Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Patient Recruitment and Sample Collection
2.3. Real-Time PCR Microarrays
2.4. Confirmation and Validation of the Up- and Downregulated Genes by Customized Real-Time PCR Microarrays
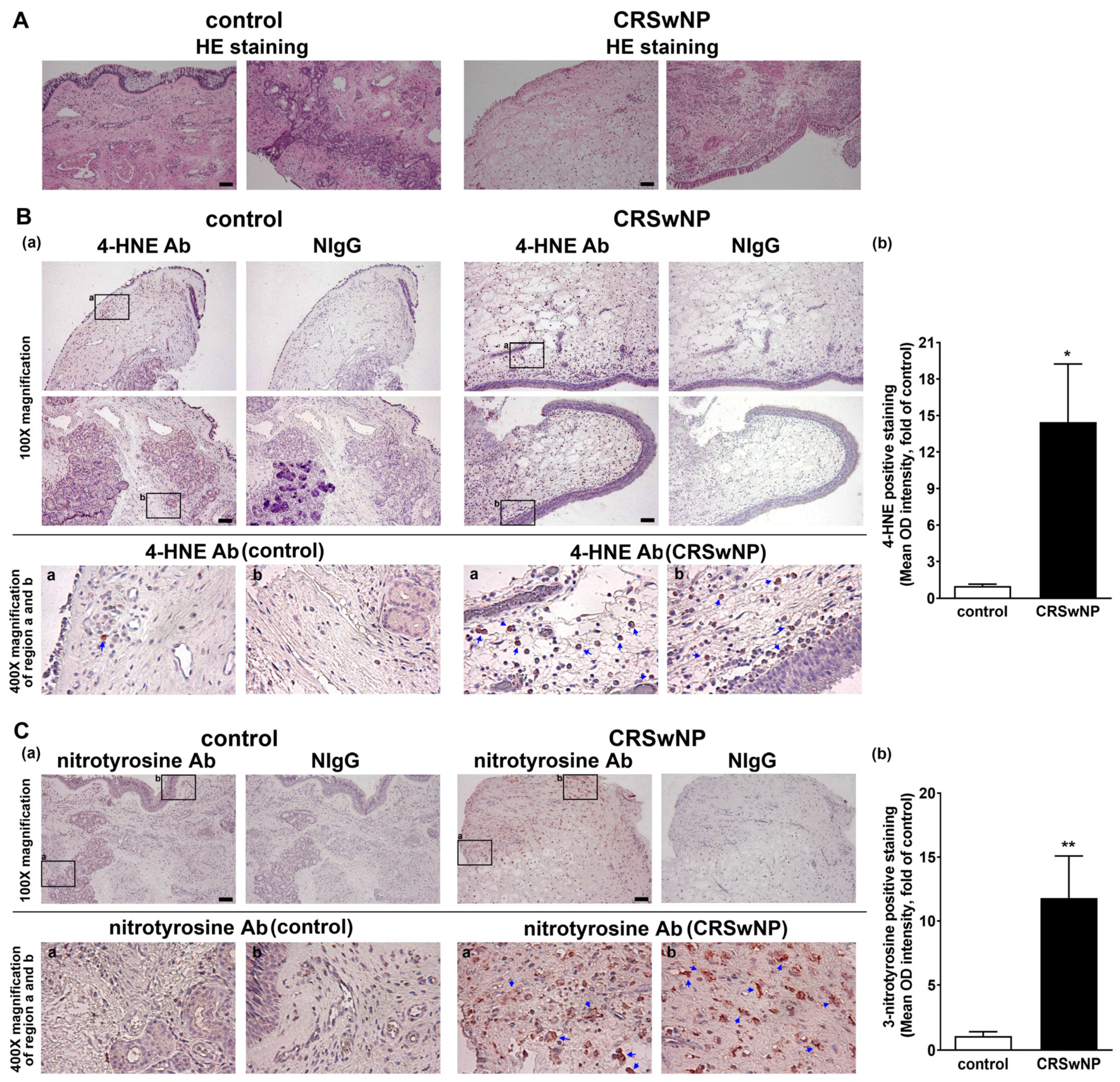
2.5. Immunohistochemistry
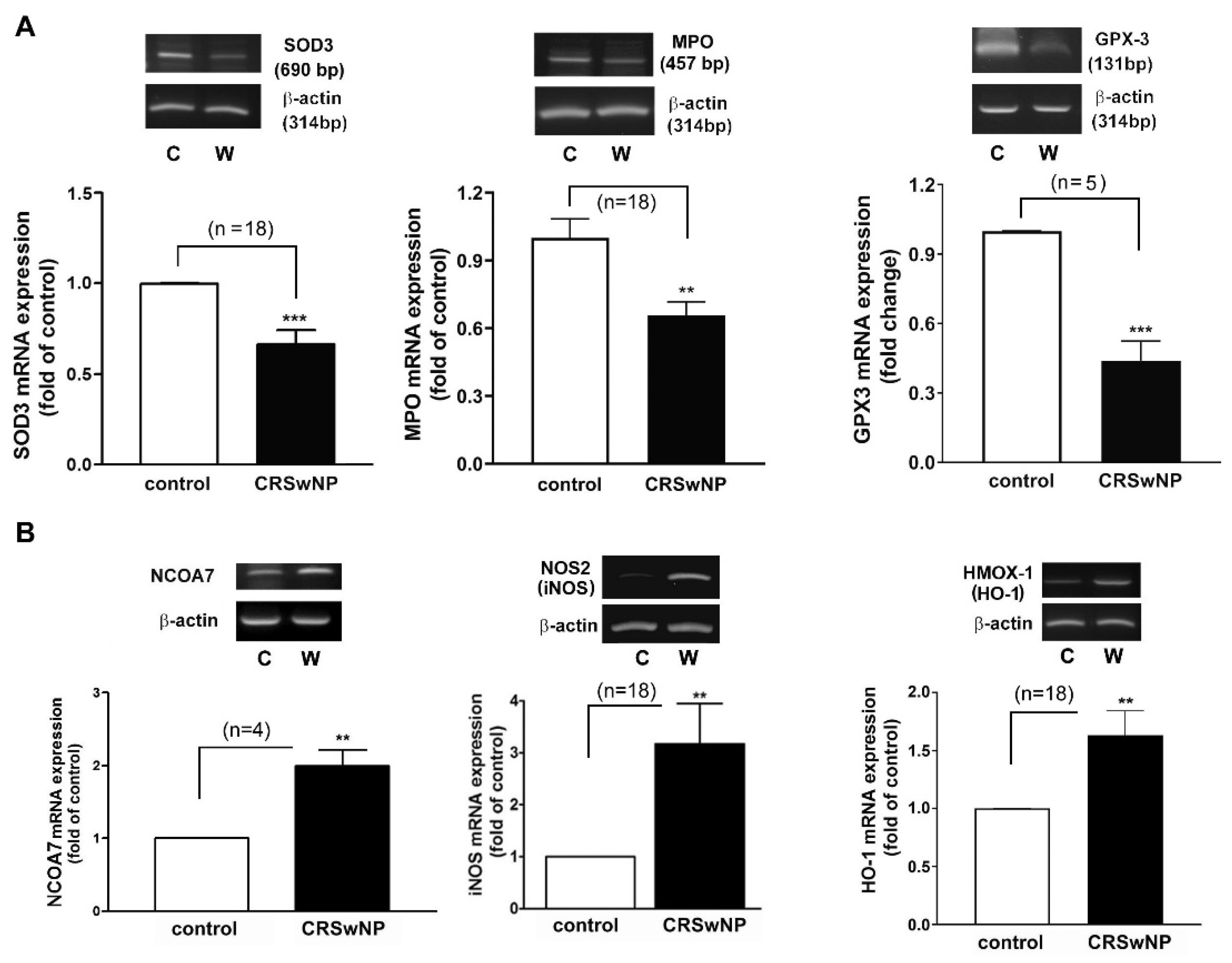
2.6. RT-PCR Analysis of mRNA Expression Level in Up- and Downregulated Genes
2.7. Tissue Lysate Preparation and Western Blot Analysis
2.8. Data Analysis
3. Results
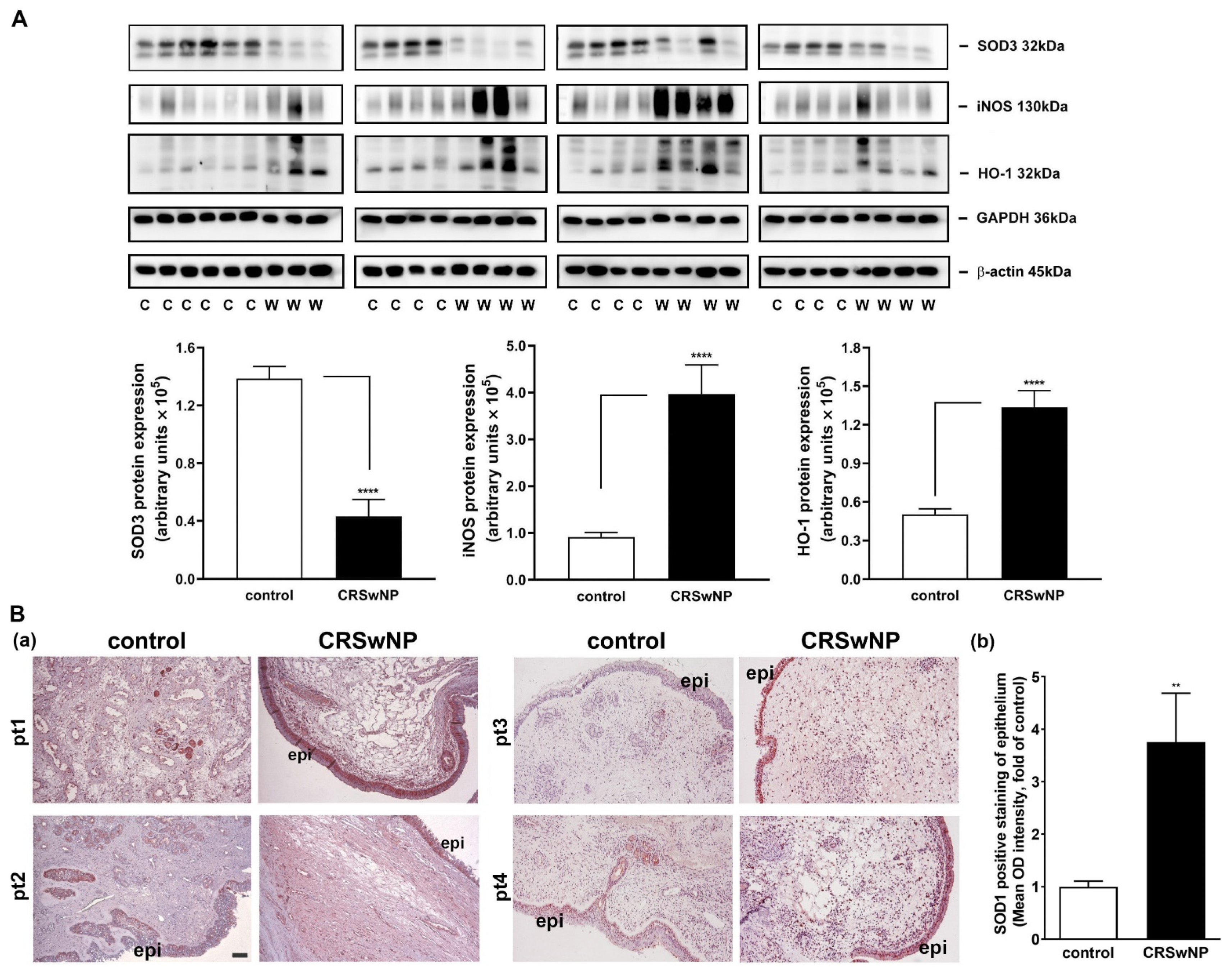
3.1. The Oxidative Damage Is Increased in the NPs of CRSwNP
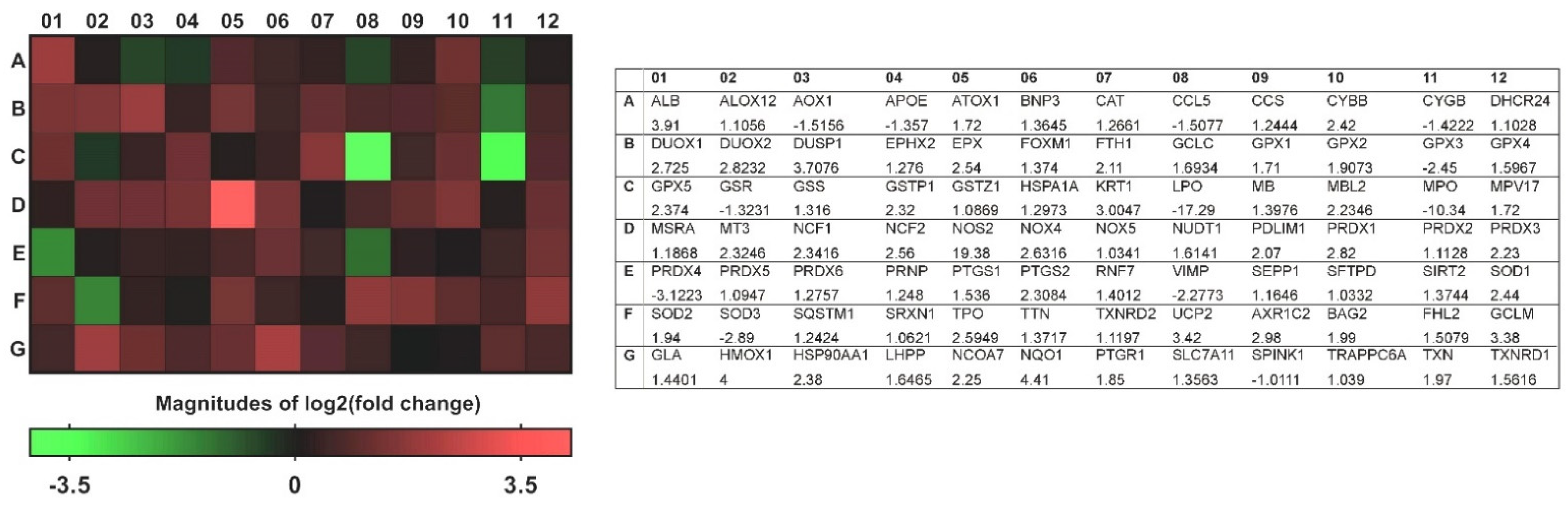
3.2. A Transcriptional Profile of Genes Involved in Oxidative Stress in CRSwNP NP Tissues
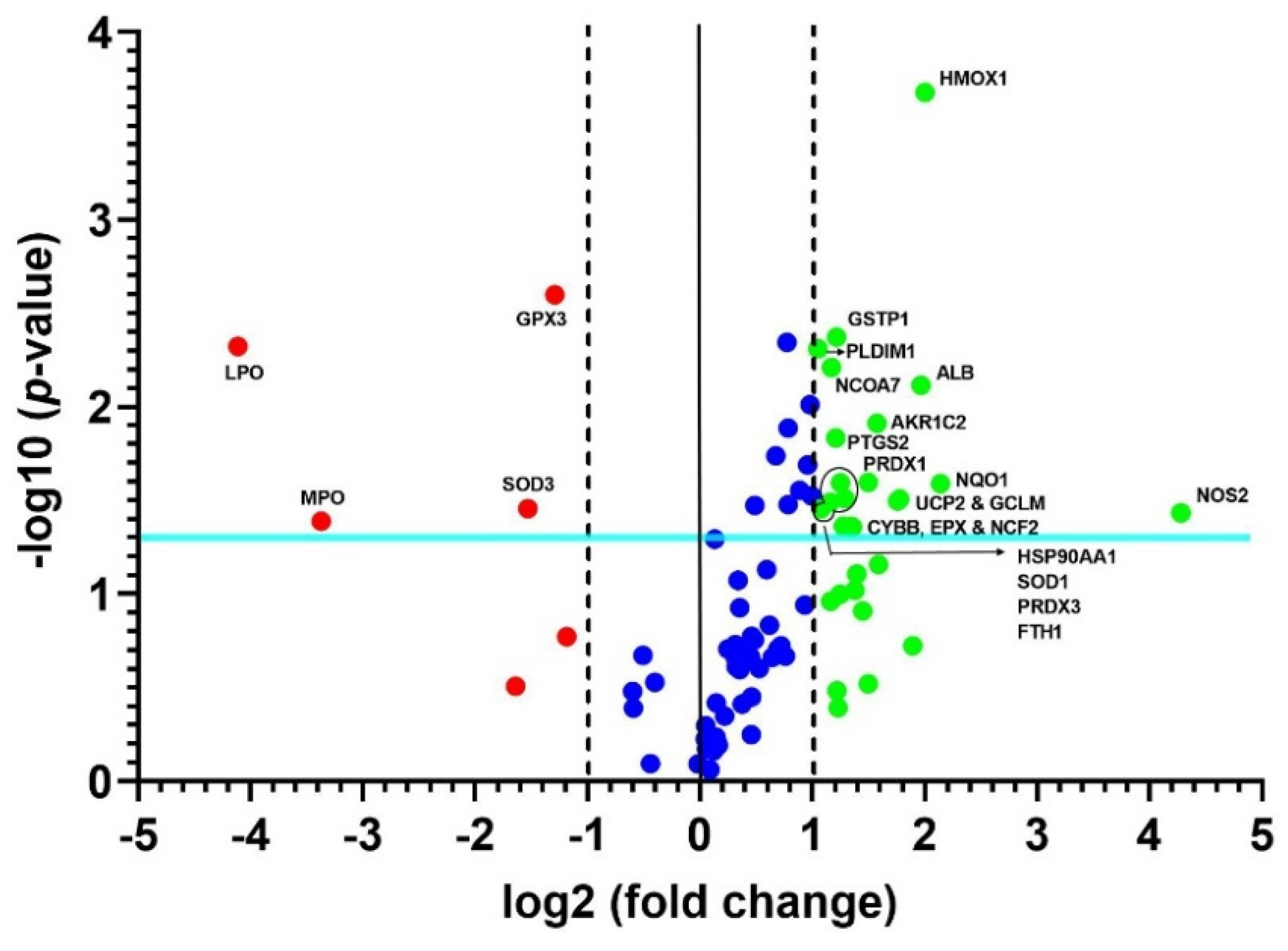
3.3. Confirmation and Validation of the Significantly Changed Genes
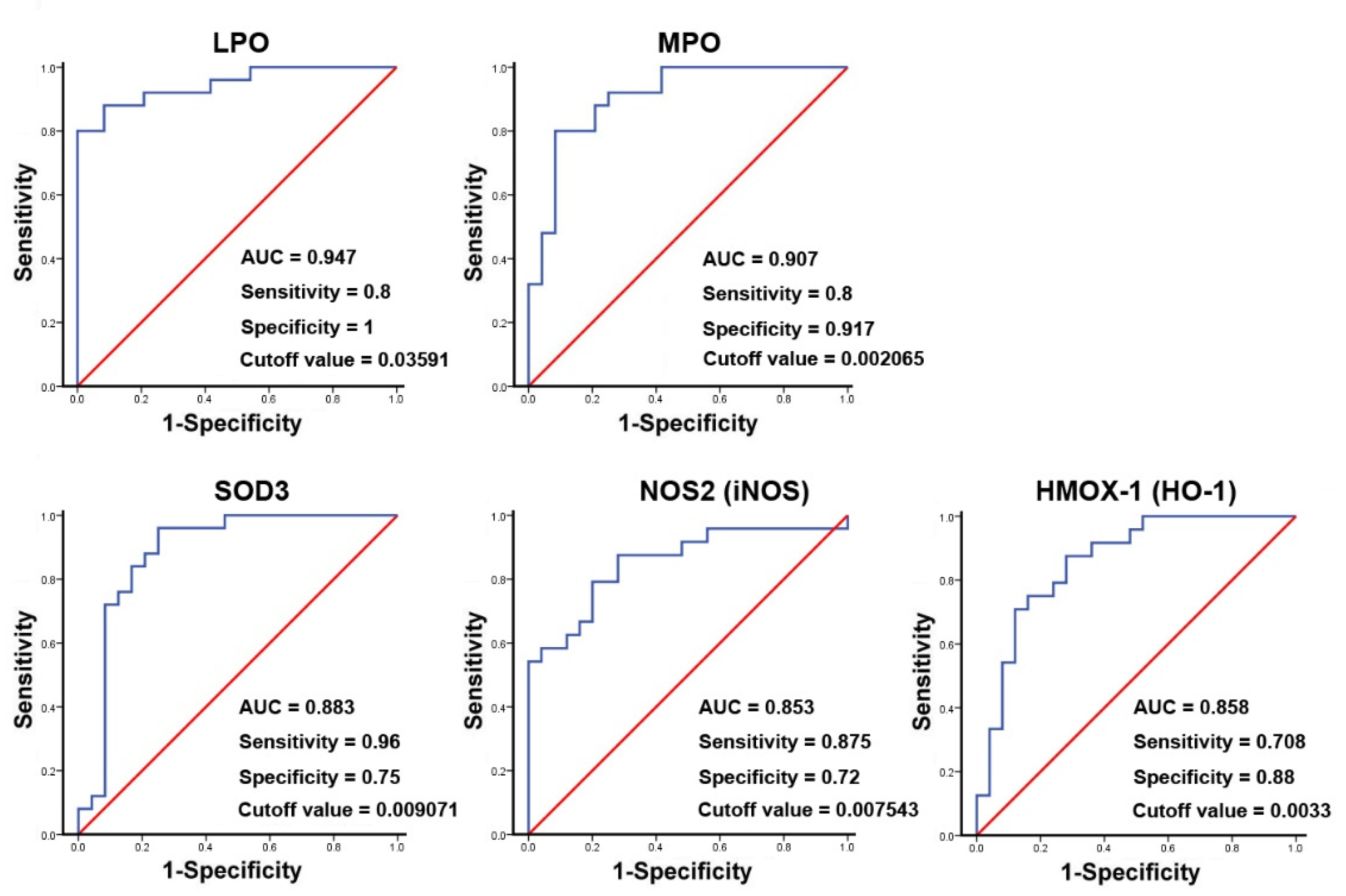
3.4. The Oxidative Gene Expression Level and Its Association with the Development of CRSwNP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, K.; Kim, S.W. Effect of Chronic Rhinosinusitis with or without Nasal Polyp on Quality of Life in South Korea: 5th Korea National Health and Nutrition Examination Survey Korean. Clin. Exp. Otorhinolaryngol. 2016, 9, 150–156. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, N.; Bo, M.; Holtappels, G.; Zheng, M.; Lou, H.; Wang, H.; Zhang, L.; Bachert, C. Diversity of T(H) cytokine profiles in patients with chronic rhinosinusitis: A multicenter study in Europe, Asia, and Oceania. J. Allergy Clin. Immunol. 2016, 138, 1344–1353. [Google Scholar] [CrossRef]
- Duracková, Z. Some current insights into oxidative stress. Physiol. Res. 2010, 59, 459–469. [Google Scholar] [CrossRef]
- Van Bruaene, N.; Derycke, L.; Perez-Novo, C.A.; Gevaert, P.; Holtappels, G.; De Ruyck, N.; Cuvelier, C.; Van Cauwenberge, P.; Bachert, C. TGF-beta signaling and collagen deposition in chronic rhinosinusitis. J. Allergy Clin. Immunol. 2009, 124, 253–259. [Google Scholar] [CrossRef]
- Takabayashi, T.; Kato, A.; Peters, A.T.; Hulse, K.E.; Suh, L.A.; Carter, R.; Norton, J.; Grammer, L.C.; Cho, S.H.; Tan, B.K.; et al. Excessive fibrin deposition in nasal polyps caused by fibrinolytic impairment through reduction of tissue plasminogen activator expression. Am. J. Respir. Crit. Care Med. 2013, 187, 49–57. [Google Scholar] [CrossRef]
