Chenodeoxycholic Acid (CDCA) Promoted Intestinal Epithelial Cell Proliferation by Regulating Cell Cycle Progression and Mitochondrial Biogenesis in IPEC-J2 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Selection of CDCA Concentration
2.3. Proliferation Assay
2.4. Measurement of Apoptosis, Cell Cycle, Mitochondrial Membrane Potential, and Intracellular Reactive Oxygen Species
2.5. Determination of Cellular Antioxidant Indices and ATP Level
2.6. RNA Extraction, Library Preparation, Sequencing, and Data Analysis
2.7. RNA Extraction and RT-qPCR
2.8. mtDNA Copy Number Determination
2.9. Statistical Analysis
3. Results
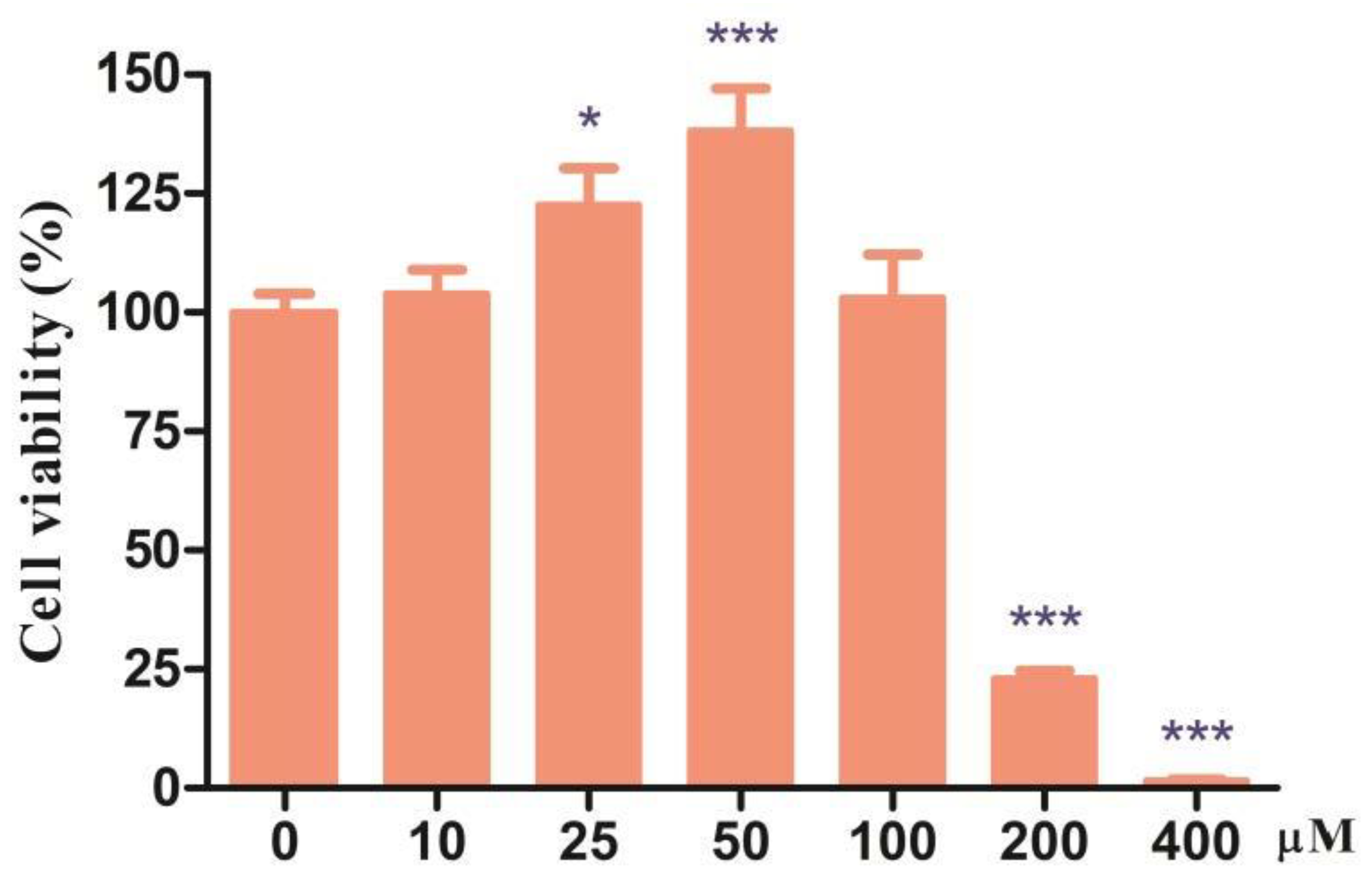
3.1. Effects of Different Doses of CDCA on Cell Viability of IPEC-J2 Cells
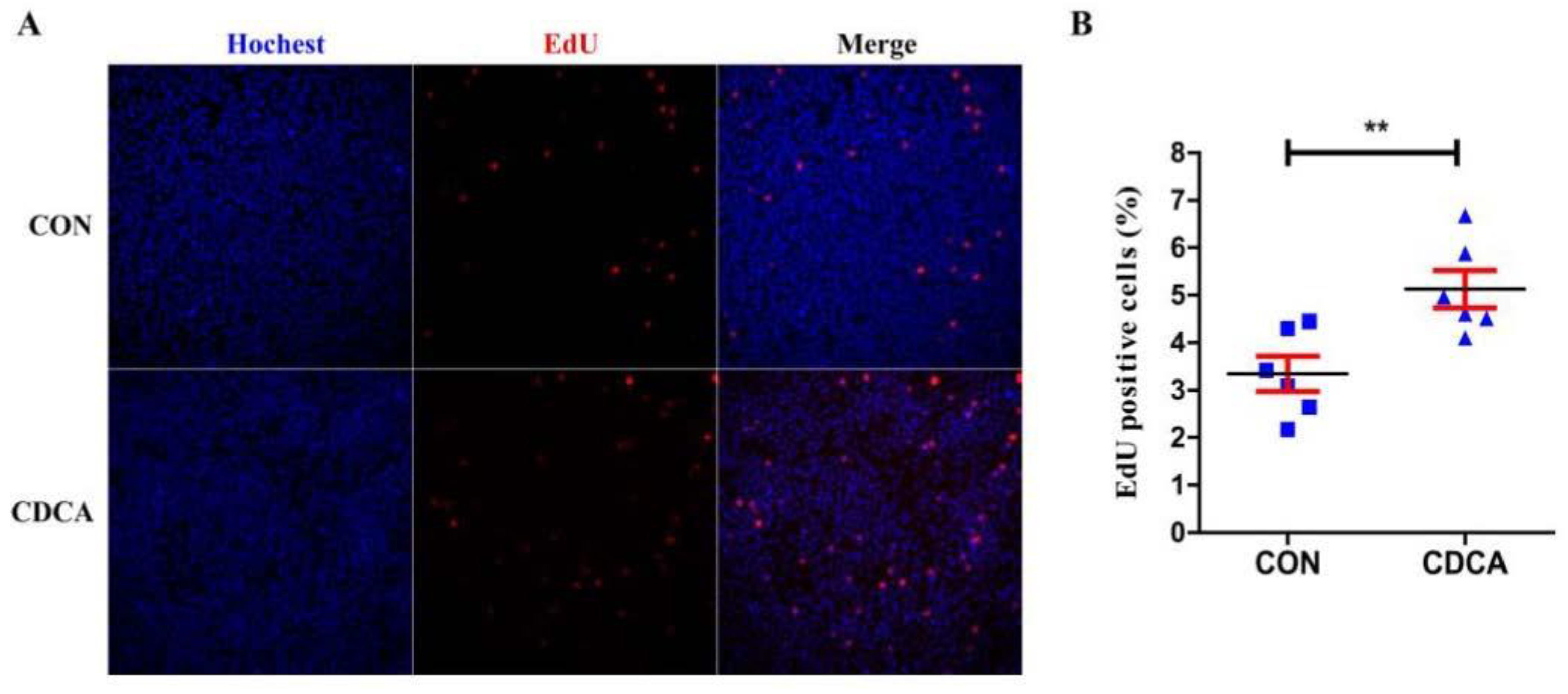
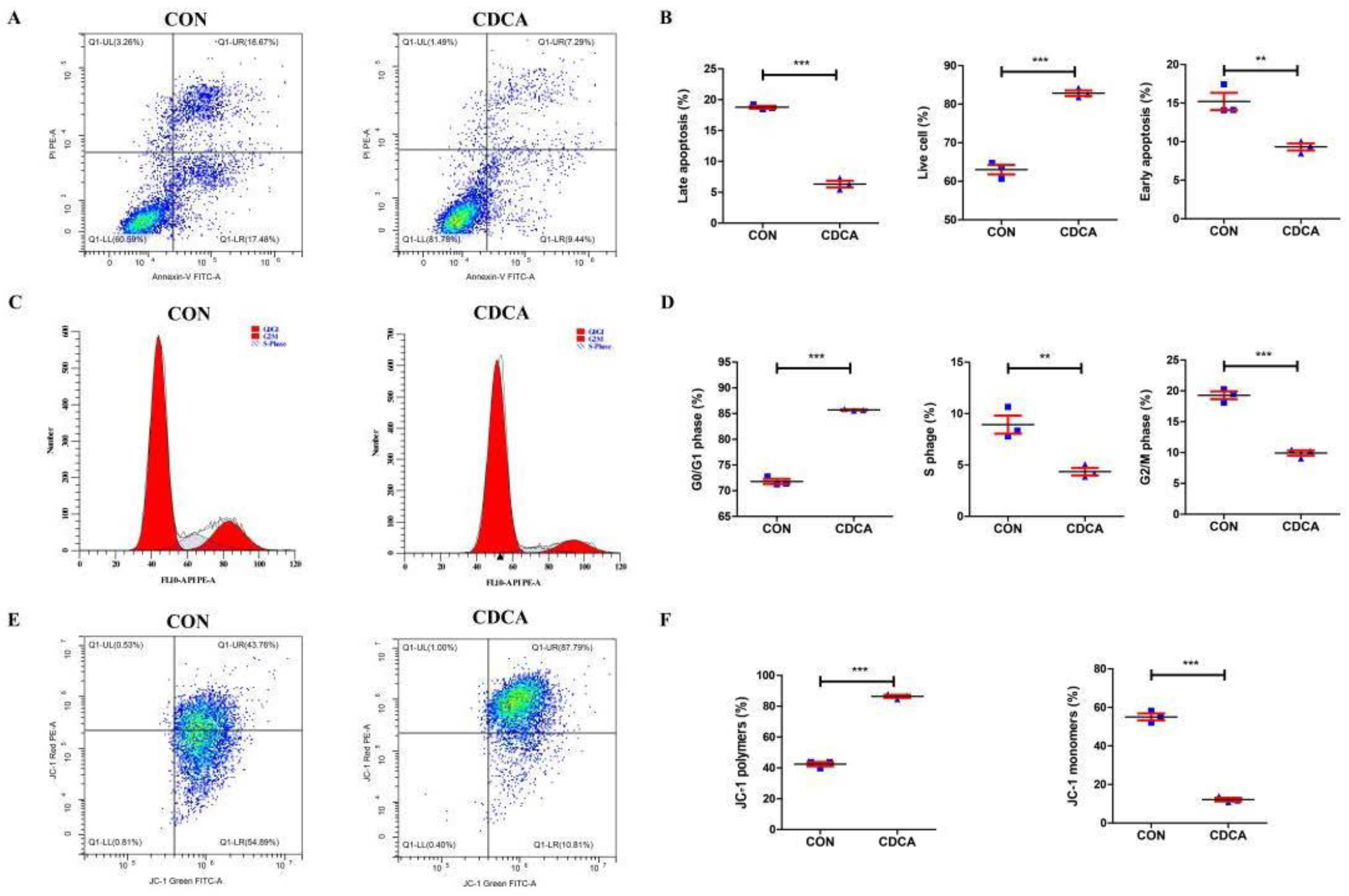
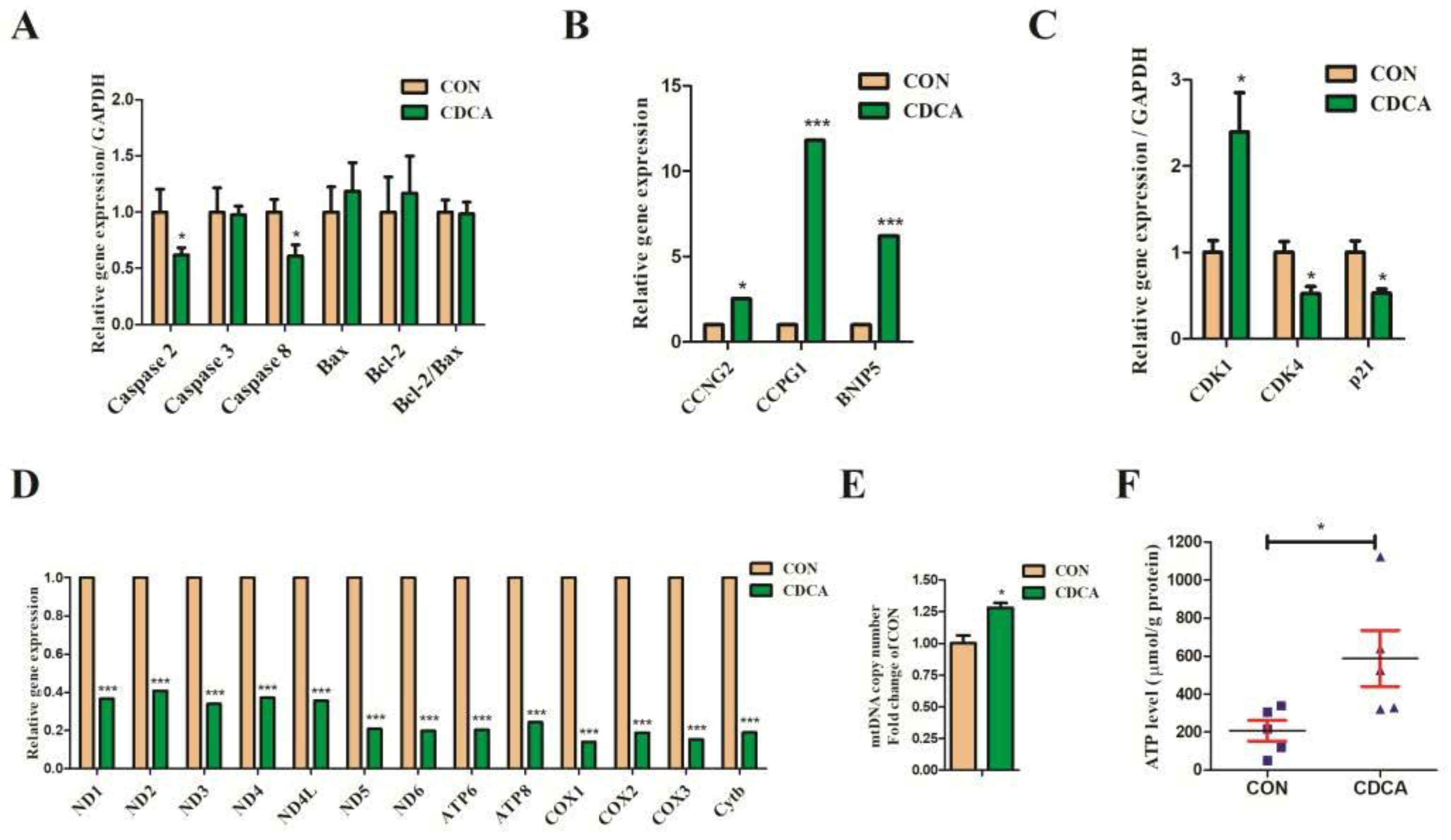
3.2. CDCA Reduces Apoptosis, Improves Mitochondrial Function, and Regulates the Cell Cycle
3.3. CDCA Could Enhance Redox Balance in IPEC-J2 Cells
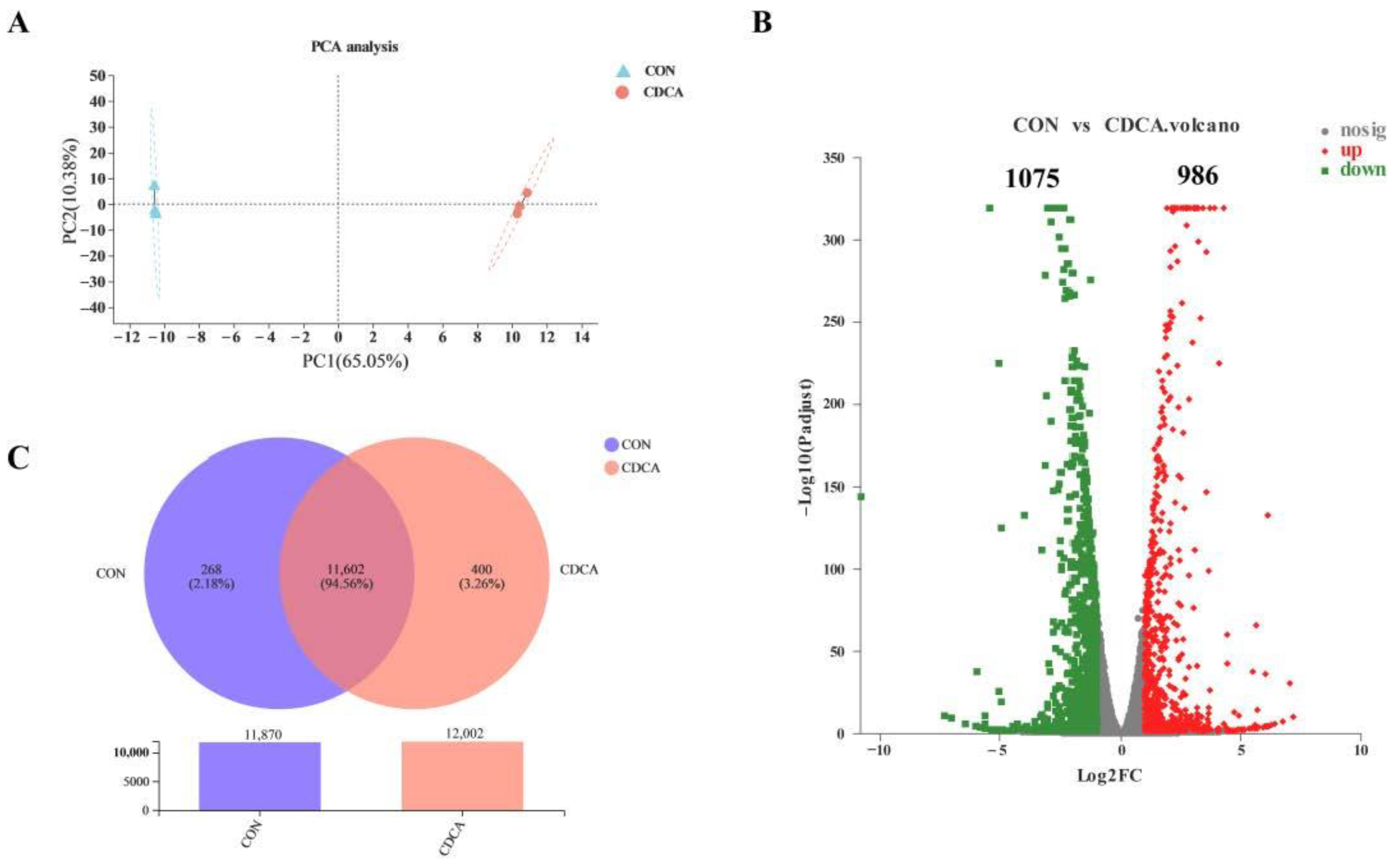
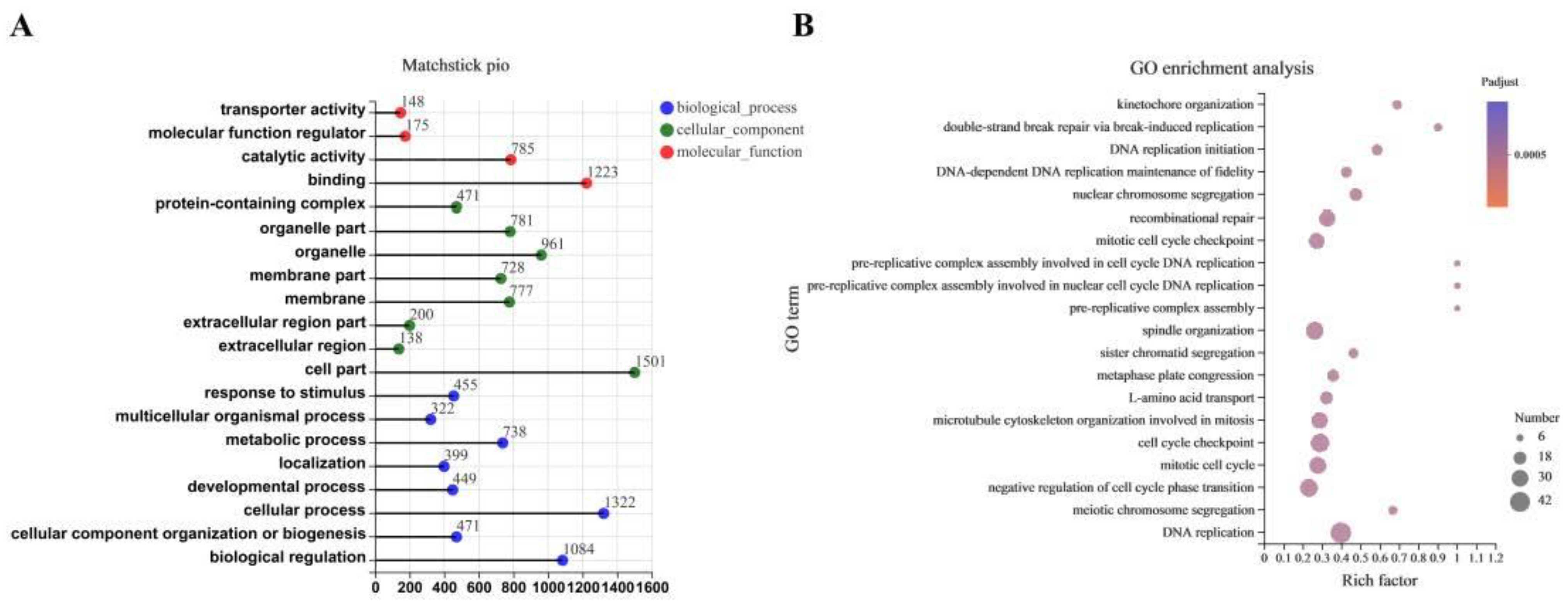
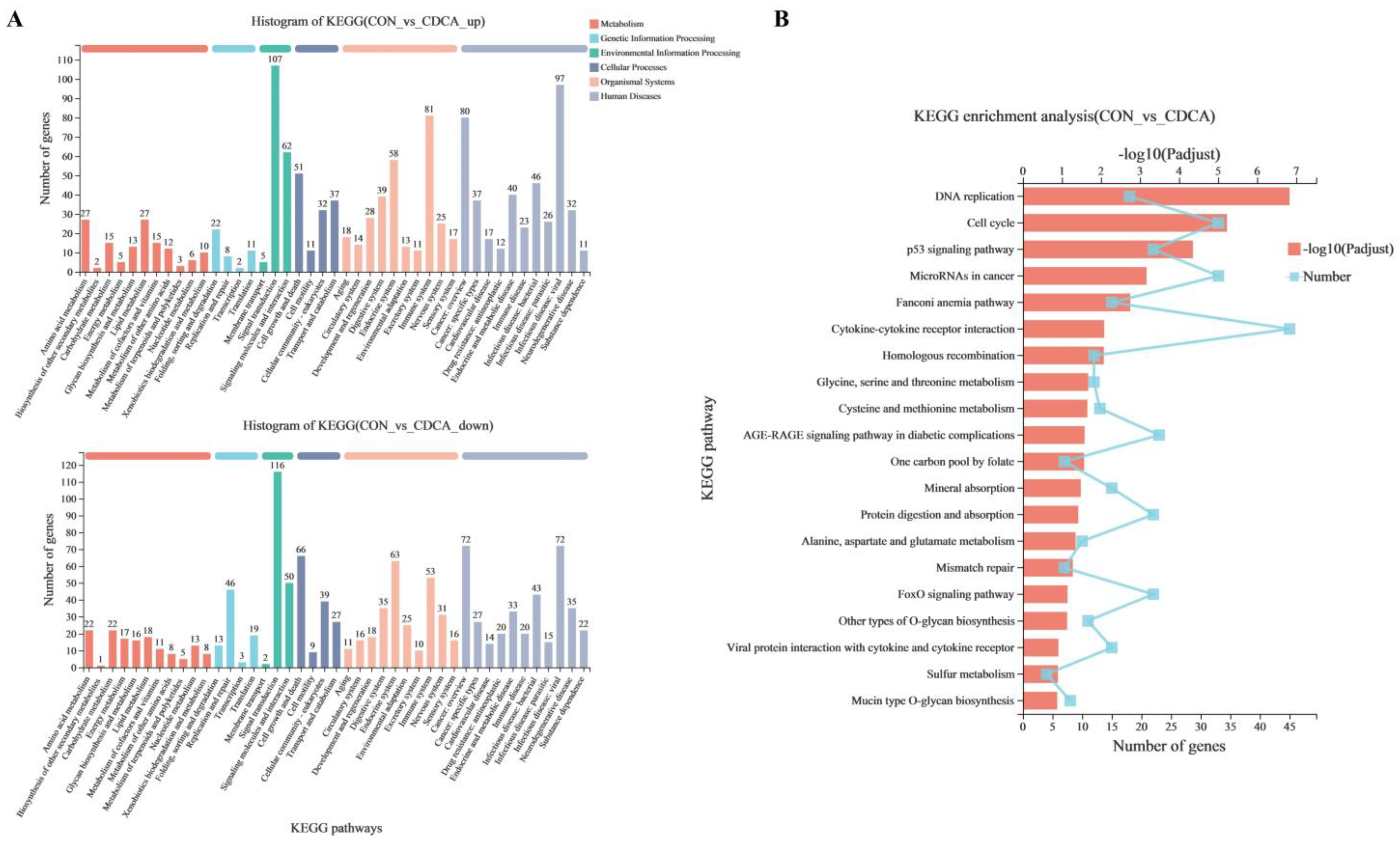
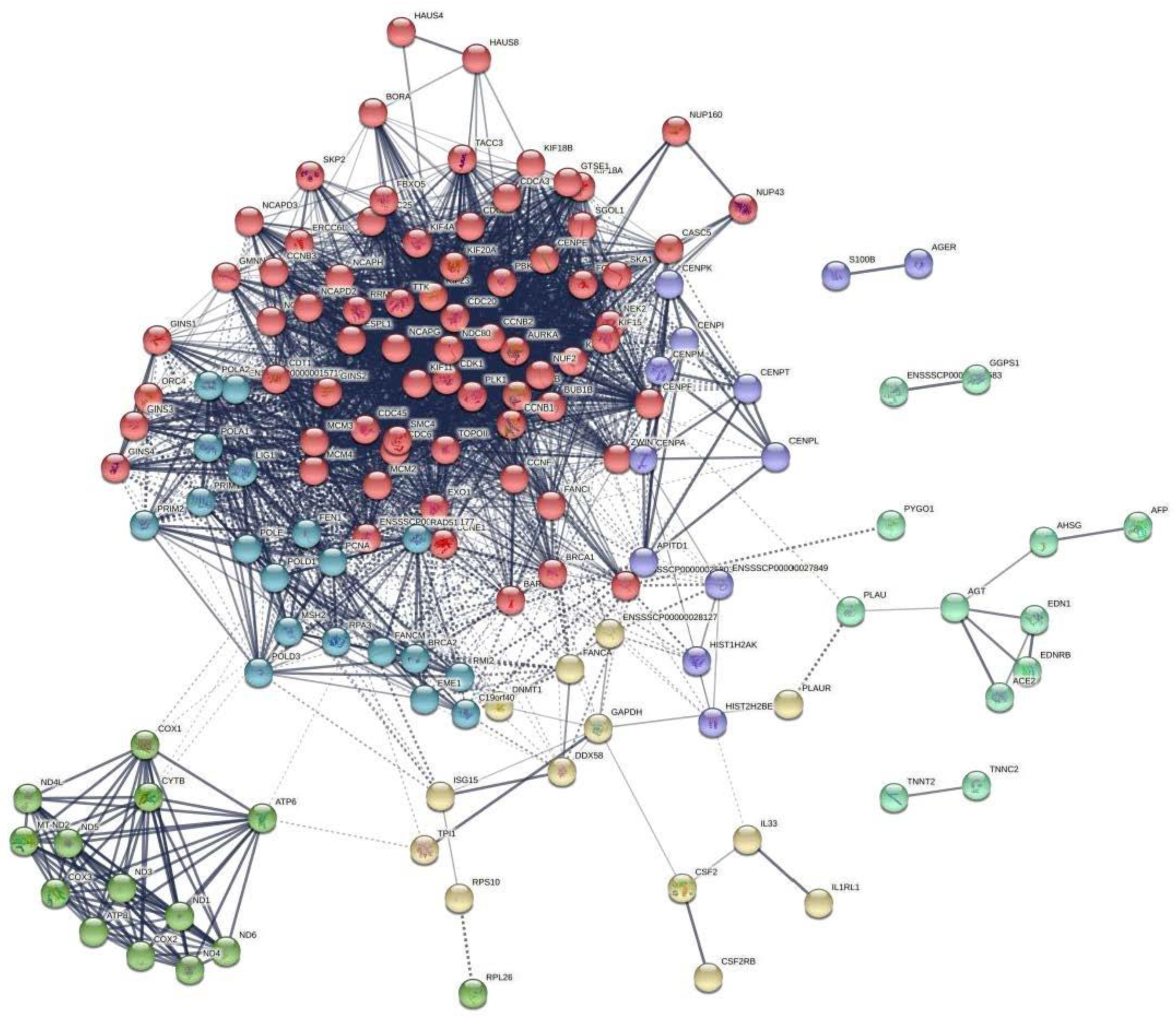
3.4. The Transcriptome Response to CDCA Treatment in IPEC-J2 Cells
3.5. Cell Apoptosis, Cell Cycle, and Mitochondrial Biogenesis-Related Gene Expression Regulated by CDCA Treatment in IPEC-J2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thomas, C.; Gioiello, A.; Noriega, L.; Strehle, A.; Oury, J.; Rizzo, G.; Macchiarulo, A.; Yamamoto, H.; Mataki, C.; Pruzanski, M.; et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009, 10, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.J.; Chen, T.L.; Jiang, R.Q.; Zhao, A.H.; Wu, Q.; Kuang, J.L.; Sun, D.N.; Ren, Z.X.; Li, M.C.; Zhao, M.L.; et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021, 33, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, F.; de Boer, J.F.; Staels, B. Microbiome modulation of the host adaptive immunity through bile acid modification. Cell Metab. 2020, 31, 445–447. [Google Scholar] [CrossRef]
