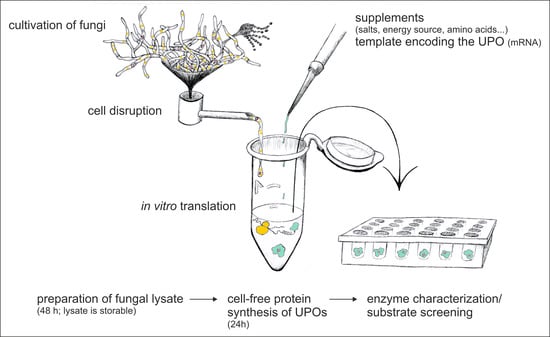
Cell-Free Protein Synthesis with Fungal Lysates for the Rapid Production of Unspecific Peroxygenases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Lysate Preparation
2.1.1. Cultivation
2.1.2. Preparation of Cell Lysates
2.2. Cell-Free Protein Synthesis
2.2.1. Preparation of mRNA Templates
2.2.2. Cell-Free Protein Synthesis
2.3. Analytics
2.3.1. Enzymatic Assays
2.3.2. High-Resolution Mass Spectrometry
2.3.3. Western Blot
2.3.4. Deglycosylation
3. Results and Discussion
3.1. Lysate Preparation from Neurospora crassa and Aspergillus niger
3.2. Cell-Free Protein Synthesis of AaeUPO
3.3. Substrate Screening with Cell-Free AaeUPO
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic synthesis for industrial applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-J.; Pan, J.; Ma, B.-D.; Xu, J.-H. Efficient biocatalytic synthesis of chiral chemicals. In Bioreactor Engineering Research and Industrial Applications I; Ye, Q., Bao, J., Zhong, J.-J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 155, pp. 55–106. [Google Scholar]
- Sheldon, R.A.; Brady, D.; Bode, M.L. The Hitchhiker’s guide to biocatalysis: Recent advances in the use of enzymes in organic synthesis. Chem. Sci. 2020, 11, 2587–2605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofrichter, M.; Kellner, H.; Pecyna, M.J.; Ullrich, R. Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. In Monooxygenase, Peroxidase and Peroxygenase Properties and Mechanisms of Cytochrome P450; Hrycay, E.G., Bandiera, S.M., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; Volume 851, pp. 341–368. [Google Scholar]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Kiebist, J.; Scheibner, K.; Ullrich, R. Peroxide-mediated oxygenation of organic compounds by fungal peroxygenases. Antioxidants 2022, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lan, D.; Durrani, R.; Hollmann, F. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Sigmund, M.-C.; Poelarends, G.J. Current state and future perspectives of engineered and artificial peroxygenases for the oxyfunctionalization of organic molecules. Nat. Catal. 2020, 3, 690–702. [Google Scholar] [CrossRef]
- Kiebist, J.; Hofrichter, M.; Zuhse, R.; Scheibner, K. Oxyfunctionalization of pharmaceuticals by fungal peroxygenases. In Pharmaceutical Biocatalysis, 1st ed.; Grunwald, P., Ed.; Jenny Stanford Publishing: Singapore, 2019; pp. 643–680. [Google Scholar]
- Hobisch, M.; Holtmann, D.; Gomez de Santos, P.; Alcalde, M.; Hollmann, F.; Kara, S. Recent developments in the use of peroxygenases—Exploring their high potential in selective oxyfunctionalisations. Biotechnol. Adv. 2021, 51, 107615. [Google Scholar] [CrossRef]
- Kiebist, J.; Holla, W.; Heidrich, J.; Poraj-Kobielska, M.; Sandvoss, M.; Simonis, R.; Gröbe, G.; Atzrodt, J.; Hofrichter, M.; Scheibner, K. One-pot synthesis of human metabolites of SAR548304 by fungal peroxygenases. Bioorg. Med. Chem. 2015, 23, 4324–4332. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecht, S.; Kiebist, J.; König, R.; Thiessen, M.; Schmidtke, K.-U.; Kammerer, S.; Küpper, J.-H.; Scheibner, K. Synthesis of cyclophosphamide metabolites by a peroxygenase from Marasmius rotula for toxicological studies on human cancer cells. AMB Express 2020, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Gomez de Santos, P.; Cañellas, M.; Tieves, F.; Younes, S.H.H.; Molina-Espeja, P.; Hofrichter, M.; Hollmann, F.; Guallar, V.; Alcalde, M. Selective synthesis of the human drug metabolite 5′-hydroxypropranolol by an evolved self-sufficient peroxygenase. ACS Catal. 2018, 8, 4789–4799. [Google Scholar] [CrossRef] [Green Version]
- Kiebist, J.; Schmidtke, K.-U.; Zimmermann, J.; Kellner, H.; Jehmlich, N.; Ullrich, R.; Zänder, D.; Hofrichter, M.; Scheibner, K. A peroxygenase from Chaetomium globosum catalyzes the selective oxygenation of testosterone. Chembiochem 2017, 18, 563–569. [Google Scholar] [CrossRef] [Green Version]