- Hulse, K.E.; Stevens, W.W.; Tan, B.K.; Schleimer, R.P. Pathogenesis of nasal polyposis. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015, 45, 328–346. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, A.; Emmanuel, B.; Thomas, K.; Guiang, H. Systematic literature review of the epidemiology and clinical burden of chronic rhinosinusitis with nasal polyposis. Curr. Med. Res. Opin. 2020, 36, 1897–1911. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.T.; Kountakis, S.E. Significance of nasal polyps in chronic rhinosinusitis: Symptoms and surgical outcomes. Laryngoscope 2004, 114, 1932–1935. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Huynh, T.M.T.; Vandeplas, G.; Joish, V.N.; Mannent, L.P.; Tomassen, P.; van Zele, T.; Cardell, L.O.; Arebro, J.; Olze, H.; et al. The GALEN rhinosinusitis cohort: Chronic rhinosinusitis with nasal polyps affects health-related quality of life. Rhinology 2019, 57, 343–351. [Google Scholar] [CrossRef]
- Hussain, T.; Tan, B.; Yin, Y.; Blachier, F.; Tossou, M.C.; Rahu, N. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us? Oxid. Med. Cell. Longev. 2016, 2016, 7432797. [Google Scholar] [CrossRef] [Green Version]
- Popa-Wagner, A.; Mitran, S.; Sivanesan, S.; Chang, E.; Buga, A.M. ROS and brain diseases: The good, the bad, and the ugly. Oxid. Med. Cell Longev. 2013, 2013, 963520. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Lee, R.J.; Schleimer, R.P.; Cohen, N.A. Chronic rhinosinusitis pathogenesis. J. Allergy Clin. Immunol. 2015, 136, 1442–1453. [Google Scholar] [CrossRef]
- Khandrika, L.; Kumar, B.; Koul, S.; Maroni, P.; Koul, H.K. Oxidative stress in prostate cancer. Cancer Lett. 2009, 282, 125–136. [Google Scholar] [CrossRef]
- Topal, O.; Kulaksızoglu, S.; Erbek, S.S. Oxidative stress and nasal polyposis: Does it affect the severity of the disease? Am. J. Rhinol. Allergy 2014, 28, e1–e4. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.Y.; Nayak, J.V.; Bravo, D.T.; Le, W.; Nguyen, A.; Edward, J.A.; Hwang, P.H.; Illek, B.; Fischer, H. Expression of dual oxidases and secreted cytokines in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2013, 3, 376–383. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Zhang, J.; Li, L.; Wu, X.; Ma, R.; Han, M.; Xu, G.; Wen, W.; Li, H. Expression of heme oxygenase-1 in eosinophilic and non-eosinophilic chronic rhinosinusitis with nasal polyps: Modulation by cytokines. Int. Forum Allergy Rhinol. 2015, 5, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Hao, J.; Xiao, L.; Wang, M.; Zhao, Y.; Fan, D.; Li, Y.; Wang, X.; Zhang, L. Expression of nicotinamide adenine dinucleotide phosphate oxidase in chronic rhinosinusitis with nasal polyps. Int. Forum Allergy Rhinol. 2020, 10, 646–655. [Google Scholar] [CrossRef]
- Cho, D.Y.; Zhang, S.; Lazrak, A.; Skinner, D.; Thompson, H.M.; Grayson, J.; Guroji, P.; Aggarwal, S.; Bebok, Z.; Rowe, S.M.; et al. LPS decreases CFTR open probability and mucociliary transport through generation of reactive oxygen species. Redox Biol. 2021, 43, 101998. [Google Scholar] [CrossRef] [PubMed]
