Exploiting Interdata Relationships in Prostate Cancer Proteomes: Clinical Significance of HO-1 Interactors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Treatments, Reagents, and Antibodies
2.2. Antibodies
2.3. PEBG-GST-HO-1 Cloning
2.4. Transfection with PEBG-GST and PEBG-GST-HO1
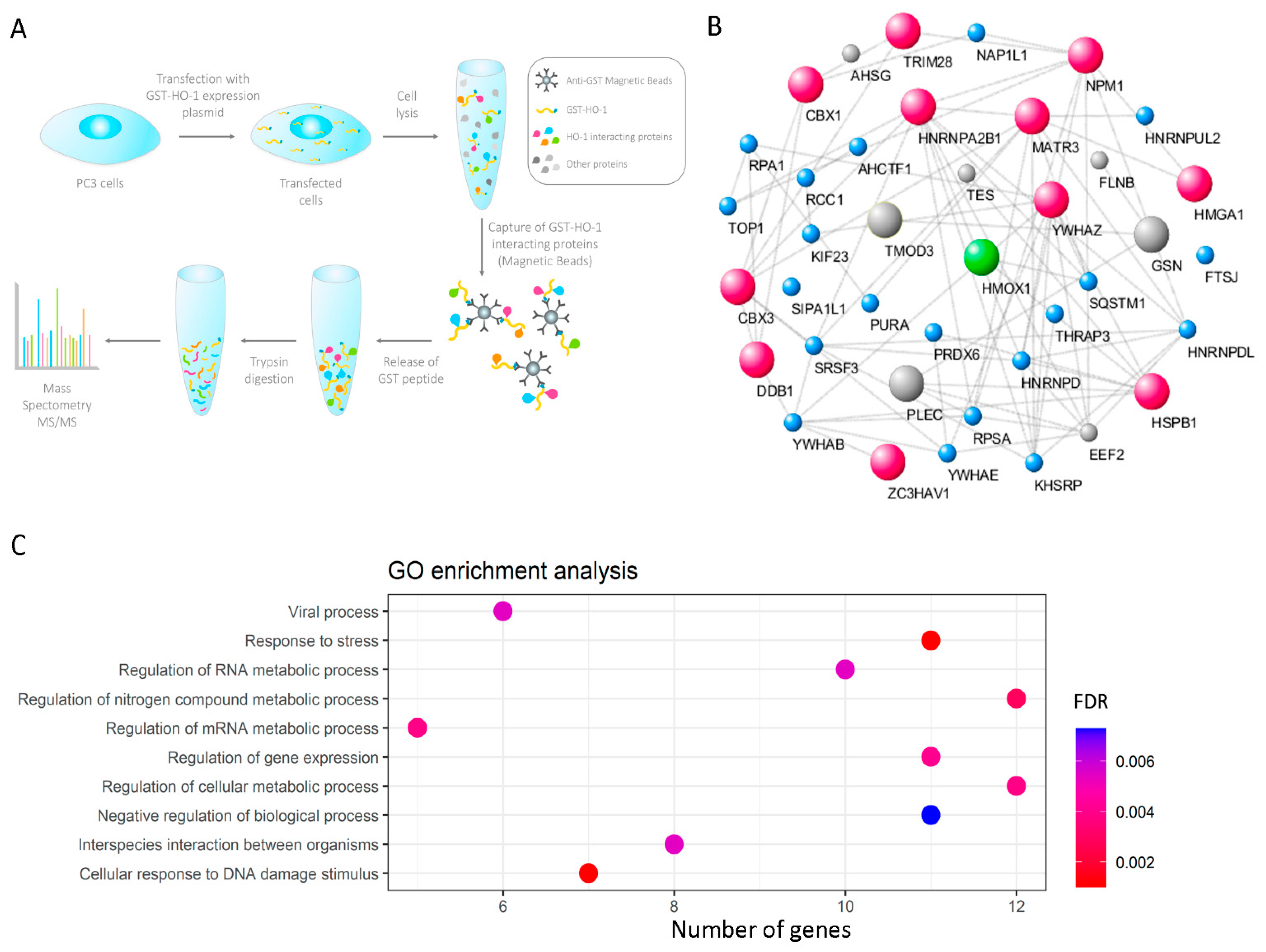
2.5. GST Immunoprecipitation Strategy
2.6. Separation of Peptides and Mass Spectrometry Analysis
2.7. Co-Immunoprecipitation (Co-IP)
2.8. Immunofluorescence (IF) Experiment
2.9. Image Processing for Presentation
2.10. Bioinformatics Analysis
2.10.1. Identification of HO-1 Interactor Proteins with Nuclear Localization and GO Enrichment Analysis
2.10.2. Information Source and Eligibility Criteria (GEO: Gene Expression Omnibus)
2.10.3. Gene Correlation
2.10.4. Risk Scoring System Analysis
2.10.5. Survival Analyses
3. Results
3.1. Proteomics Profile of HO-1 Interactors in PCa Cells
3.2. Gene Correlation between HMOX1 and the Genes Encoding for HO-1 Interactors with Nuclear Localization in PCa Cells
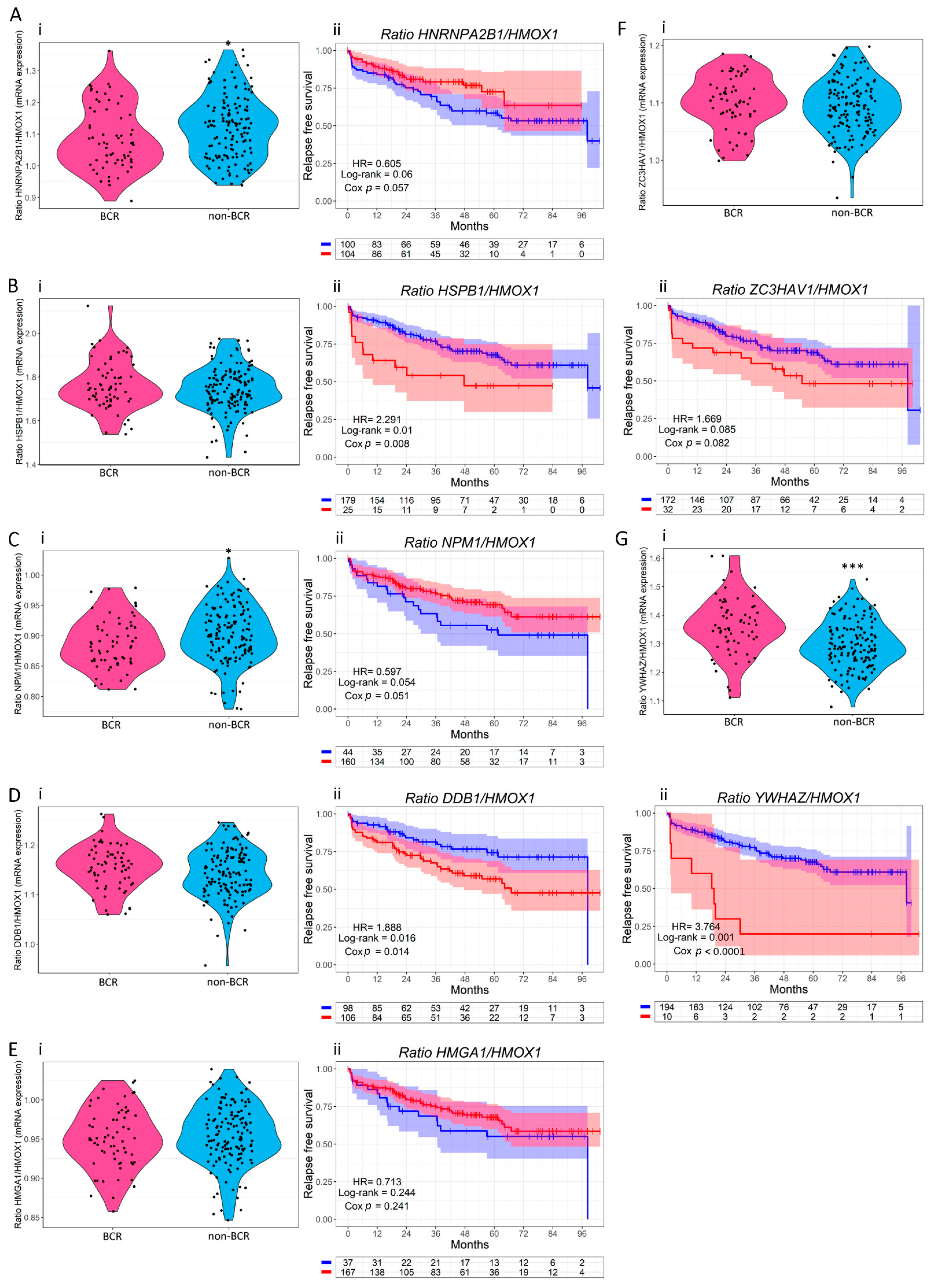
3.3. Clinical Relevance of HO-1 Interactors with Nuclear Localization in PCa
3.4. Validation of the Interaction between HO-1 and 14-3-3ζ/δ in PCa Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, H.; Wartenberg, M.; Hescheler, J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell. Physiol. Biochem. 2001, 11, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.M.; Papa, N.; Ta, A.D.; Millar, J.; Davidson, A.-J.; Pedersen, J.; Syme, R.; Patel, M.I.; Giles, G.G. Predictors of prostate cancer specific mortality after radical prostatectomy: 10 year oncologic outcomes from the Victorian Radical Prostatectomy Registry. BJU Int. 2015, 116, 66–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Marzo, A.M.; Platz, E.A.; Sutcliffe, S.; Xu, J.; Gronberg, H.; Drake, C.G.; Nakai, Y.; Isaacs, W.B.; Nelson, W.G. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 2007, 7, 256–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labanca, E.; De Luca, P.; Gueron, G.; Paez, A.; Moiola, C.P.; Massillo, C.; Porretti, J.; Giudice, J.; Zalazar, F.; Navone, N.; et al. Association of HO-1 and BRCA1 Is Critical for the Maintenance of Cellular Homeostasis in Prostate Cancer. Mol. Cancer Res. 2015, 13, 1455–1464. [Google Scholar] [CrossRef] [Green Version]
- Gueron, G.; De Siervi, A.; Vazquez, E. Advanced prostate cancer: Reinforcing the strings between inflammation and the metastatic behavior. Prostate Cancer Prostatic Dis. 2012, 15, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Jozkowicz, A.; Was, H.; Dulak, J. Heme oxygenase-1 in tumors: Is it a false friend? Antioxid. Redox Signal. 2007, 9, 2099–2117. [Google Scholar] [CrossRef] [Green Version]