- Ullrich, R.; Poraj-Kobielska, M.; Scholze, S.; Halbout, C.; Sandvoss, M.; Pecyna, M.J.; Scheibner, K.; Hofrichter, M. Side chain removal from corticosteroids by unspecific peroxygenase. J. Inorg. Biochem. 2018, 183, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Kiebist, J.; Schmidtke, K.-U.; Schramm, M.; König, R.; Quint, S.; Kohlmann, J.; Zuhse, R.; Ullrich, R.; Hofrichter, M.; Scheibner, K. Biocatalytic syntheses of antiplatelet metabolites of the thienopyridines clopidogrel and prasugrel using fungal peroxygenases. J. Fungi 2021, 7, 752. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, R.; Karich, A.; Hofrichter, M. Fungal peroxygenases—A versatile tool for biocatalysis. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 260–280. [Google Scholar]
- Ullrich, R.; Nüske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anh, D.H.; Ullrich, R.; Benndorf, D.; Svatoś, A.; Muck, A.; Hofrichter, M. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 2007, 73, 5477–5485. [Google Scholar] [CrossRef] [Green Version]
- Gröbe, G.; Ullrich, R.; Pecyna, M.J.; Kapturska, D.; Friedrich, S.; Hofrichter, M.; Scheibner, K. High-yield production of aromatic peroxygenase by the agaric fungus Marasmius rotula. AMB Express 2011, 1, 31. [Google Scholar] [CrossRef] [Green Version]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Kimani, V.W.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 369–403. [Google Scholar]
- Kinner, A.; Rosenthal, K.; Lütz, S. Identification and expression of new unspecific peroxygenases—Recent advances, challenges and opportunities. Front. Bioeng. Biotechnol. 2021, 9, 705630. [Google Scholar] [CrossRef]
- Rotilio, L.; Swoboda, A.; Ebner, K.; Rinnofner, C.; Glieder, A.; Kroutil, W.; Mattevi, A. Structural and biochemical studies enlighten the unspecific peroxygenase from Hypoxylon sp. EC38 as an efficient oxidative biocatalyst. ACS Catal. 2021, 11, 11511–11525. [Google Scholar] [CrossRef]
- Miyazaki-Imamura, C.; Oohira, K.; Kitagawa, R.; Nakano, H.; Yamane, T.; Takahashi, H. Improvement of H2O2 stability of manganese peroxidase by combinatorial mutagenesis and high-throughput screening using in vitro expression with protein disulfide isomerase. Protein Eng. Des. Sel. 2003, 16, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Ninomiya, R.; Zhu, B.; Kojima, T.; Iwasaki, Y.; Nakano, H. Role of disulfide bond isomerase DsbC, calcium ions, and hemin in cell-free protein synthesis of active manganese peroxidase isolated from Phanerochaete chrysosporium. J. Biosci. Bioeng. 2014, 117, 652–657. [Google Scholar] [CrossRef]
- Kwon, Y.-C.; Oh, I.-S.; Lee, N.; Lee, K.-H.; Yoon, Y.J.; Lee, E.Y.; Kim, B.-G.; Kim, D.-M. Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng. 2013, 110, 1193–1200. [Google Scholar] [CrossRef]
- Katayama, Y.; Shimokata, K.; Suematsu, M.; Ogura, T.; Tsukihara, T.; Yoshikawa, S.; Shimada, H. Cell-free synthesis of cytochrome c oxidase, a multicomponent membrane protein. J. Bioenerg. Biomembr. 2010, 42, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Mizoguchi, T.; Kojima, T.; Nakano, H. Ultra-high-throughput screening of an in vitro-synthesized horseradish peroxidase displayed on microbeads using cell sorter. PLoS ONE 2015, 10, e0127479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolf, J.; Rosenthal, K.; Lütz, S. Application of cell-free protein synthesis for faster biocatalyst development. Catalysts 2019, 9, 190. [Google Scholar] [CrossRef] [Green Version]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A user’s guide to cell-free protein synthesis. Methods Protoc. 