- Nishida, M.; Takeno, S.; Takemoto, K.; Takahara, D.; Hamamoto, T.; Ishino, T.; Kawasumi, T. Increased Tissue Expression of Lectin-Like Oxidized LDL Receptor-1 (LOX-1) Is Associated with Disease Severity in Chronic Rhinosinusitis with Nasal Polyps. Diagnostics 2020, 10, 246. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Tharakan, A.; London, N.R.; Lane, A.P.; Ramanathan, M., Jr. Bactericidal antibiotics promote oxidative damage and programmed cell death in sinonasal epithelial cells. Int. Forum Allergy Rhinol. 2017, 7, 359–364. [Google Scholar] [CrossRef] [Green Version]
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J. Executive summary of EPOS 2020 including integrated care pathways. Rhinology 2020, 58, 82–111. [Google Scholar] [CrossRef]
- Tsai, Y.J.; Hao, C.Y.; Chen, C.L.; Wu, P.H.; Wu, W.B. Expression of long pentraxin 3 in human nasal mucosa fibroblasts, tissues, and secretions of chronic rhinosinusitis without nasal polyps. J. Mol. Med. 2020, 98, 673–689. [Google Scholar] [CrossRef]
- Lai, T.-H.; Shieh, J.-M.; Tsou, C.-J.; Wu, W.-B. Gold nanoparticles induce heme oxygenase-1 expression through Nrf2 activation and Bach1 export in human vascular endothelial cells. Int. J. Nanomed. 2015, 10, 5925–5939. [Google Scholar] [CrossRef]
- Tsai, Y.-J.; Chi, J.C.-Y.; Hao, C.-Y.; Wu, W.-B. Peptidoglycan induces bradykinin receptor 1 expression through Toll-like receptor 2 and NF-κB signaling pathway in human nasal mucosa-derived fibroblasts of chronic rhinosinusitis patients. J. Cell. Physiol. 2018, 233, 7226–7238. [Google Scholar] [CrossRef]
- Csala, M.; Kardon, T.; Legeza, B.; Lizák, B.; Mandl, J.; Margittai, É.; Puskás, F.; Száraz, P.; Szelényi, P.; Bánhegyi, G. On the role of 4-hydroxynonenal in health and disease. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2015, 1852, 826–838. [Google Scholar] [CrossRef]
- Bandookwala, M.; Sengupta, P. 3-Nitrotyrosine: A versatile oxidative stress biomarker for major neurodegenerative diseases. Int. J. Neurosci. 2020, 130, 1047–1062. [Google Scholar] [CrossRef]
- Asher, B.F.; Guilford, F.T. Oxidative Stress and Low Glutathione in Common Ear, Nose, and Throat Conditions: A Systematic Review. Altern. Ther. Health Med. 2016, 22, 44–50. [Google Scholar] [PubMed]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Gornicka, A.; Morris-Stiff, G.; Thapaliya, S.; Papouchado, B.G.; Berk, M.; Feldstein, A.E. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxid. Redox Signal. 2011, 15, 437–445. [Google Scholar] [CrossRef]
- Anavi, S.; Tirosh, O. iNOS as a metabolic enzyme under stress conditions. Free. Radic. Biol. Med. 2020, 146, 16–35. [Google Scholar] [CrossRef]