- Cai, J.; Sun, L.L.; Gonzalez, F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Dossa, A.Y.; Escobar, O.; Golden, J.; Frey, M.R.; Ford, H.R.; Gayer, C.P. Bile acids regulate intestinal cell proliferation by modulating EGFR and FXR signaling. Am. J. Physiol.-Gastrointest. Liver Physiol. 2016, 310, G81–G92. [Google Scholar] [CrossRef] [Green Version]
- Booth, D.M.; Murphy, J.A.; Mukherjee, R.; Awais, M.; Neoptolemos, J.P.; Gerasimenko, O.V.; Tepikin, A.V.; Petersen, O.H.; Sutton, R.; Criddle, D.N. Reactive oxygen species induced by bile acid induce apoptosis and protect against necrosis in pancreatic acinar cells. Gastroenterology 2011, 140, 2116–2125. [Google Scholar] [CrossRef]
- Mu, C.L.; Pi, Y.; Zhang, C.J.; Zhu, W.Y. Microbiomes in the intestine of developing pigs: Implications for nutrition and health. In Recent Advances in Animal Nutrition and Metabolism; Wu, G., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2022; pp. 161–176. [Google Scholar]
- Petroni, M.L.; Jazrawi, R.P.; Pazzi, P.; Lanzini, A.; Zuin, M.; Pigozzi, M.G.; Fracchia, M.; Galatola, G.; Alvisi, V.; Heaton, K.W.; et al. Ursodeoxycholic acid alone or with chenodeoxycholic acid for dissolution of cholesterol gallstones: A randomized multicentre trial. Aliment. Pharmacol. Ther. 2001, 15, 123–128. [Google Scholar]
- Molinaro, A.; Wahlstrom, A.; Marschall, H.U. Role of bile acids in metabolic control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef]
- Teodoro, J.S.; Machado, I.F.; Castela, A.C.; Amorim, J.A.; Jarak, I.; Carvalho, R.A.; Palmeira, C.M.; Rolo, A.P. Chenodeoxycholic acid has non-thermogenic, mitodynamic anti-obesity effects in an in vitro CRISPR/Cas9 model of bile acid receptor TGR5 knockdown. Int. J. Mol. Sci. 2021, 22, 11738. [Google Scholar] [CrossRef]
- Broeders, E.P.M.; Nascimento, E.B.M.; Havekes, B.; Brans, B.; Roumans, K.H.M.; Tailleux, A.; Schaart, G.; Kouach, M.; Charton, J.; Deprez, B.; et al. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 2015, 22, 418–426. [Google Scholar] [CrossRef] [Green Version]
- Shihabudeen, M.S.; Roy, D.; James, J.; Thirumurugan, K. Chenodeoxycholic acid, an endogenous FXR ligand alters adipokines and reverses insulin resistance. Mol. Cell Endocrinol. 2015, 414, 19–28. [Google Scholar] [CrossRef]
