Selection of Potential Probiotic Yeasts from Dry-Cured Xuanwei Ham and Identification of Yeast-Derived Antioxidant Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Ham Sampling and Yeast Isolation
2.3. Determination of Probiotic Properties
2.3.1. Growth at pH 2.5 and in Bile Salts
2.3.2. Proteinase Activity
2.3.3. Auto-Aggregation Capacity
2.3.4. Antimicrobial Activity
2.3.5. Antioxidant Activity of Yeast
2.4. Yeast Identification
2.5. Peptide Content
2.6. Determination of Antioxidant Activity
2.6.1. DPPH Radical Scavenging Activity
2.6.2. Hydroxyl Radical Scavenging Activity
2.6.3. ABTS Radical-Scavenging Activity
2.6.4. Reducing Power
2.7. Separation and Identification of Antioxidant Peptides
2.7.1. Peptide Isolation and Purification by Ultrafiltration (UF)
2.7.2. Gel Filtration Chromatography (GFC)
2.7.3. Preparative Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.7.4. Peptide Identification by LC-MS/MS
2.8. Statistical Analysis
3. Results and Discussion
3.1. Probiotic Potential of Yeast
3.1.1. Ability to Grow at pH 2.5 and in Bile Salts
3.1.2. The Proteinase Activity of Yeasts
3.1.3. The Auto-Aggregation Ability of Yeasts
3.1.4. The Antibacterial Properties of Yeasts
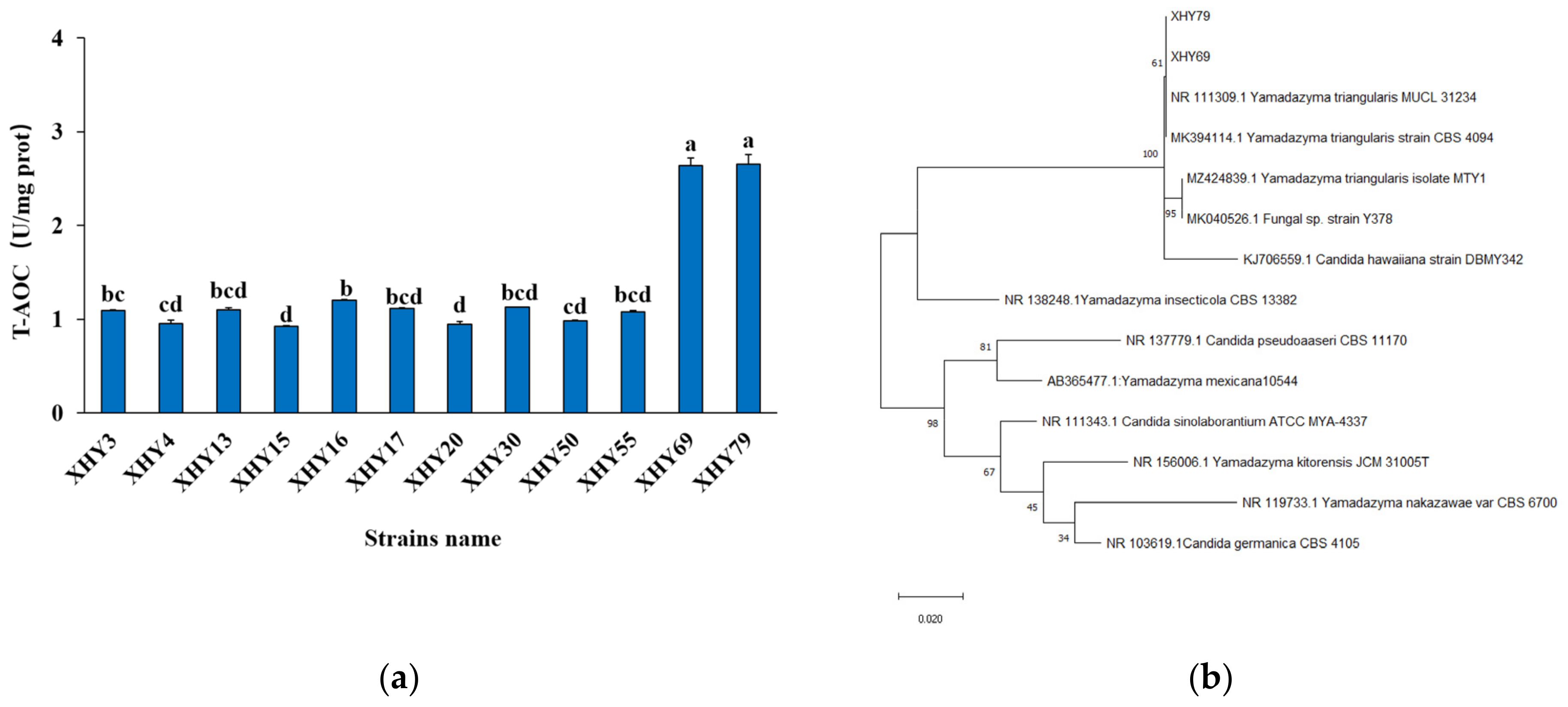
3.1.5. The Antioxidant Activity of Yeasts
3.1.6. Strain Identification
3.2. Purification and Analysis of Yeast-Derived Peptides
3.2.1. Selection of Yeast-Derived Peptides by UF
3.2.2. The Antioxidant Activity of XHY69AP
3.2.3. Separation of XHY69AP by GFC
3.2.4. Purification of AP-D by Reverse Phase-HPLC
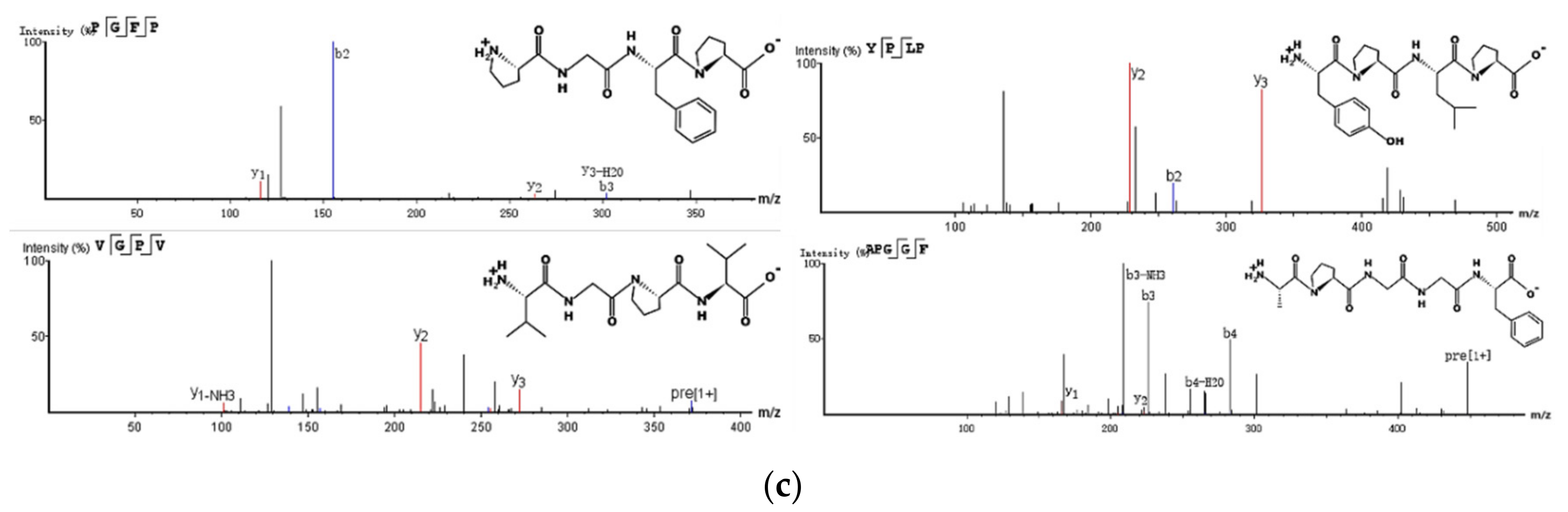
3.2.5. Identification of AP-D10 by LC-MS/MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, X.H.; Ma, P.; Jiang, D.F.; Peng, Q.; Yang, H.Y. The natural microflora of Xuanwei ham and the no-mouldy ham production. J. Food Eng. 2006, 77, 103–111. [Google Scholar] [CrossRef]
- Núñez, F.; Rodríguez, M.M.; Córdoba, J.J.; Bermúdez, M.E.; Asensio, M.A. Yeast population during ripening of dry-cured Iberian ham. Int. J. Food Microbiol. 1996, 29, 271–280. [Google Scholar] [CrossRef]
- FAO/WHO. Evaluation of Health and Nutritional Properties of Powder Milk and Live Lactic Acid Bacteria; FAO/WHO: Cordoba, Argentina, 2001; pp. 1–4. [Google Scholar]