- Gueron, G.; De Siervi, A.; Ferrando, M.; Salierno, M.; De Luca, P.; Elguero, B.; Meiss, R.; Navone, N.; Vazquez, E.S. Critical role of endogenous heme oxygenase 1 as a tuner of the invasive potential of prostate cancer cells. Mol. Cancer Res. 2009, 7, 1745–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrando, M.; Gueron, G.; Elguero, B.; Giudice, J.; Salles, A.; Leskow, F.C.; Jares-Erijman, E.A.; Colombo, L.; Meiss, R.; Navone, N.; et al. Heme oxygenase 1 (HO-1) challenges the angiogenic switch in prostate cancer. Angiogenesis 2011, 14, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Elguero, B.; Gueron, G.; Giudice, J.; Toscani, M.A.; De Luca, P.; Zalazar, F.; Coluccio-Leskow, F.; Meiss, R.; Navone, N.; De Siervi, A.; et al. Unveiling the association of STAT3 and HO-1 in prostate cancer: Role beyond heme degradation. Neoplasia 2012, 14, 1043–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gueron, G.; Giudice, J.; Valacco, P.; Paez, A.; Elguero, B.; Toscani, M.; Jaworski, F.; Leskow, F.C.; Cotignola, J.; Marti, M.; et al. Heme-oxygenase-1 implications in cell morphology and the adhesive behavior of prostate cancer cells. Oncotarget 2014, 5, 4087–4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaworski, F.M.; Gentilini, L.D.; Gueron, G.; Meiss, R.P.; Ortiz, E.G.; Berguer, P.M.; Ahmed, A.; Navone, N.; Rabinovich, G.A.; Compagno, D.; et al. In Vivo Hemin Conditioning Targets the Vascular and Immunologic Compartments and Restrains Prostate Tumor Development. Clin. Cancer Res. 2017, 23, 5135–5148. [Google Scholar] [CrossRef] [Green Version]
- Ferrando, M.; Wan, X.; Meiss, R.; Yang, J.; De Siervi, A.; Navone, N.; Vazquez, E. Heme oxygenase-1 (HO-1) expression in prostate cancer cells modulates the oxidative response in bone cells. PLoS ONE 2013, 8, e80315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Weis, S.; Yang, G.; Weng, Y.-H.; Helston, R.; Rish, K.; Smith, A.; Bordner, J.; Polte, T.; Gaunitz, F.; et al. Heme oxygenase-1 protein localizes to the nucleus and activates transcription factors important in oxidative stress. J. Biol. Chem. 2007, 282, 20621–20633. [Google Scholar] [CrossRef] [Green Version]
- Sacca, P.; Meiss, R.; Casas, G.; Mazza, O.; Calvo, J.C.; Navone, N.; Vazquez, E. Nuclear translocation of haeme oxygenase-1 is associated to prostate cancer. Br. J. Cancer 2007, 97, 1683–1689. [Google Scholar] [CrossRef]
- Lin, Q.S.; Weis, S.; Yang, G.; Zhuang, T.; Abate, A.; Dennery, P.A. Catalytic inactive heme oxygenase-1 protein regulates its own expression in oxidative stress. Free Radic. Biol. Med. 2008, 44, 847–855. [Google Scholar] [CrossRef]