- Takeno, S.; Taruya, T.; Ueda, T.; Noda, N.; Hirakawa, K. Increased exhaled nitric oxide and its oxidation metabolism in eosinophilic chronic rhinosinusitis. Auris Nasus Larynx 2013, 40, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Nesi, R.T.; Barroso, M.V.; Muniz, V.S.; de Arantes, A.C.; Martins, M.A.; Gitirana, L.B.; Neves, J.S.; Benjamim, C.F.; Lanzetti, M.; Valenca, S.S. Pharmacological modulation of reactive oxygen species (ROS) improves the airway hyperresponsiveness by shifting the Th1 response in allergic inflammation induced by ovalbumin. Free. Radic. Res. 2017, 51, 708–722. [Google Scholar] [CrossRef]
- Sadek, A.A.; Abdelwahab, S.; Eid, S.Y.; Almaimani, R.A.; Althubiti, M.A.; El-Readi, M.Z. Overexpression of Inducible Nitric Oxide Synthase in Allergic and Nonallergic Nasal Polyp. Oxid. Med. Cell Longev. 2019, 2019, 7506103. [Google Scholar] [CrossRef] [PubMed]
- Begleiter, A.; Fourie, J. Induction of NQO1 in cancer cells. Methods Enzymol. 2004, 382, 320–351. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Ham, W.; Kim, J. Roles of NAD(P)H:quinone Oxidoreductase 1 in Diverse Diseases. Life 2021, 11, 1301. [Google Scholar] [CrossRef] [PubMed]
- Nioi, P.; Hayes, J.D. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat. Res. 2004, 555, 149–171. [Google Scholar] [CrossRef]
- Luo, S.; Lei, K.; Xiang, D.; Ye, K. NQO1 is Regulated by PTEN in Glioblastoma, Mediating Cell Proliferation and Oxidative Stress. Oxid. Med. Cell. Longev. 2018, 2018, 9146528. [Google Scholar] [CrossRef]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef]
- Ono, N.; Kusunoki, T.; Miwa, M.; Hirotsu, M.; Shiozawa, A.; Ikeda, K. Reduction in superoxide dismutase expression in the epithelial mucosa of eosinophilic chronic rhinosinusitis with nasal polyps. Int. Arch. Allergy Immunol. 2013, 162, 173–180. [Google Scholar] [CrossRef]
- Lin, H.; Ba, G.; Tang, R.; Li, M.; Li, Z.; Li, D.; Ye, H.; Zhang, W. Increased Expression of TXNIP Facilitates Oxidative Stress in Nasal Epithelial Cells of Patients with Chronic Rhinosinusitis with Nasal Polyps. Am. J. Rhinol. Allergy 2021, 35, 607–614. [Google Scholar] [CrossRef]
- Arnhold, J. Heme Peroxidases at Unperturbed and Inflamed Mucous Surfaces. Antioxidants 2021, 10, 1805. [Google Scholar] [CrossRef] [PubMed]
- Sarr, D.; Tóth, E.; Gingerich, A.; Rada, B. Antimicrobial actions of dual oxidases and lactoperoxidase. J. Microbiol. 2018, 56, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Aratani, Y. Myeloperoxidase: Its role for host defense, inflammation, and neutrophil function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef]