- Zullo, B.A.; Ciafardini, G. Evaluation of physiological properties of yeast strains isolated from olive oil and their in vitro probiotic trait. Food Microbiol. 2019, 78, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Hatoum, R.; Labrie, S.; Fliss, I. Antimicrobial and probiotic properties of yeasts: From fundamental to novel applications. Front. Microbiol. 2012, 3, 421. [Google Scholar] [CrossRef]
- Gil-Rodríguez, A.M.; Carrascosa, A.V.; Requena, T. Yeasts in foods and beverages: In vitro characterisation of probiotic traits. LWT-Food Sci. Technol. 2015, 64, 1156–1162. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Marini, E.; Zannini, E.; Ciani, M.; Comitini, F. Potential Probiotic Yeasts sourced from natural environmental and spontaneous processed foods. Foods 2020, 9, 287. [Google Scholar] [CrossRef]
- Klemashevich, C.; Wu, C.; Howsmon, D.; Alaniz, R.C.; Lee, K.; Jayaraman, A. Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Curr. Opin. Biotechnol. 2014, 26, 85–90. [Google Scholar] [CrossRef]
- Aguilar-Toala, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; Gonzalez-Cordova, A.F.; Vallejo-Cordoba, B.; Hernandez-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Canani, R.B.; De Filippis, F.; Nocerino, R.; Laiola, M.; Paparo, L.; Calignano, A.; De Caro, C.; Coretti, L.; Chiariotti, L.; Gilbert, J.A.; et al. Specific signatures of the gut microbiota and increased levels of butyrate in children treated with fermented cow’s milk containing heat-killed Lactobacillus paracasei CBA L74. Appl. Environ. Microbiol. 2017, 83, e01206-17. [Google Scholar]
- Gosalbez, L.; Ramon, D. Probiotics in transition: Novel strategies. Trends Biotechnol. 2015, 33, 195–196. [Google Scholar] [CrossRef]
- Martorell, P.; Alvarez, B.; Llopis, S.; Navarro, V.; Ortiz, P.; Gonzalez, N.; Balaguer, F.; Rojas, A.; Chenoll, E.; Ramon, D.; et al. Heat-treated Bifidobacterium longum CECT-7347: A whole-cell postbiotic with antioxidant, anti-inflammatory, and gut-barrier protection properties. Antioxidants 2021, 10, 536. [Google Scholar] [CrossRef]
- De Almada, C.N.; Almada, C.N.; Martinez, R.C.R.; Sant’Ana, A.S. Paraprobiotics: Evidences on their ability to modify biological responses, inactivation methods and perspectives on their application in foods. Trends Food Sci. Technol. 2016, 58, 96–114. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef]
- Delgado, M.C.O.; Nardo, A.; Pavlovic, M.; Rogniaux, H.; Anon, M.C.; Tironi, V.A. Identification and characterization of antioxidant peptides obtained by gastrointestinal digestion of amaranth proteins. Food Chem. 2016, 197, 1160–1167. [Google Scholar] [CrossRef]