- Anselmino, N.; Bizzotto, J.; Sanchis, P.; Lage-Vickers, S.; Ortiz, E.; Valacco, P.; Paez, A.; Labanca, E.; Meiss, R.; Navone, N.; et al. HO-1 Interactors Involved in the Colonization of the Bone Niche: Role of ANXA2 in Prostate Cancer Progression. Biomolecules 2020, 10, 467. [Google Scholar] [CrossRef] [Green Version]
- Sacca, P.; Caballero, F.; Batlle, A.; Vazquez, E. Cell cycle arrest and modulation of HO-1 expression induced by acetyl salicylic acid in hepatocarcinogenesis. Int. J. Biochem. Cell Biol. 2004, 36, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Yu, G.; Hu, E. Package ‘Enrichplot’: Visualization of Functional Enrichment Result. 2021. Available online: https://www.bioconductor.org/packages/devel/bioc/manuals/enrichplot/man/enrichplot.pdf (accessed on 10 October 2021).
- Ross-Adams, H.; Lamb, A.; Dunning, M.; Halim, S.; Lindberg, J.; Massie, C.; Egevad, L.; Russell, R.; Ramos-Montoya, A.; Vowler, S.; et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine 2015, 2, 1133–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff Finder: A comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kassambara, A.; Kosinski, M.; Biecek, P. Survminer: Drawing Survival Curves Using “ggplot2”. 2019. Available online: https://cran.r-project.org/web/packages/survminer/survminer.pdf (accessed on 10 October 2021).
- Paez, A.V.; Pallavicini, C.; Schuster, F.; Valacco, M.P.; Giudice, J.; Ortiz, E.G.; Anselmino, N.; Labanca, E.; Binaghi, M.; Salierno, M.; et al. Heme oxygenase-1 in the forefront of a multi-molecular network that governs cell-cell contacts and filopodia-induced zippering in prostate cancer. Cell Death Dis. 2016, 7, e2570. [Google Scholar] [CrossRef]
- Kan, C.; Vargas, G.; Pape, F.L.; Clezardin, P. Cancer Cell Colonisation in the Bone Microenvironment. Int. J. Mol. Sci. 2016, 17, 1674. [Google Scholar] [CrossRef] [Green Version]
- Christensen, M.V.; Høgdall, C.K.; Jochumsen, K.M.; Høgdall, E.V.S. Annexin A2 and cancer: A systematic review. Int. J. Oncol. 2018, 52, 5–18. [Google Scholar] [CrossRef]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Investig. Urol. 1979, 17, 16–23. [Google Scholar]
- Nishimura, K.; Ting, H.-J.; Harada, Y.; Tokizane, T.; Nonomura, N.; Kang, H.-Y.; Chang, H.-C.; Yeh, S.; Miyamoto, H.; Shin, M.; et al. Modulation of androgen receptor transactivation by gelsolin: A newly identified androgen receptor coregulator. Cancer Res. 2003, 63, 4888–4894. [Google Scholar]