- Ndrepepa, G. Myeloperoxidase—A bridge linking inflammation and oxidative stress with cardiovascular disease. Clin. Chim. Acta Int. J. Clin. Chem. 2019, 493, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Carsuzaa, F.; Béquignon, É.; Dufour, X.; de Bonnecaze, G.; Lecron, J.C.; Favot, L. Cytokine Signature and Involvement in Chronic Rhinosinusitis with Nasal Polyps. Int. J. Mol. Sci. 2021, 23, 417. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-W.; Kim, D.W.; Kim, J.-W.; Lee, C.H.; Rhee, C.-S. Classification of chronic rhinosinusitis according to a nasal polyp and tissue eosinophilia: Limitation of current classification system for Asian population. Asia Pac. Allergy 2017, 7, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.-P.; Li, H.-B.; Wang, B.-F.; Wang, S.-B.; You, X.-J.; Cui, Y.-H.; Wang, D.-Y.; Desrosiers, M.; Liu, Z. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J. Allergy Clin. Immunol. 2009, 124, 478–484.e472. [Google Scholar] [CrossRef] [PubMed]
- Haegens, A.; Heeringa, P.; van Suylen, R.J.; Steele, C.; Aratani, Y.; O’Donoghue, R.J.; Mutsaers, S.E.; Mossman, B.T.; Wouters, E.F.; Vernooy, J.H. Myeloperoxidase deficiency attenuates lipopolysaccharide-induced acute lung inflammation and subsequent cytokine and chemokine production. J. Immunol. 2009, 182, 7990–7996. [Google Scholar] [CrossRef] [PubMed]
- Prokopowicz, Z.; Marcinkiewicz, J.; Katz, D.R.; Chain, B.M. Neutrophil myeloperoxidase: Soldier and statesman. Arch. Immunol. Ther. Exp. 2012, 60, 43–54. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilska-Wilkosz, A.; Iciek, M.; Górny, M. Chemistry and Biochemistry Aspects of the 4-Hydroxy-2,3-trans-nonenal. Biomolecules 2022, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Mihalj, H.; Butković, J.; Tokić, S.; Štefanić, M.; Kizivat, T.; Bujak, M.; Lončar, M.B.; Mihalj, M. Expression of Oxidative Stress and Inflammation-Related Genes in Nasal Mucosa and Nasal Polyps from Patients with Chronic Rhinosinusitis. Int. J. Mol. Sci. 2022, 23, 5521. [Google Scholar] [CrossRef] [PubMed]
- Bakhshaee, M.; Sharifian, M.R.; Ghazizadeh, A.H.; Nahid, K.; Samani, K.J. Smell Decline as a good Predictor of Sinonasal Polyposis Recurrence after Endoscopic Surgery. Iran. J. Otorhinolaryngol. 2016, 28, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Braskett, M.; Riedl, M.A. Novel antioxidant approaches to the treatment of upper airway inflammation. Curr. Opin. Allergy Clin. Immunol. 2010, 10, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Pace, E.; Ferraro, M.; Di Vincenzo, S.; Gerbino, S.; Bruno, A.; Lanata, L.; Gjomarkaj, M. Oxidative stress and innate immunity responses in cigarette smoke stimulated nasal epithelial cells. Toxicol. Vitr. Int. J. Publ. Assoc. BIBRA 2014, 28, 292–299. [Google Scholar] [CrossRef]
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | Product Size |
|---|---|---|---|