- Meyer-Gerspach, A.C.; Steinert, R.E.; Keller, S.; Malarski, A.; Schulte, F.H.; Beglinger, C. Effects of chenodeoxycholic acid on the secretion of gut peptides and fibroblast growth factors in healthy humans. J. Clin. Endocrinol. Metab. 2013, 98, 3351–3358. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Zhang, F.L.; Chen, L.; Yang, Q.; Su, H.; Yang, X.H.; He, H.W.; Ling, M.F.; Zheng, J.S.; Duan, C.; et al. Dietary chenodeoxycholic acid improves growth performance and intestinal health by altering serum metabolic profiles and gut bacteria in weaned piglets. Anim. Nutr. 2021, 7, 365–375. [Google Scholar] [CrossRef]
- van der Meer, Y.; Gerrits, W.J.J.; van den Bosch, M.; Holst, J.J.; Moreto, M.; Buurman, W.A.; Kulik, W.; van Kempen, T.A.T.G. Chenodeoxycholic acid reduces intestinal permeability in newly weaned piglets. J. Anim. Sci. 2012, 90, 302–304. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Ye, J.Y.; Zhang, F.L.; Su, H.; Yang, X.H.; He, H.W.; Liu, F.F.; Zhu, X.T.; Wang, L.N.; Gao, P.; et al. Chenodeoxycholic acid (CDCA) protects against the lipopolysaccharide-induced impairment of the intestinal epithelial barrier function via the FXR-MLCK pathway. J. Agr. Food Chem. 2019, 67, 8868–8874. [Google Scholar] [CrossRef]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef]
- Chen, Z.G.; Yuan, Q.L.; Xu, G.R.; Chen, H.Y.; Lei, H.Y.; Su, J.M. Effects of quercetin on proliferation and H2O2-induced apoptosis of intestinal porcine enterocyte cells. Molecules 2018, 23, 2012. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.L.; Fuller, C.R.; Dieleman, L.A.; DaCosta, C.M.; Haldeman, K.M.; Sartor, R.B.; Lund, P.K. Enhanced survival and mucosal repair after dextran sodium sulfate—Induced colitis in transgenic mice that overexpress growth hormone. Gastroenterology 2001, 120, 925–937. [Google Scholar] [CrossRef]
- Wang, M.; Wu, H.; Lu, L.; Jiang, L.; Yu, Q. Lactobacillus reuteri promotes intestinal development and regulates mucosal immune function in newborn piglets. Front. Vet. Sci. 2020, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Ishizuka, S.; Shiwaku, M.; Hagio, M.; Suzuki, T.; Hira, T.; Hara, H. Glycochenodeoxycholic acid promotes proliferation of intestinal epithelia via reduction of cyclic