- Vieira, E.F.; das Neves, J.; Vitorino, R.; da Silva, D.D.; Carmo, H.; Ferreira, I.M.P.L.V.O. Impact of in vitro gastrointestinal digestion and transepithelial transport on antioxidant and ACE-inhibitory activities of brewer’s spent yeast autolysate. J. Agr. Food Chem. 2016, 64, 7335–7341. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Safavi, M.; Soleymanzadeh, N. The stability of antioxidant and ACE-inhibitory peptides as influenced by peptide sequences. LWT-Food Sci. Technol. 2020, 130, 109710. [Google Scholar] [CrossRef]
- Mirzaei, M.; Shavandi, A.; Mirdamadi, S.; Soleymanzadeh, N.; Motahari, P.; Mirdamadi, N.; Moser, M.; Subra, G.; Alimoradi, H.; Goriely, S. Bioactive peptides from yeast: A comparative review on production methods, bioactivity, structure-function relationship, and stability. Trends Food Sci. Technol. 2021, 118, 297–315. [Google Scholar] [CrossRef]
- Zoumpopoulou, G.; Foligne, B.; Christodoulou, K.; Grangette, C.; Pot, B.; Tsakalidou, E. Lactobacillus fermentum ACA-DC 179 displays probiotic potential in vitro and protects against trinitrobenzene sulfonic acid (TNBS)-induced colitis and Salmonella infection in murine models. Int. J. Food Microbiol. 2008, 121, 18–26. [Google Scholar] [CrossRef]
- Xing, L.J.; Hu, Y.Y.; Hu, H.Y.; Ge, Q.F.; Zhou, G.H.; Zhang, W.G. Purification and identification of antioxidative peptides from dry-cured Xuanwei ham. Food Chem. 2016, 194, 951–958. [Google Scholar] [CrossRef]
- Ge, Q.; Yang, B.; Liu, R.; Jiang, D.; Yu, H.; Wu, M.; Zhang, W. Antioxidant activity of Lactobacillus plantarum NJAU-01 in an animal model of aging. BMC Microbiol. 2021, 21, 182. [Google Scholar] [CrossRef]
- Staniszewski, A.; Kordowska-Wiater, M. Probiotic and potentially probiotic yeasts-characteristics and food application. Foods 2021, 10, 1306. [Google Scholar] [CrossRef]
- Sambrani, R.; Abdolalizadeh, J.; Kohan, L.; Jafari, B. Recent advances in the application of probiotic yeasts, particularly Saccharomyces, as an adjuvant therapy in the management of cancer with focus on colorectal cancer. Mol. Biol. Rep. 2021, 48, 951–960. [Google Scholar] [CrossRef]
- Buzzini, P.; Martini, A. Extracellular enzymatic activity profiles in yeast and yeast-like strains isolated from tropical environments. J. Appl. Microbiol. 2002, 93, 1020–1025. [Google Scholar] [CrossRef]