| SOD3 | ATGCTGGCGCTACTGTGTT | GCTTCTTGCGCTCTGAGT | 690 bp |
| GPX3 | GCCGGGGACAAGAGAAGT | GAGGACGTATTTGCCAGCAT | 131 bp |
| MPO | CGCCAACGTCTTCACCAATG | GATGTCGATGTTGTTGGGCG | 457 bp |
| NCOA7 | GCCACACTTCTCACTGCTCA | TAGGACAGGCAGCACCTCTT | 181 bp |
| iNOS | GCAGAATGTGACCATCATGG | ACAACCTTGGGGTTGAAGGC | 426 bp |
| HMOX1 | GAGACGGCTTCAAGCTGGTGATG | GTTGAGCAGGAACGCAGTCTTGG | 500 bp |
| β-ACTIN | ATCATGTTTGAGACCTTCAA | CATCTCTTGCTCGAAGTCCA | 314 bp |
| Gene Ref No. | Gene Name | Fold Regulation | p-Value |
|---|---|---|---|
| NM_000581 | Glutathione peroxidase 1 (GPX1) | 1.71 | 0.00453 |
| NM_002083 | Glutathione peroxidase 2 (gastrointestinal) (GPX2) | 1.9073 | 0.11468 |
| NM_002084 | Glutathione peroxidase 3 (plasma) (GPX3) | −2.45 | 0.00253 |
| NM_002085 | Glutathione peroxidase 4 (phospholipid hydroperoxidase) (GPX4) | 1.5967 | 0.01834 |
| NM_001509 | Glutathione peroxidase 5 (epididymal androgen-related protein) (GPX5) | 2.374 | 0.10027 |
| NM_000637 | Glutathione reductase (GSR) | −1.3231 | 0.29685 |
| NM_000178 | Glutathione synthetase (GSS) | 1.316 | 0.18993 |
| NM_000852 | Glutathione S-transferase pi 1 (GSTP1) | 2.32 | 0.00425 |
| NM_001513 | Glutathione transferase zeta 1 (GSTZ1) | 1.0869 | 0.68955 |
| NM_001979 | Epoxide hydrolase 2, cytoplasmic (EPHX2) | 1.276 | 0.25403 |
| NM_002574 | Peroxiredoxin 1 (PRDX1) | 2.82 | 0.02537 |
| NM_005809 | Peroxiredoxin 2 (PRDX2) | 1.1128 | 0.62476 |
| NM_006793 | Peroxiredoxin 3 (PRDX3) | 2.23 | 0.0324 |
| NM_006406 | Peroxiredoxin 4 (PRDX4) | −3.1223 | 0.31044 |
| NM_181652 | Peroxiredoxin 5 (PRDX5) | 1.0947 | 0.05096 |
| NM_004905 | Peroxiredoxin 6 (PRDX6) | 1.2757 | 0.11847 |
| NM_001752 | Catalase (CAT) | 1.2661 | 0.08469 |
| NM_000397 | Cytochrome b-245, beta polypeptide (CYBB) | 2.42 | 0.04344 |
| NM_134268 | Cytoglobin (CYGB) | −1.4222 | 0.21308 |
| NM_175940 | Dual oxidase 1 (DUOX1) | 2.725 | 0.1233 |
| NM_014080 | Dual oxidase 2 (DUOX2) | 2.8232 | 0.30205 |
| NM_004417 | Dual specificity phosphatase 1 (DUSP1) | 3.7076 | 0.1896 |
| NM_000502 | Eosinophil peroxidase (EPX) | 2.54 | 0.04358 |
| NM_006151 | Lactoperoxidase (LPO) | −17.29 | 0.00476 |
| NM_000250 | Myeloperoxidase (MPO) | −10.34 | 0.04083 |
| NM_000962 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (PTGS1) | 1.536 | 0.14707 |
| NM_000963 | Prostaglandin–endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (PTGS2) | 2.3084 | 0.01467 |
| NM_000547 | Thyroid peroxidase (TPO) | 2.5949 | 0.09569 |
| NM_003319 | Titin (TTN) | 1.3717 | 0.5637 |
| NM_000477 | Albumin (ALB) | 3.91 | 0.0077 |
| NM_000041 | Apolipoprotein E (APOE) | −1.357 | 0.80674 |
| NM_005954 | Metallothionein 3 (MT3) | 2.3246 | 0.32864 |
| NM_080725 | Sulfiredoxin 1 (SRXN1) | 1.0621 | 0.86638 |
