, A. An efficient method for enzyme immobilization evidenced by atomic force microscopy. Protein Eng. Des. Sel. 2012, 25, 715–723. [Google Scholar] [CrossRef] [Green Version]
- Alsteens, D.; Dupres, V.; Yunus, S.; Latge, J.-P.; Heinisch, J.J.; Dufrene, Y.F. High-Resolution Imaging of Chemical and Biological Sites on Living Cells Using Peak Force Tapping Atomic Force Microscopy. Langmuir 2012, 28, 16738–16744. [Google Scholar] [CrossRef]
- Meinander, K.; Jensen, T.N.; Simonsen, S.B.; Helveg, S.; Lauritsen, J.V. Quantification of tip-broadening in non-contact atomic force microscopy with carbon nanotube tips. Nanotechnology 2012, 23, 405705. [Google Scholar] [CrossRef] [Green Version]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef] [Green Version]
- Horcas, I.; Fernandez, R.; Gomez-Rodriguez, J.M.; Colchero, J.; Gomez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 13705. [Google Scholar] [CrossRef]
- Lostao, A.; Peleato, M.L.; Gomez-Moreno, C.; Fillat, M.F. Oligomerization properties of FurA from the cyanobacterium Anabaena sp PCC 7120: Direct visualization by in situ atomic force microscopy under different redox conditions. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1723–1729. [Google Scholar] [CrossRef]
- Marcuello, C.; Arilla-Luna, S.; Medina, M.; Lostao, A. Detection of a quaternary organization into dimer of trimers of Corynebacterium ammoniagenes FAD synthetase at the single-molecule level and at the in cell level. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.W.; Larmer, J.; Henderson, R.M.; Oberleithner, H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflügers Arch. 1998, 435, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Bukharina, N.S.; Archakov, A.I.; Ivanov, Y.D. Atomic Force Microscopy for Protein Detection and Their Physicochemical Characterization. Int. J. Mol. Sci. 2018, 19, 1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svergun, D.; Barberato, C.; Koch, M.H.J. CRYSOL—A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Gari, R.R.S.; Frey, N.C.; Mao, C.; Randall, L.L.; King, G.M. Dynamic Structure of the Translocon SecYEG in Membrane: Direct Single Molecule Observations. J. Biol. Chem. 2013, 288, 16848–16854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lira-Navarrete, E.; De Las Rivas, M.; Compañón, I.; Pallarés, M.C.; Kong, Y.; Iglesias-Fernández, J.; Bernardes, G.J.L.; Peregrina, J.M.; Rovira, C.; Bernadó, P.; et al. Dynamic Interplay between Catalytic and Lectin Domains of GalNAc-Transferases Modulates Protein O-Glycosylation. Nat. Commun. 2015, 6, 6937. [Google Scholar] [CrossRef] [Green Version]
- Lira-Navarrete, E.; Pallares, M.C.; Castello, F.; Ruedas-Rama, M.J.; Orte, A.; Lostao, A.; Hurtado-Guerrero, R. Protein O-Fucosyltransferase 1 Undergoes Interdomain Flexibility in Solution. Molecules 2021, 26, 2105. [Google Scholar] [CrossRef]
- Wagner, P. Immobilization strategies for biological scanning probe microscopy. FEBS Lett. 1998, 430, 112–115. [Google Scholar] [CrossRef] [Green Version]
- El Kirat, K.; Burton, I.; Dupres, V.; Dufrene, Y.F. Sample preparation procedures for biological atomic force microscopy. J. Microsc. 2005, 218, 199–207. [Google Scholar] [CrossRef]
- Ferreira, P.; Villanueva, R.; Martinez-Julvez, M.; Herguedas, B.; Marcuello, C.; Fernandez-Silva, P.; Cabon, L.; Hermoso, J.A.; Lostao, A.; Susin, S.A.; et al. Structural Insights into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor. Biochemistry 2014, 53, 4204–4215. [Google Scholar] [CrossRef]
- Jensenius, H.; Klein, D.C.G.; van Hecke, M.; Oosterkamp, T.H.; Schmidt, T.; Jensenius, J.C. Mannan-Binding Lectin: Structure, Oligomerization, and Flexibility Studied by Atomic Force Microscopy. J. Mol. Biol. 2009, 391, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Perez, M.J.; de Miguel, R.; Carbonera, C.; Martinez-Julvez, M.; Lostao, A.; Piquer, C.; Gomez-Moreno, C.; Bartolome, J.; Luis, F. Size-dependent properties of magnetoferritin. Nanotechnology 2010, 21, 465707. [Google Scholar] [CrossRef]
- Gomes, S.; Numata, K.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kapan, D.L. AFM Study of Morphology and Mechanical Properties of a Chimeric Spider Silk and Bone Sialoprotein Protein for Bone Regeneration. Biomacromolecules 2011, 12, 1675–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnally, S.M.; Edwardson, J.M.; Barrera, N.P. Imaging the Spatial Orientation of Subunits Within Membrane Receptors by Atomic Force Microscopy. Methods Mol. Biol. 2011, 736, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Pallarés, M.C.; Marcuello, C.; Botello-Morte, L.; González, A.; Fillat, M.F.; Lostao, A. Sequential Binding of FurA from Anabaena Sp. PCC 7120 to Iron Boxes: Exploring Regulation at the Nanoscale. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2014, 1844, 623–631. [Google Scholar] [CrossRef]
- Lostao, A.; Medina, M. Atomic Force Microscopy: Single-Molecule Imaging and Force Spectroscopy in the Study of Flavoproteins Ligand Binding and Reaction Mechanisms. Methods Mol. Biol. 2021, 2280, 157–178. [Google Scholar] [CrossRef]
- Main, K.; Provan, J.I.; Haynes, P.J.; Wells, G.; Hartley, J.A.; Pyne, A. Atomic Force Microscopy-A Tool for Structural and Translational DNA Research. APL Bioeng. 2021, 5, 031504. [Google Scholar] [CrossRef]
- Marcuello, C.; de Miguel, R.; Martinez-Julvez, M.; Gomez-Moreno, C.; Lostao, A. Mechanostability of the Single-Electron-Transfer Complexes of Anabaena Ferredoxin-NADP(+) Reductase. Chemphyschem 2015, 16, 3161–3169. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcuello, C.; Frempong, G.A.; Balsera, M.; Medina, M.; Lostao, A. Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. Antioxidants 2021, 10, 1437. https://doi.org/10.3390/antiox10091437
Marcuello C, Frempong GA, Balsera M, Medina M, Lostao A. Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. Antioxidants. 2021; 10(9):1437. https://doi.org/10.3390/antiox10091437
Chicago/Turabian StyleMarcuello, Carlos, Gifty Animwaa Frempong, Mónica Balsera, Milagros Medina, and Anabel Lostao. 2021. "Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases" Antioxidants 10, no. 9: 1437. https://doi.org/10.3390/antiox10091437
APA StyleMarcuello, C., Frempong, G. A., Balsera, M., Medina, M., & Lostao, A. (2021). Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. Antioxidants, 10(9), 1437. https://doi.org/10.3390/antiox10091437