2019, 2, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczin-Skorupa, E.; Filipowicz, W.; Paszewski, A. The cell-free protein synthesis system from the ‘slime’ mutant of Neurospora crassa. Preparation and characterisation of importance of 7-methylguanosine for translation of viral and cellular mRNAs. Eur. J. Biochem. 1981, 121, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Sachs, M.S. Arginine-specific regulation mediated by the Neurospora crassa arg-2 upstream open reading frame in a homologous, cell-free in vitro translation system. J. Biol. Chem. 1997, 272, 255–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Dasgupta, A.; Shen, L.; Bell-Pedersen, D.; Sachs, M.S. The cell free protein synthesis system from the model filamentous fungus Neurospora crassa. Methods 2018, 137, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Devchand, M.; Gwynne, D.; Buxton, F.P.; Davies, R.W. An efficient cell-free translation system from Aspergillus nidulans and in vitro translocation of prepro-a-factor across Aspergillus microsomes. Curr. Genet. 1988, 14, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Nieland, S.; Stahmann, K.-P. A developmental stage of hyphal cells shows riboflavin overproduction instead of sporulation in Ashbya gossypii. Appl. Microbiol. Biotechnol. 2013, 97, 10143–10153. [Google Scholar] [CrossRef] [PubMed]
- Nieland, S.; Barig, S.; Salzmann, J.; Gehrau, F.; Zamani, A.I.; Richter, A.; Ibrahim, J.; Gräser, Y.; Ng, C.L.; Stahmann, K.P. Aspergillus fumigatus AR04 obeys Arrhenius’ rule in cultivation temperature shifts from 30 to 40 °C. Microb. Biotechnol. 2021, 14, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Hodgman, C.E.; Jewett, M.C. Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis: Improved Yeast CFPS. Biotechnol. Bioeng. 2013, 110, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.; Jewett, M.C. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthe. Biotechnol. J. 2014, 9, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Pecyna, M.J.; Ullrich, R.; Bittner, B.; Clemens, A.; Scheibner, K.; Schubert, R.; Hofrichter, M. Molecular characterization of aromatic peroxygenase from Agrocybe aegerita. Appl. Microbiol. Biotechnol. 2009, 84, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Kinne, M.; Poraj-Kobielska, M.; Aranda, E.; Ullrich, R.; Hammel, K.E.; Scheibner, K.; Hofrichter, M. Regioselective preparation of 5-hydroxypropranolol and 4′-hydroxydiclofenac with a fungal peroxygenase. Bioorg. Med. Chem. Lett. 2009, 19, 3085–3087. [Google Scholar] [CrossRef]
- Thoring, L.; Dondapati, S.K.; Stech, M.; Wüstenhagen, D.A.; Kubick, S. High-yield production of “difficult-to-express” proteins in a continuous exchange cell-free system based on CHO cell lysates. Sci. Rep. 2017, 7, 11710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichty, J.J.; Malecki, J.L.; Agnew, H.D.; Michelson-Horowitz, D.J.; Tan, S. Comparison of affinity tags for protein purification. Protein Expr. Purif. 2005, 41, 98–105. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Hodgman, C.E.; Anderson, M.J.; Jewett, M.C. Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J. 2014, 9, 630–640. [Google Scholar] [CrossRef]
- Molina-Espeja, P.; Garcia-Ruiz, E.; Gonzalez-Perez, D.; Ullrich, R.; Hofrichter, M.; Alcalde, M. Directed evolution of unspecific peroxygenase from Agrocybe aegerita. Appl. Environ. Microbiol. 2014, 80, 3496–3507. [Google Scholar] [CrossRef] [Green Version]
- Molina-Espeja, P.; Ma, S.; Mate, D.M.; Ludwig, R.; Alcalde, M. Tandem-yeast expression system for engineering and producing unspecific peroxygenase. Enzyme Microb. Technol. 2015, 73–74, 29–33. [Google Scholar] [CrossRef]