AMP and increase in H2AX phosphorylation after exposure to gamma-rays. Biomed. Res. 2012, 33, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Krishna-Subramanian, S.; Hanski, M.L.; Loddenkemper, C.; Choudhary, B.; Pages, G.; Zeitz, M.; Hanski, C. UDCA slows down intestinal cell proliferation by inducing high and sustained ERK phosphorylation. Int. J. Cancer 2012, 130, 2771–2782. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Yang, Q.; Zhang, F.L.; Chen, L.; Su, H.; Yang, X.H.; He, H.W.; Liu, F.F.; Zheng, J.S.; Ling, M.F.; et al. Hyodeoxycholic acid (HDCA) suppresses intestinal epithelial cell proliferation through FXR-PI3K/AKT pathway, accompanied by alteration of bile acids metabolism profiles induced by gut bacteria. FASEB J. 2020, 34, 7103–7117. [Google Scholar] [CrossRef] [PubMed]
- Schierack, P.; Nordhoff, M.; Pollmann, M.; Weyrauch, K.D.; Amasheh, S.; Lodemann, U.; Jores, J.; Tachu, B.; Kleta, S.; Blikslager, A.; et al. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 2006, 125, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramirez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A python library for gene ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Luo, Z.; Luo, W.; Li, S.; Zhao, S.; Sho, T.; Xu, X.; Zhang, J.; Xu, W.; Xu, J.X. Reactive oxygen species mediated placental oxidative stress, mitochondrial content, and cell cycle progression through mitogen-activated protein kinases in intrauterine growth restricted pigs. Reprod. Biol. 2018, 18, 422–431. [Google Scholar] [CrossRef]
- Huber, K.; Mestres-Arenas, A.; Fajas, L.; Leal-Esteban, L.C. The multifaceted role of cell cycle regulators in the coordination of growth and metabolism. FEBS J. 2020, 288, 3813–3833. [Google Scholar] [CrossRef]
- Marzec, M.; Kasprzycka, M.; Lai, R.; Gladden, A.B.; Wlodarski, P.; Tomczak, E.; Nowell, P.; Deprimo, S.E.; Sadis, S.; Eck, S.; et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood 2006, 108, 1744–1750. [Google Scholar] [CrossRef]
- Nurse, P. Universal control mechanism regulating onset of M-phase. Nature 1990, 344, 503–508. [Google Scholar] [CrossRef]