- McFarland, L.V. From yaks to yogurt: The history, development, and current use of probiotics. Clin. Infect. Dis. 2015, 60, S85–S90. [Google Scholar] [CrossRef]
- Ilango, S.; Antony, U. Probiotic microorganisms from non-dairy traditional fermented foods. Trends Food Sci. Technol. 2021, 118, 617–638. [Google Scholar] [CrossRef]
- Hojjati, M.; Behabahani, B.A.; Falah, F. Aggregation, adherence, anti-adhesion and antagonistic activity properties relating to surface charge of probiotic Lactobacillus brevis gp104 against Staphylococcus aureus. Microb. Pathogenes. 2020, 147, 104420. [Google Scholar] [CrossRef]
- Binetti, A.; Carrasco, M.; Reinheimer, J.; Suarez, V. Yeasts from autochthonal cheese starters: Technological and functional properties. J. Appl. Microbiol. 2013, 115, 434–444. [Google Scholar] [CrossRef]
- Goktas, H.; Dikmen, H.; Demirbas, F.; Sagdic, O.; Dertli, E. Characterisation of probiotic properties of yeast strains isolated from kefir samples. Int. J. Dairy Technol. 2021, 74, 715–722. [Google Scholar] [CrossRef]
- Amaretti, A.; di Nunzio, M.; Pompei, A.; Raimondi, S.; Rossi, M.; Bordoni, A. Antioxidant properties of potentially probiotic bacteria: In vitro and in vivo activities. Appl. Microbiol. Biot. 2013, 97, 809–817. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant properties of probiotic bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Gallardo, G.; Ruiz-Moyano, S.; Hernández, A.; Benito, M.J.; Córdoba, M.G.; Pérez-Nevado, F.; Martín, A. Application of ISSR-PCR for rapid strain typing of Debaryomyces hansenii isolated from dry-cured Iberian ham. Food Microbiol. 2014, 42, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, C.; Johansen, P.G.; Petersen, M.A.; Poojary, M.M.; Lund, M.N.; Jespersen, L.; Arneborg, N. The utilisation of amino acids by Debaryomyces hansenii and Yamadazyma triangularis associated with cheese. Int. Dairy J. 2021, 121, 105135. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Ehsani, M.R.; Aminlari, M. Production of antioxidant and ACE-inhibitory peptides from Kluyveromyces marxianus protein hydrolysates: Purification and molecular docking. J. Food Drug Anal. 2018, 26, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.N.; Tain, Y.L. Regulation of nitric oxide production in the developmental programming of hypertension and kidney disease. Int. J. Mol. Sci. 2019, 20, 681. [Google Scholar] [CrossRef]