| NM_003330 | Thioredoxin reductase 1 (TXNRD1) | 1.5616 | 0.21785 |
| NM_006440 | Thioredoxin reductase 2 (TXNRD2) | 1.1197 | 0.64345 |
| NM_005410 | Selenoprotein P, plasma, 1 (SEPP1) | 1.1646 | 0.4487 |
| NM_000454 | Superoxide dismutase 1, soluble (SOD1) | 2.44 | 0.03099 |
| NM_000636 | Superoxide dismutase 2, mitochondrial (SOD2) | 1.94 | 0.02045 |
| NM_003102 | Superoxide dismutase 3, extracellular (SOD3) | −2.89 | 0.03504 |
| NM_000697 | Arachidonate 12-lipoxygenase (ALOX12) | 1.1056 | 0.38209 |
| NM_005125 | Copper chaperone for superoxide dismutase (CCS) | 1.2444 | 0.21894 |
| NM_000265 | Neutrophil cytosolic factor 1 (NCF1) | 2.3416 | 0.40425 |
| NM_000433 | Neutrophil cytosolic factor 2 (NCF2) | 2.56 | 0.04391 |
| NM_000625 | Nitric oxide synthase 2, inducible (NOS2) | 19.38 | 0.037 |
| NM_016931 | NADPH oxidase 4 (NOX4) | 2.6316 | 0.07836 |
| NM_024505 | NADPH oxidase, EF-hand calcium-binding domain 5 (NOX5) | 1.0341 | 0.50355 |
| NM_003355 | Uncoupling protein 2 (mitochondrial, proton carrier) (UCP2) | 3.42 | 0.03109 |
| NM_001159 | Aldehyde oxidase 1 (AOX1) | −1.5156 | 0.33213 |
| NM_004052 | BCL2/adenovirus E1B 19 kDa-interacting protein 3 (BNIP3) | 1.3645 | 0.21708 |
| NM_002437 | MpV17 mitochondrial inner membrane protein (MPV17) | 1.72 | 0.01303 |
| NM_003019 | Surfactant protein D (SFTPD) | 1.0332 | 0.59598 |
| NM_004045 | ATX1 antioxidant protein 1 homolog (yeast) (ATOX1) | 1.72 | 0.03331 |
| NM_002985 | Chemokine (C-C motif) ligand 5 (CCL5) | −1.5077 | 0.4059 |
| NM_014762 | 24-dehydrocholesterol reductase (DHCR24) | 1.1028 | 0.58043 |
| NM_021953 | Forkhead box M1 (FOXM1) | 1.374 | 0.35409 |
| NM_002032 | Ferritin, heavy polypeptide 1 (FTH1) | 2.11 | 0.03497 |
| NM_001498 | Glutamate–cysteine ligase, catalytic subunit (GCLC) | 1.6934 | 0.21463 |
| NM_002061 | Glutamate–cysteine ligase, modifier subunit (GCLM) | 3.38 | 0.03198 |
| NM_002133 | Heme oxygenase (decycling) 1 (HMOX1) | 4 | 0.00021 |
| NM_005345 | Heat shock 70 kDa protein 1A (HSPA1A) | 1.2973 | 0.38672 |
| NM_006121 | Keratin 1 (KRT1) | 3.0047 | 0.06958 |
| NM_000242 | Mannose-binding lectin (protein C) 2, soluble (MBL2) | 2.2346 | 0.10945 |
| NM_012331 | Methionine sulfoxide reductase A (MSRA) | 1.1868 | 0.19668 |
| NM_000903 | NAD(P)H dehydrogenase, quinone 1 (NQO1) | 4.41 | 0.02578 |
| NM_002452 | Nudix (nucleoside diphosphate linked moiety X)-type motif 1 (NUDT1) | 1.6141 | 0.19677 |
| NM_020992 | PDZ and LIM domain 1 (PDLIM1) | 2.07 | 0.0049 |
| NM_183079 | Prion protein (PRNP) | 1.248 | 0.24491 |
| NM_014245 | Ring finger protein 7 (RNF7) | 1.4012 | 0.03359 |
| NM_203472 | Selenoprotein S (VIMP) | −2.2773 | 0.16892 |
| NM_012237 | Sirtuin 2 (SIRT2) | 1.3744 | 0.16827 |