- Narain, N.R.; Diers, A.R.; Lee, A.; Lao, S.; Chan, J.Y.; Schofield, S.; Andreazi, J.; Ouro-Djobo, R.; Jimenez, J.J.; Friss, T.; et al. Identification of Filamin-A and -B as potential biomarkers for prostate cancer. Future Sci. OA 2016, 3, FSO161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Cui, L.; Zeng, Y.; Song, W.; Gaur, U.; Yang, M. 14-3-3 Proteins Are on the Crossroads of Cancer, Aging, and Age-Related Neurodegenerative Disease. Int. J. Mol. Sci. 2019, 20, 3518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popiel, A.; Kobierzycki, C.; Dzięgiel, P. The Role of Testin in Human Cancers. Pathol. Oncol. Res. 2019, 25, 1279–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, K.; Zhao, J.C.; Song, B.; Zheng, B.; Yu, J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat. Commun. 2018, 9, 5007. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Vacas, J.M.; Herrero-Aguayo, V.; Montero-Hidalgo, A.J.; Gómez-Gómez, E.; Fuentes-Fayos, A.C.; León-González, A.J.; Sáez-Martínez, P.; Alors-Pérez, E.; Pedraza-Arévalo, S.; González-Serrano, T.; et al. Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer. EBioMedicine 2020, 51, 102547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lage-Vickers, S.; Bizzotto, J.; Valacco, M.P.; Sanchis, P.; Nemirovsky, S.; Labanca, E.; Scorticati, C.; Mazza, O.; Mitrofanova, A.; Navone, N.; et al. The expression of YWHAZ and NDRG1 predicts aggressive outcome in human prostate cancer. Commun. Biol. 2021, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.A.; Dodson, A.R.; Parsons, K.F.; Desmond, A.D.; Woolfenden, A.; Fordham, M.; Neoptolemos, J.P.; Ke, Y.; Foster, C.S. Heat shock protein expression independently predicts clinical outcome in prostate cancer. Cancer Res. 2000, 60, 7099–7105. [Google Scholar]
- Foster, C.S.; Dodson, A.R.; Ambroisine, L.; Fisher, G.; Møller, H.; Clark, J.; Attard, G.; De-Bono, J.; Scardino, P.; Reuter, V.E.; et al. Hsp-27 expression at diagnosis predicts poor clinical outcome in prostate cancer independent of ETS-gene rearrangement. Br. J. Cancer 2009, 101, 1137–1144. [Google Scholar] [CrossRef] [Green Version]
- Zoubeidi, A.; Zardan, A.; Beraldi, E.; Fazli, L.; Sowery, R.; Rennie, P.; Nelson, C.; Gleave, M. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res. 2007, 67, 10455–10465. [Google Scholar] [CrossRef] [Green Version]
- Yoon, T.; Chakrabortty, A.; Franks, R.; Valli, T.; Kiyokawa, H.; Raychaudhuri, P. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene 2005, 24, 469–478. [Google Scholar] [CrossRef] [Green Version]