- Sung, W.W.; Lin, Y.M.; Wu, P.R.; Yen, H.H.; Lai, H.W.; Su, T.C.; Huang, R.H.; Wen, C.K.; Chen, C.Y.; Chen, C.J.; et al. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer 2014, 14, 951. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Hannon, G.J.; Zhang, H.; Casso, D.; Kobayashi, R.; Beach, D. p21 is a universal inhibitor of cyclin kinases. Nature 1993, 366, 701–704. [Google Scholar] [CrossRef]
- Zimmermann, M.; Arachchige-Don, A.S.; Donaldson, M.S.; Dallapiazza, R.F.; Cowan, C.E.; Horne, M.C. Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin. J. Biol. Chem. 2012, 287, 22838–22853. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Hsueh, C.Y.; Guo, Y.; Wu, X.F.; Li, J.Y.; Zhou, L. Lack of miR-1246 in small extracellular vesicle blunts tumorigenesis of laryngeal carcinoma cells by regulating Cyclin G2. IUBMB Life 2020, 72, 1491–1503. [Google Scholar] [CrossRef]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef] [Green Version]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Bomzon, A.; Holt, S.; Moore, K. Bile acids, oxidative stress, and renal function in biliary obstruction. Semin. Nephrol. 1997, 17, 549–562. [Google Scholar]
- Rolo, A.P.; Oliveira, P.J.; Moreno, A.J.M.; Palmeira, C.M. Bile acids affect liver mitochondrial bioenergetics: Possible relevance for cholestasis therapy. Toxicol. Sci. 2000, 57, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Palmeira, C.M.; Rolo, A.P. Mitochondrially-mediated toxicity of bile acids. Toxicology 2004, 203, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrigo, J.; Olguín, H.; Gutierrez, D.; Tacchi, F.; Arrese, M.; Cabrera, D.; Valero-Breton, M.; Elorza, A.A.; Simon, F.; Cabello-Verrugio, C. Bile acids induce alterations in mitochondrial function in skeletal muscle fibers. Antioxidants 2022, 11, 1706. [Google Scholar] [CrossRef] [PubMed]
- Rolo, A.P.; Palmeira, C.M.; Wallace, K.B. Mitochondrially mediated synergistic cell killing by bile acids. BBA-Mol. Basis Dis. 2003, 1637, 127–132. [Google Scholar] [CrossRef] [Green Version]