| NM_003900 | Sequestosome 1 (SQSTM1) | 1.2424 | 0.18601 |
| NM_003329 | Thioredoxin (TXN) | 1.97 | 0.00976 |
| NM_005368 | Myoglobin (MB) | 1.3976 | 0.17586 |
| NM_001354 | Aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid-binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (AKR1C2) | 2.98 | 0.0123 |
| NM_004282 | BCL2-associated athanogene 2 (BAG2) | 1.99 | 0.03 |
| NM_001450 | Four and a half LIM domains 2 (FHL2) | 1.5079 | 0.07401 |
| NM_000169 | Galactosidase, alpha (GLA) | 1.4401 | 0.24986 |
| NM_001017963 | Heat shock protein 90 kDa alpha (cytosolic), class A member 1 (HSP90AA1) | 2.38 | 0.0255 |
| NM_022126 | Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) | 1.6465 | 0.18936 |
| NM_181782 | Nuclear receptor coactivator 7 (NCOA7) | 2.25 | 0.00618 |
| NM_012212 | Prostaglandin reductase 1 (PTGR1) | 1.85 | 0.02804 |
| NM_014331 | Solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 (SLC7A11) | 1.3563 | 0.22685 |
| NM_003122 | Serine peptidase inhibitor, Kazal type 1 (SPINK1) | −1.0111 | 0.8077 |
| NM_024108 | Trafficking protein particle complex 6A (TRAPPC6A) | 1.039 | 0.67335 |
| No. | Gene Name | Fold Regulation | p-Value |
|---|---|---|---|
| 1 | Glutathione peroxidase 2 (gastrointestinal) (GPX2) | 1.2269 | 0.53943 |
| 2 | Peroxiredoxin 4 (PRDX4) | −1.6239 | 0.00016 |
| 3 | Dual specificity phosphatase 1 (DUSP1) | −1.1024 | 0.8687 |
| 4 | Lactoperoxidase (LPO) | −16.662 | <1 × 10−6 |
| 5 | Myeloperoxidase (MPO) | −3.5766 | 0.00017 |
| 6 | Superoxide dismutase 3, extracellular (SOD3) | −3.2295 | 0.000095 |
| 7 | Nitric oxide synthase 2, inducible (NOS2) | 5.6641 | 0.024785 |
| 8 | Glutamate–cysteine ligase, modifier subunit (GCLM) | 2.0229 | 0.000028 |
| 9 | Heme oxygenase (decycling) 1 (HMOX1) | 2.3368 | 0.000017 |
| 10 | Aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid-binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (AKR1C2) | 2.3061 | 0.000751 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, Y.-J.; Hsu, Y.-T.; Ma, M.-C.; Wu, C.-K.; Luo, S.-D.; Wu, W.-B. Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis. Antioxidants 2022, 11, 1899. https://doi.org/10.3390/antiox11101899
Tsai Y-J, Hsu Y-T, Ma M-C, Wu C-K, Luo S-D, Wu W-B. Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis. Antioxidants. 2022; 11(10):1899. https://doi.org/10.3390/antiox11101899
Chicago/Turabian StyleTsai, Yih-Jeng, Yu-Ting Hsu, Ming-Chieh Ma, Chun-Kuang Wu, Sheng-Dean Luo, and Wen-Bin Wu. 2022. "Transcriptomic Analysis of Genes Associated with Oxidative Stress in Chronic Rhinosinusitis Patients with Nasal Polyps: Identifying Novel Genes Involved in Nasal Polyposis" Antioxidants 11, no. 10: 1899. https://doi.org/10.3390/antiox11101899