- Alekseev, S.; Kool, H.; Rebel, H.; Fousteri, M.; Moser, J.; Backendorf, C.; de Gruijl, F.R.; Vrieling, H.; Mullenders, L.H.F. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 2005, 65, 10298–10306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.-W.; Su, C.-H.; Chen, H.-H.; Huang, C.-W.; Tsao, L.-P.; Tsao, Y.-P.; Chen, S.-L. DDB2 is a novel AR interacting protein and mediates AR ubiquitination/degradation. Int. J. Biochem. Cell Biol. 2012, 44, 1952–1961. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.K. The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009, 19, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, M.K.; Morrison, D.K. Unlocking the code of 14-3-3. J. Cell Sci. 2004, 117, 1875–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzivion, G.; Gupta, V.S.; Kaplun, L.; Balan, V. 14-3-3 proteins as potential oncogenes. Semin. Cancer Biol. 2006, 16, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Neal, C.L.; Yu, D. 14-3-3ζ as a prognostic marker and therapeutic target for cancer. Expert Opin. Ther. Targets 2010, 14, 1343–1354. [Google Scholar] [CrossRef] [Green Version]
- Matta, A.; Siu, K.W.M.; Ralhan, R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert Opin. Ther. Targets 2012, 16, 515–523. [Google Scholar] [CrossRef]
- Banerjee, S.; Lin, C.-F.L.; Skinner, K.A.; Schiffhauer, L.M.; Peacock, J.; Hicks, D.G.; Redmond, E.M.; Morrow, D.; Huston, A.; Shayne, M.; et al. Heat shock protein 27 differentiates tolerogenic macrophages that may support human breast cancer progression. Cancer Res. 2011, 71, 318–327. [Google Scholar] [CrossRef] [Green Version]
- Barrio, S.; Munawar, U.; Zhu, Y.X.; Giesen, N.; Shi, C.-X.; Da Viá, M.; Sanchez, R.; Bruins, L.; Demler, T.; Müller, N.; et al. IKZF1/3 and CRL4(CRBN) E3 ubiquitin ligase mutations and resistance to immunomodulatory drugs in multiple myeloma. Haematologica 2020, 105, e237–e241. [Google Scholar] [CrossRef] [Green Version]
- Chan, N.; Pires, I.M.; Bencokova, Z.; Coackley, C.; Luoto, K.R.; Bhogal, N.; Lakshman, M.; Gottipati, P.; Oliver, F.J.; Helleday, T.; et al. Contextual Synthetic Lethality of Cancer Cell Kill Based on the Tumor Microenvironment. Cancer Res. 2010, 70, 8045–8054. [Google Scholar] [CrossRef] [Green Version]
- Teyssonneau, D.; Margot, H.; Cabart, M.; Anonnay, M.; Sargos, P.; Vuong, N.-S.; Soubeyran, I.; Sevenet, N.; Roubaud, G. Prostate cancer and PARP inhibitors: Progress and challenges. J. Hematol. Oncol. 2021, 14, 51. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-C.; Li, C.-F.; Chen, I.-H.; Lai, M.-T.; Lin, Z.-J.; Korla, P.K.; Chai, C.-Y.; Ko, G.; Chen, C.-M.; Hwang, T.; et al. YWHAZ amplification/overexpression defines aggressive bladder cancer and contributes to chemo-/radio-resistance by suppressing caspase-mediated apoptosis. J. Pathol. 