- Liu, R.; Xing, L.J.; Fu, Q.Q.; Zhou, G.H.; Zhang, W.G. A review of antioxidant peptides derived from meat muscle and by-products. Antioxidants 2016, 5, 32. [Google Scholar] [CrossRef]
- Mirzaei, M.; Mirdamadi, S.; Ehsani, M.R.; Aminlari, M.; Hosseini, E. Purification and identification of antioxidant and ACE-inhibitory peptide from Saccharomyces cerevisiae protein hydrolysate. J. Funct. Foods 2015, 19, 259–268. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef]
- Yang, W.S.; Hao, X.Y.; Zhang, X.X.; Zhang, G.X.; Li, X.D.; Liu, L.; Sun, Y.; Pan, Y. Identification of antioxidant peptides from cheddar cheese made with Lactobacillus helveticus. LWT-Food Sci. Technol. 2021, 141, 110866. [Google Scholar] [CrossRef]
- You, L.J.; Zhao, M.M.; Regenstein, J.M.; Ren, J.Y. Purification and identification of antioxidative peptides from loach (Misgurnus anguillicaudatus) protein hydrolysate by consecutive chromatography and electrospray ionization-mass spectrometry. Food Res. Int. 2010, 43, 1167–1173. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Wang, Z.Y.; Chen, S.W.; Luo, Y.K. Production and identification of antioxidant and angiotensin-converting enzyme inhibition and dipeptidyl peptidase IV inhibitory peptides from bighead carp (Hypophthalmichthys nobilis) muscle hydrolysate. J. Funct. Foods 2017, 35, 224–235. [Google Scholar] [CrossRef]
- Mirdamadi, S.; Mirzaei, M.; Soleymanzadeh, N.; Safavi, M.; Bakhtiari, N.; Zandi, M. Antioxidant and cytoprotective effects of synthetic peptides identified from Kluyveromyces marxianus protein hydrolysate: Insight into the molecular mechanism. LWT-Food Sci. Technol. 2021, 148, 111792. [Google Scholar] [CrossRef]
- Fu, Y.; Young, J.F.; Rasmussen, M.K.; Dalsgaard, T.K.; Lametsch, R.; Aluko, R.E.; Therkildsen, M. Angiotensin I-converting enzyme-inhibitory peptides from bovine collagen: Insights into inhibitory mechanism and transepithelial transport. Food Res. Int. 2016, 89, 373–381. [Google Scholar] [CrossRef]
- Fu, L.; Xing, L.; Hao, Y.; Yang, Z.; Teng, S.; Wei, L.; Zhang, W. The anti-inflammatory effects of dry-cured ham derived peptides in RAW264.7 macrophage cells. J. Funct. Foods 2021, 87, 104827. [Google Scholar] [CrossRef]



| Yeast Strain Name | Growth at pH 2.5 | Growth at 1% Bile Salt | Proteinase Activity | Yeast Strain Name | Growth at pH 2.5 | Growth at 1% Bile Salt | Proteinase Activity |