- Spivey, J.R.; Bronk, S.F.; Gores, G.J. Glycochenodeoxycholate-induced lethal hepatocellular injury in rat hepatocytes. Role of ATP depletion and cytosolic free calcium. J. Clin. Investig. 1993, 92, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidari, R.; Abdoli, N.; Ommati, M.M.; Jamshidzadeh, A.; Niknahad, H. Mitochondrial impairment induced by chenodeoxycholic acid: The protective effect of taurine and carnosine supplementation. Trends Pharm. Sci. 2018, 4, 99–108. [Google Scholar]
- Detloff, S.J.; Khan, N.; Ao, M.; Movva, B.; Nair, T.; Sirajuddin, H.; Domingue, J.; Rao, M.; Sarathy, J. The Yin and Yang of bile acid (BA) action on tight junctions (TJ) in colonic epithelia: A putative role for pro-inflammatory cytokine (PiC). FASEB J. 2016, 30, 1223–1229. [Google Scholar]
- Raimondi, F.; Santoro, P.; Barone, M.V.; Pappacoda, S.; Barretta, M.L.; Nanayakkara, M.; Apicella, C.; Capasso, L.; Paludetto, R. Bile acids modulate tight junction structure and barrier function of Caco-2 monolayers via EGFR activation. Am. J. Physiol.-Gastrointest. Liver Physiol. 2008, 294, G906–G913. [Google Scholar] [CrossRef] [Green Version]
- Kehrein, K.; Bonnefoy, N.; Ott, M. Mitochondrial protein synthesis: Efficiency and accuracy. Antioxid. Redox Sign. 2013, 19, 1928–1939. [Google Scholar] [CrossRef]
- Szczepanowska, J.; Malinska, D.; Wieckowski, M.R.; Duszynski, J. Effect of mtDNA point mutations on cellular bioenergetics. BBA-Bioenerg. 2012, 1817, 1740–1746. [Google Scholar] [CrossRef] [Green Version]
- Lenaz, G.; Baracca, A.; Carelli, V.; D’Aurelio, M.; Sgarbi, G.; Solaini, G. Bioenergetics of mitochondrial diseases associated with mtDNA mutations. BBA-Bioenerg. 2004, 1658, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, G.; Cortopassi, G. Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med. 2015, 88, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.Y.; Xiong, Y.; Han, D.D.; Pi, Y.; Zhang, H.L.; Wang, J.J. N-(3-oxododecanoyl)-l-homoserine lactone disrupts intestinal epithelial barrier through triggering apoptosis and collapsing extracellular matrix and tight junction. J. Cell Physiol. 2021, 236, 5771–5784. [Google Scholar] [CrossRef]