- Miguez, A.M.; McNerney, M.P.; Styczynski, M.P. Metabolic profiling of Escherichia coli -based cell-free expression systems for process optimization. Ind. Eng. Chem. Res. 2019, 58, 22472–22482. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, O.; Koch, M.; Zettor, A.; Pandi, A.; Batista, A.C.; Soudier, P.; Faulon, J.-L. Large scale active-learning-guided exploration for in vitro protein production optimization. Nat. Commun. 2020, 11, 1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgman, C.E.; Jewett, M.C. Characterizing IGR IRES-mediated translation initiation for use in yeast cell-free protein synthesis. New Biotechnol. 2014, 31, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Le Fourn, V.; Girod, P.-A.; Buceta, M.; Regamey, A.; Mermod, N. CHO cell engineering to prevent polypeptide aggregation and improve therapeutic protein secretion. Metab. Eng. 2014, 21, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Quast, R.B.; Sachse, R.; Schulze, C.; Wüstenhagen, D.A.; Kubick, S. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS ONE 2014, 9, e96635. [Google Scholar] [CrossRef] [Green Version]
- Krainer, F.W.; Capone, S.; Jäger, M.; Vogl, T.; Gerstmann, M.; Glieder, A.; Herwig, C.; Spadiut, O. Optimizing cofactor availability for the production of recombinant heme peroxidase in Pichia pastoris. Microb. Cell Fact. 2015, 14, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poraj-Kobielska, M.; Atzrodt, J.; Holla, W.; Sandvoss, M.; Grobe, G.; Scheibner, K.; Hofrichter, M. Preparation of labeled human drug metabolites and drug-drug interaction-probes with fungal peroxygenases. J. Label. Compd. Rad. 2013, 56, 513–519. [Google Scholar] [CrossRef]
- Makky, E.A.; AlMatar, M.; Mahmood, M.H.; Ting, O.W.; Qi, W.Z. Evaluation of the antioxidant and antimicrobial activities of ethyl acetate extract of Saccharomyces cerevisiae. Food Technol. Biotechnol. 2021, 59, 127–136. [Google Scholar] [CrossRef]
- Bormann, S.; Burek, B.O.; Ulber, R.; Holtmann, D. Immobilization of unspecific peroxygenase expressed in Pichia pastoris by metal affinity binding. Mol. Catal. 2020, 492, 110999. [Google Scholar] [CrossRef]
- Pullmann, P.; Knorrscheidt, A.; Munch, J.; Palme, P.R.; Hoehenwarter, W.; Marillonnet, S.; Alcalde, M.; Westermann, B.; Weissenborn, M.J. A modular two yeast species secretion system for the production and preparative application of unspecific peroxygenases. Commun. Biol. 2021, 4, 562. [Google Scholar] [CrossRef]





| Origin of AaeUPO | Homologous Expression (wtAaeUPO) | Heterologous Expression 1 (rAaeUPO) | Cell-Free Expression (cfAaeUPO) |
|---|---|---|---|
| Activity (veratryl alcohol U L−1) | 1550 | <1 | 105 |
| Production time | 10 days | 6 days | 24 h |
| Reference | [17] | [44] | present study |
| wtAaeUPO | cfAaeUPO | |
|---|---|---|
| ABTS (a) | 414 ± 35 | 229 ± 24 |
| 5-Nitrobenzodioxole (a) | 90 ± 12 | 93 ± 8 |
| Naphthalene (a) | 209 ± 14 | 212 ± 22 |
| Propranolol (b) | 7505 ± 103 | 9972 ± 69 |
| Diclofenac (b) | 10,701 ± 73 | 4875 ± 134 |
| Clopidogrel (b) | 8949 ± 496 | 2616 ± 86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schramm, M.; Friedrich, S.; Schmidtke, K.-U.; Kiebist, J.; Panzer, P.; Kellner, H.; Ullrich, R.; Hofrichter, M.; Scheibner, K. Cell-Free Protein Synthesis with Fungal Lysates for the Rapid Production of Unspecific Peroxygenases. Antioxidants 2022, 11, 284. https://doi.org/10.3390/antiox11020284
Schramm M, Friedrich S, Schmidtke K-U, Kiebist J, Panzer P, Kellner H, Ullrich R, Hofrichter M, Scheibner K. Cell-Free Protein Synthesis with Fungal Lysates for the Rapid Production of Unspecific Peroxygenases. Antioxidants. 2022; 11(2):284. https://doi.org/10.3390/antiox11020284
Chicago/Turabian StyleSchramm, Marina, Stephanie Friedrich, Kai-Uwe Schmidtke, Jan Kiebist, Paul Panzer, Harald Kellner, René Ullrich, Martin Hofrichter, and Katrin Scheibner. 2022. "Cell-Free Protein Synthesis with Fungal Lysates for the Rapid Production of Unspecific Peroxygenases" Antioxidants 11, no. 2: 284. https://doi.org/10.3390/antiox11020284