|---|---|---|---|---|---|---|---|
| XHY1 | - | - | - | XHY55 | - | ++ | + |
| XHY2 | ++ | - | - | XHY56 | - | ++ | - |
| XHY3 | + | ++ | + | XHY57 | - | - | - |
| XHY4 | + | ++ | + | XHY58 | - | ++ | + |
| XHY5 | ++ | - | - | XHY59 | + | + | + |
| XHY6 | - | - | + | XHY60 | - | - | + |
| XHY7 | + | - | + | XHY61 | - | - | - |
| XHY8 | ++ | - | - | XHY62 | - | - | + |
| XHY9 | - | ++ | - | XHY63 | - | + | - |
| XHY10 | - | - | - | XHY64 | - | - | + |
| XHY11 | + | - | - | XHY65 | - | - | - |
| XHY12 | + | - | - | XHY66 | - | - | - |
| XHY13 | + | ++ | + | XHY67 | - | - | + |
| XHY14 | - | - | - | XHY68 | - | - | + |
| XHY15 | + | ++ | + | XHY69 | + | ++ | + |
| XHY16 | + | + | + | XHY70 | - | ++ | ++ |
| XHY17 | ++ | ++ | + | XHY71 | - | - | - |
| XHY18 | + | ++ | ++ | XHY72 | + | - | + |
| XHY19 | - | - | - | XHY73 | - | - | - |
| XHY20 | + | ++ | - | XHY74 | - | - | + |
| XHY21 | + | ++ | + | XHY75 | - | - | - |
| XHY22 | - | - | -- | XHY76 | - | + | - |
| XHY23 | ++ | ++ | + | XHY77 | - | - | - |
| XHY24 | - | - | - | XHY78 | - | - | - |
| XHY25 | - | - | - | XHY79 | + | ++ | + |
| XHY26 | - | + | + | XHY80 | - | - | + |
| XHY27 | - | + | ++ | XHY81 | - | ++ | + |
| XHY28 | ++ | ++ | + | XHY82 | - | - | + |
| XHY29 | - | ++ | + | XHY83 | - | ++ | ++ |
| XHY30 | + | + | + | XHY84 | - | ++ | - |
| XHY31 | - | ++ | - | XHY85 | - | - | ++ |
| XHY32 | - | ++ | - | XHY86 | - | - | - |
| XHY33 | - | - | + | XHY87 | - | - | + |
| XHY34 | - | - | - | XHY88 | ++ | - | + |
| XHY35 | + | ++ | + | XHY89 | + | + | + |
| XHY36 | - | + | - | XHY90 | ++ | - | + |
| XHY37 | - | - | - | XHY91 | - | - | - |
| XHY38 | - | - | + | XHY92 | + | ++ | ++ |
| XHY39 | - | + | - | XHY93 | + | + | ++ |
| XHY40 | - | - | - | XHY94 | - | - | ++ |
| XHY41 | - | - | - | XHY95 | - | + | - |
| XHY42 | - | + | - | XHY96 | - | + | - |
| XHY43 | + | + | + | XHY97 | ++ | - | - |
| XHY44 | + | - | ++ | XHY98 | + | - | + |
| XHY45 | - | + | - | XHY99 | - | - | - |
| XHY46 | + | - | + | XHY100 | + | - | ++ |
| XHY47 | - | - | - | XHY101 | + | - | - |
| XHY48 | - | ++ | - | XHY102 | - | + | - |
| XHY49 | + | ++ | + | XHY103 | - | + | - |
| XHY50 | ++ | ++ | + | XHY104 | ++ | - | - |
| XHY51 | - | - | + | XHY105 | + | - | + |
| XHY52 | - | ++ | + | XHY106 | - | - | - |
| XHY53 | - | ++ | - | XHY107 | + | - | ++ |
| XHY54 | - | + | + | XHY108 | + | - | - |