2019, 248, 476–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Zhang, X.; Liao, Z.; Liang, H.; Chu, L.; Dong, W.; Zhang, X.; Ge, Q.; Liu, Q.; Fan, P.; et al. 14-3-3zeta inhibits heme oxygenase-1 (HO-1) degradation and promotes hepatocellular carcinoma proliferation: Involvement of STAT3 signaling. J. Exp. Clin. Cancer Res. 2019, 38, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aitken, A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin. Cell Dev. Biol. 2011, 22, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.; Wu, D.; Lee, N.; Shibata, E.; Dutta, A. 14-3-3 proteins play a role in the cell cycle by shielding cdt2 from ubiquitin-mediated degradation. Mol. Cell. Biol. 2014, 34, 4049–4061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-H.; Chuang, S.-M.; Yang, M.-F.; Liao, J.-W.; Yu, S.-L.; Chen, J.J.W. A novel function of YWHAZ/beta-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Mol. Cancer Res. 2012, 10, 1319–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.G.; Yang, J.; Vazquez, E.S.; Rose, D.; Vakar-Lopez, F.; Mathew, P.; Lopez, A.; Logothetis, C.J.; Lin, S.-H.; Navone, N.M. Low-density lipoprotein receptor-related protein 5 (LRP5) mediates the prostate cancer-induced formation of new bone. Oncogene 2008, 27, 596–603. [Google Scholar] [CrossRef] [Green Version]



| Gene | Coeff. | [95% CI] | p-Value |
|---|---|---|---|
| HMOX1 | −0.44 | −1.04–0.16 | 0.147 |
| YWHAZ | 1.43 | 0.82–2.04 | <0.001 |
| DDB1 | −0.48 | −1.01–0.05 | 0.08 |
| HSPB1 | −0.86 | −1.47–−0.25 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lage-Vickers, S.; Sanchis, P.; Bizzotto, J.; Toro, A.; Sabater, A.; Lavignolle, R.; Anselmino, N.; Labanca, E.; Paez, A.; Navone, N.; et al. Exploiting Interdata Relationships in Prostate Cancer Proteomes: Clinical Significance of HO-1 Interactors. Antioxidants 2022, 11, 290. https://doi.org/10.3390/antiox11020290
Lage-Vickers S, Sanchis P, Bizzotto J, Toro A, Sabater A, Lavignolle R, Anselmino N, Labanca E, Paez A, Navone N, et al. Exploiting Interdata Relationships in Prostate Cancer Proteomes: Clinical Significance of HO-1 Interactors. Antioxidants. 2022; 11(2):290. https://doi.org/10.3390/antiox11020290
Chicago/Turabian StyleLage-Vickers, Sofia, Pablo Sanchis, Juan Bizzotto, Ayelen Toro, Agustina Sabater, Rosario Lavignolle, Nicolas Anselmino, Estefania Labanca, Alejandra Paez, Nora Navone, and et al. 2022. "Exploiting Interdata Relationships in Prostate Cancer Proteomes: Clinical Significance of HO-1 Interactors" Antioxidants 11, no. 2: 290. https://doi.org/10.3390/antiox11020290