- Yang, Y.; Zong, M.; Xu, W.; Zhang, Y.; Wang, B.; Yang, M.; Tao, L. Natural pyrethrins induces apoptosis in human hepatocyte cells via Bax- and Bcl-2-mediated mitochondrial pathway. Chem. Biol. Interact. 2017, 262, 38–45. [Google Scholar] [CrossRef]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes Cytochrome c—Dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. Bcl-2-regulated apoptosis: Mechanism and therapeutic potential. Curr. Opin. Immunol. 2007, 19, 488–496. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef]
- Lee, J.; You, J.; Lee, G.S.; Hyun, S.H.; Lee, E. Pig Oocytes with a Large Perivitelline Space Matured In Vitro Show Greater Developmental Competence After Parthenogenesis and Somatic Cell Nuclear Transfer. Mol. Reprod. Dev. 2013, 80, 753–762. [Google Scholar] [CrossRef]
- Yin, B.X.; Ren, H.Y.; Cai, H.; Jiang, Y.Q.; Zhao, S.H.; Wang, H. Dynamics of cardiomyocyte and muscle stem cell proliferation in pig. Exp. Cell Res. 2020, 388, 111854. [Google Scholar] [CrossRef]
- Ding, H.X.; Yang, Y.Z.; Wei, S.L.; Spicer, L.J.; Kenez, A.; Xu, W.; Liu, Y.; Feng, T. Influence of N-acetylcysteine on steroidogenesis and gene expression in porcine placental trophoblast cells. Theriogenology 2021, 161, 49–56. [Google Scholar] [CrossRef]
- Zhu, L.H.; Cai, X.; Guo, Q.; Chen, X.L.; Zhu, S.W.; Xu, J.X. Effect of N-acetyl cysteine on enterocyte apoptosis and intracellular signalling pathways’ response to oxidative stress in weaned piglets. Br. J. Nutr. 2013, 110, 1938–1947. [Google Scholar] [CrossRef] [Green Version]
- Shibutani, M.; Mori, T.; Miyano, T.; Miyake, M. Removal of O-GlcNAcylation is important for pig preimplantation development. J. Reprod. Dev. 2015, 61, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Wei, Z.X.; Zhao, X.M.; Li, Y.P.; Li, X.L.; Jiang, X.R. Gallic acid mitigates LPS-induced inflammatory response via suppressing NF-kappa B signalling pathway in IPEC-J2 cells. J. Anim. Physiol. Anim. Nutr. 2022, 106, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Wu, Z.H.; Li, T.T.; Liu, C.; Han, D.D.; Tao, S.Y.; Pi, Y.; Li, N.; Wang, J.J. Perturbation of the lipid metabolism and intestinal inflammation in growing pigs with low birth weight is associated with the alterations of gut microbiota. Sci. Total Environ. 2020, 719, 137382. [Google Scholar] [CrossRef] [PubMed]