| Strain Name | Bacteriostatic | Auto-Aggregation (%) | DPPH Scavenging Activity (%) | ||||
|---|---|---|---|---|---|---|---|
| E. coli | S. aureus | Salmonella sp. | 2 h | 4 h | 24 h | ||
| XHY3 | - | + | - | 80.49 ± 0.45 fgh | 86.87 ± 0.4 cde | 91.3 ± 0.29 e | 84.83 ± 0.1 ab |
| XHY4 | + | - | + | 84.7 ± 0.36 cd | 87.94 ± 0.5 cd | 91.68 ± 0.07 d | 84.85 ± 0.08 ab |
| XHY13 | - | + | ++ | 86.08 ± 0.19 abc | 90.04 ± 0.28 ab | 93.01 ± 0.16 c | 84.97 ± 0.03 a |
| XHY15 | - | - | + | 88.17±0.18 a | 90.19 ± 0.17 ab | 92.89 ± 0.01 c | 84.46 ± 0.32 abc |
| XHY16 | + | + | + | 80.81 ± 1.17 fg | 81.28 ± 0.05 j | 90.24 ± 0.01 fg | 84.98 ± 0.08 a |
| XHY17 | - | - | + | 82.7 ± 0.29 def | 81.22 ± 0.69 j | 91.73 ± 0.05 d | 84.94 ± 0.08 a |
| XHY18 | - | + | - | 81.37 ± 0.4 efg | 83.76 ± 1.05 ghi | 87.6 ± 0.32 lm | 75.54 ± 0.31 fghi |
| XHY20 | + | - | + | 69.59 ± 0.06 j | 81.27 ± 0.39 j | 95.44 ± 0.04 a | 84.49 ± 0.14 bc |
| XHY21 | - | - | - | 84.34 ± 0.71 cd | 86.75 ± 0.58 de | 89.82 ± 0.09 hi | 77.35 ± 0.1 fg |
| XHY23 | - | + | + | 78.71 ± 1.76 ghi | 83.86 ± 0.41 ghi | 87.25 ± 0.06 m | 77.82 ± 0.13 f |
| XHY27 | - | + | + | 71.17 ± 0.57 f | 74.17 ± 1.73 l | 82.38 ± 0.03 o | 79.53 ± 0.18 e |
| XHY28 | - | - | - | 79.75 ± 1.04 gh | 83.47 ± 0.15 hi | 89.97 ± 0.03 gh | 76.87 ± 0.09 ghi |
| XHY30 | - | + | - | 78.67 ± 0.08 ghi | 84.85 ± 0.82 fgh | 88.78 ± 0.11 k | 80.32 ± 0.38 d |
| XHY35 | + | - | ++ | 69.93 ± 1.25 j | 75.67 ± 0.68 l | 77.62 ± 0.1 q | 74.45 ± 0.31 k |
| XHY43 | + | + | + | 79.81 ± 0.75 cde | 85.51 ± 0.44 efg | 89.9 ± 0.06 gh | 76.53 ± 0.16 hi |
| XHY44 | - | - | + | 83.87 ± 0.52 abc | 87.59 ± 0.89 cd | 91.76 ± 0.18 d | 76.5 ± 0.13 hi |
| XHY49 | + | - | + | 71.43 ± 0.42 j | 78.77 ± 2 k | 79.49 ± 0.36 p | 76.43 ± 0.14 i |
| XHY50 | + | + | + | 87.99 ± 0.02 a | 90.48 ± 0.65 a | 93.92 ± 0.03 b | 83.87 ± 0.03 c |
| XHY55 | + | - | + | 84.94 ± 1.4 bcd | 88.59 ± 0.23 bc | 91.64 ± 0.39 de | 85.03 ± 0.01 a |
| XHY69 | + | + | + | 87.62 ± 0.32 ab | 89.88 ± 0.24 ab | 93.85 ± 0.03 b | 85.08 ± 0.04 a |
| XHY70 | - | - | - | 84.22 ± 0.54 cde | 87.83 ± 0.34 cd | 91.46 ± 0.37 de | 84.1 ± 0.2 abc |
| XHY79 | - | + | + | 83.07 ± 0.39 def | 87.74 ± 0.23 cd | 90.49 ± 0.07 f | 84.82 ± 0.05 ab |
| XHY83 | - | - | + | 77.58 ± 0.22 hi | 83 ± 0.4 i | 86.01 ± 0.08 n | 77.27 ± 0.20 fgh |
| XHY88 | + | + | + | 83.28 ± 0.57 cdef | 85.64 ± 1.5 ef | 89.52 ± 0.08 ij | 77.27 ± 0.12 ghi |
| XHY89 | - | - | - | 78.55 ± 2.88 ghi | 83.77 ± 0.8 fghi | 87.51 ± 0.05 m | 76.84 ± 0.01 ghi |
| XHY92 | - | - | + | 76.71 ± 4.57 i | 84.21 ± 0.71 ghi | 87.93 ± 0.02 l | 77.03 ± 0.06 ghi |
| XHY93 | - | + | + | 79.57 ± 1.2 ghi | 86.82 ± 0.2 cde | 89.26 ± 0.09 j | 76.99 ± 0.02 jk |
| ACL (%) | Sequences | Molecular Weight (Da) | Isoelectric Point (pI) | Hydrophobicity (Kcal × mol −1) | Presumptive Parent Protein |
|---|---|---|---|---|---|
| 99 | FPPQ | 487.55 | 5.38 | 7.24 | Serine/threonine-protein kinase ATG1 |
| 99 | VGPF | 418.49 | 5.56 | 7.02 | Autophagy-related protein 22 |
| 99 | AGPL | 356.42 | 5.60 | 8.44 | Mannose-1-phosphate guanyltransferase |
| 99 | YPLP | 488.58 | 5.48 | 6.22 | Aminopeptidase |
| 98 | VGPV | 370.44 | 5.63 | 8.27 | 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase |
| 98 | GPFP | 416.47 | 5.65 | 7.62 | High-affinity K+ transporter |
| 97 | PGFP | 416.47 | 5.25 | 7.62 | Homoaconitase, mitochondrial |
| 96 | APGGF | 447.48 | 5.53 | 9.13 | Heat shock protein 70 1; vacuolar amino acid transporter 3 |
| Peptides | FPPQ | VGPF | AGPL | YPLP | VGPV | GPFP | PGFP | APGGF |
|---|---|---|---|---|---|---|---|---|
| ABTS scavenging activity (%) | 12.83 ± 1.55 d | 12.78 ± 0.11 d | 12.79 ± 1.34 d | 75.48 ± 0.23 a | 14.76 ± 0.96 ce | 34.41 ± 1.34 b | 13.83 ± 0.15 d | 18.44 ± 1.47 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, J.; Xing, L.; Zhang, W.; Fu, L.; Zhang, J. Selection of Potential Probiotic Yeasts from Dry-Cured Xuanwei Ham and Identification of Yeast-Derived Antioxidant Peptides. Antioxidants 2022, 11, 1970. https://doi.org/10.3390/antiox11101970
Cai J, Xing L, Zhang W, Fu L, Zhang J. Selection of Potential Probiotic Yeasts from Dry-Cured Xuanwei Ham and Identification of Yeast-Derived Antioxidant Peptides. Antioxidants. 2022; 11(10):1970. https://doi.org/10.3390/antiox11101970
Chicago/Turabian StyleCai, Jiaming, Lujuan Xing, Wangang Zhang, Lijuan Fu, and Jian Zhang. 2022. "Selection of Potential Probiotic Yeasts from Dry-Cured Xuanwei Ham and Identification of Yeast-Derived Antioxidant Peptides" Antioxidants 11, no. 10: 1970. https://doi.org/10.3390/antiox11101970
APA StyleCai, J., Xing, L., Zhang, W., Fu, L., & Zhang, J. (2022). Selection of Potential Probiotic Yeasts from Dry-Cured Xuanwei Ham and Identification of Yeast-Derived Antioxidant Peptides. Antioxidants, 11(10), 1970. https://doi.org/10.3390/antiox11101970