- Radtke, J.; Geissler, S.; Schutkowski, A.; Brandsch, C.; Kluge, H.; Duranti, M.M.; Keller, S.; Jahreis, G.; Hirche, F.; Stangl, G.I. Lupin protein isolate versus casein modifies cholesterol excretion and mRNA expression of intestinal sterol transporters in a pig model. Nutr. Metab. 2014, 11, 1–11. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, B.L.; Yu, C.N.; Li, J.L.; Zhang, L.; Sun, H.; Gao, F.; Zhou, G.H. L-Glutamate supplementation improves small intestinal architecture and enhances the expressions of jejunal mucosa amino acid receptors and transporters in weaning piglets. PLoS ONE 2014, 9, e111950. [Google Scholar] [CrossRef] [Green Version]
- WWeller, M.M.D.C.A.; Alebrante, L.; Campos, P.H.R.F.; Saraiva, A.; Silva, B.A.N.; Donzele, J.L.; Oliveira, R.F.M.; Silva, F.F.; Gasparino, E.; Lopes, P.S.; et al. Effect of heat stress and feeding phosphorus levels on pig electron transport chain gene expression. Animal 2013, 7, 1985–1993. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Song, H.; Gao, G.; Cai, D.; Yang, X.; Zhao, R. Maternal betaine supplementation during gestation enhances expression of mtDNA-encoded genes through D-loop DNA hypomethylation in the skeletal muscle of newborn piglets. J. Agric. Food Chem. 2015, 63, 10152–10160. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Li, Y.; Wei, Z.; Bai, R.; Gao, G.; Sun, W.; Jiang, X.; Wang, J.; Li, X.; Pi, Y. Chenodeoxycholic Acid (CDCA) Promoted Intestinal Epithelial Cell Proliferation by Regulating Cell Cycle Progression and Mitochondrial Biogenesis in IPEC-J2 Cells. Antioxidants 2022, 11, 2285. https://doi.org/10.3390/antiox11112285
Xu L, Li Y, Wei Z, Bai R, Gao G, Sun W, Jiang X, Wang J, Li X, Pi Y. Chenodeoxycholic Acid (CDCA) Promoted Intestinal Epithelial Cell Proliferation by Regulating Cell Cycle Progression and Mitochondrial Biogenesis in IPEC-J2 Cells. Antioxidants. 2022; 11(11):2285. https://doi.org/10.3390/antiox11112285
Chicago/Turabian StyleXu, Lei, Yanpin Li, Zixi Wei, Rong Bai, Ge Gao, Wenjuan Sun, Xianren Jiang, Junjun Wang, Xilong Li, and Yu Pi. 2022. "Chenodeoxycholic Acid (CDCA) Promoted Intestinal Epithelial Cell Proliferation by Regulating Cell Cycle Progression and Mitochondrial Biogenesis in IPEC-J2 Cells" Antioxidants 11, no. 11: 2285. https://doi.org/10.3390/antiox11112285





