Functional Characterization of Serotonin N-Acetyltransferase in Archaeon Thermoplasma volcanium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Archaeal GNAT Genes
2.2. Vector Construction and Purification of Recombinant Proteins
2.3. Homology and Phylogenetic Analysis
2.4. SNAT Enzyme Kinetics Measurements
2.5. Transgenic Rice Plants Overexpressing TvSNAT
2.6. Quantification of Melatonin and Radical Scavenging Activity Using the DPPH Method
2.7. Statistical Analyses
3. Results
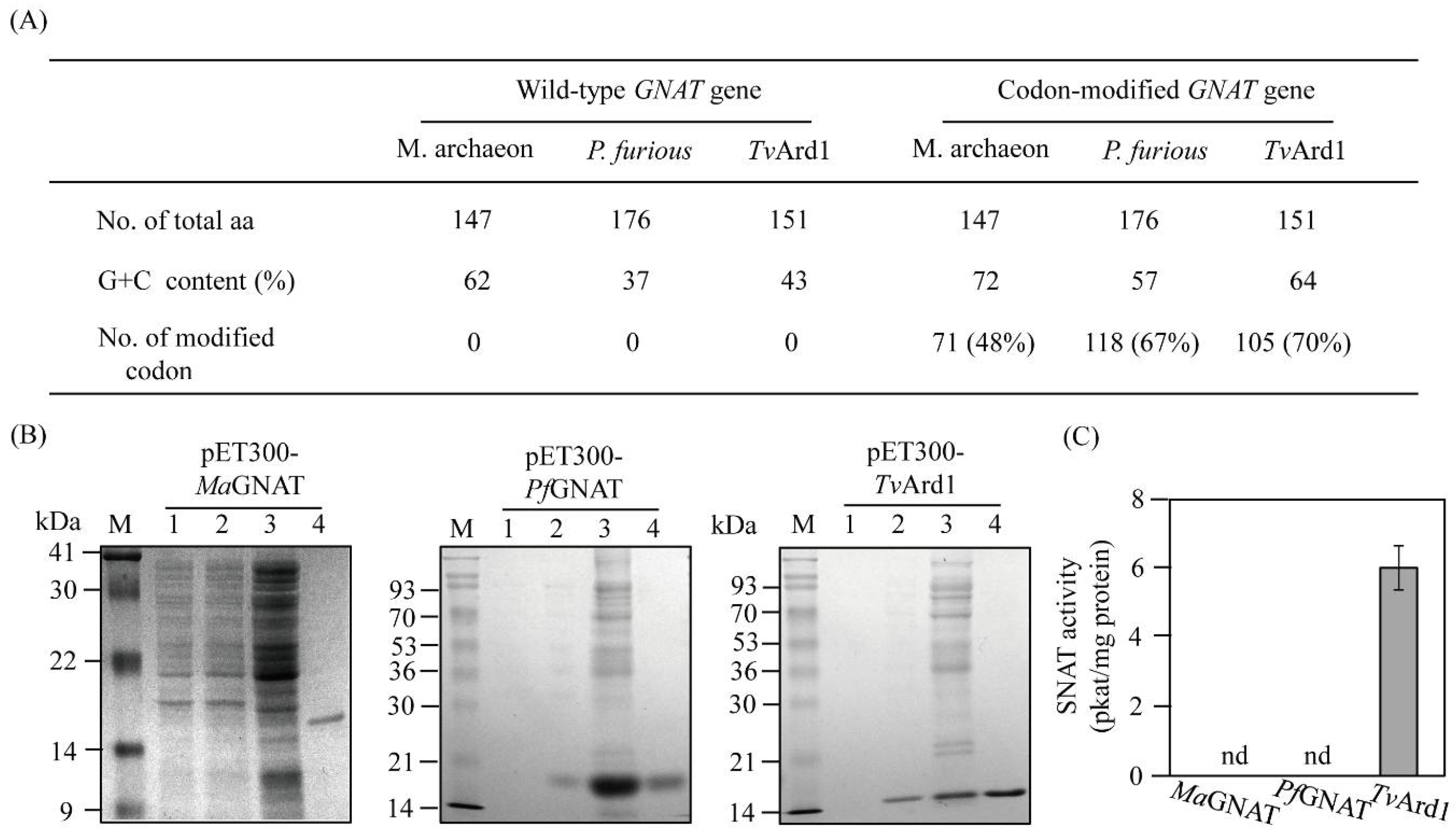
3.1. Codon-Optimized Synthesis of Three Archaeal GNAT Genes
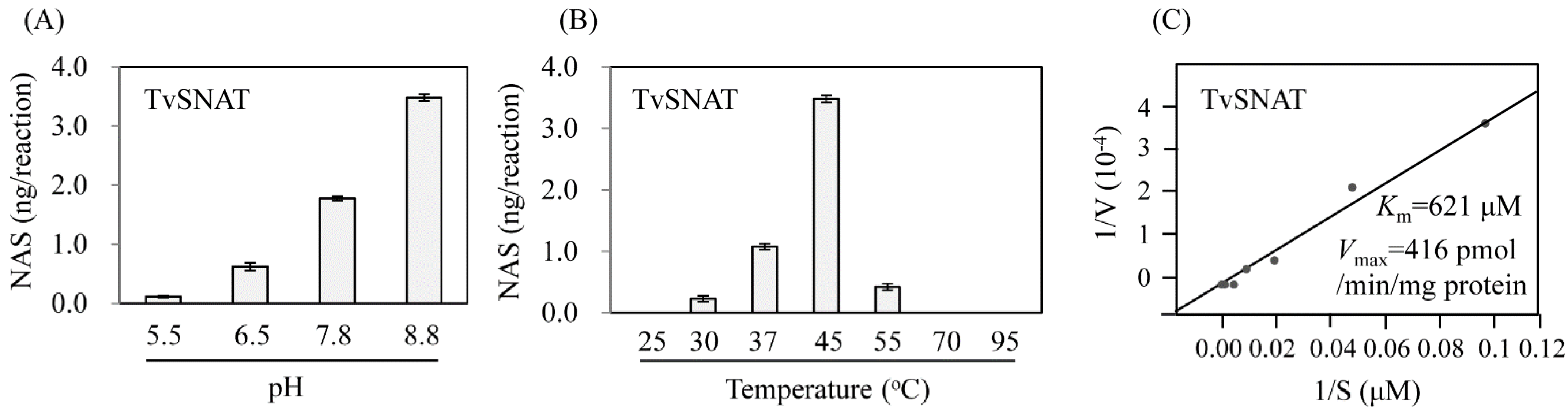
3.2. Enzyme Kinetics of TvSNAT
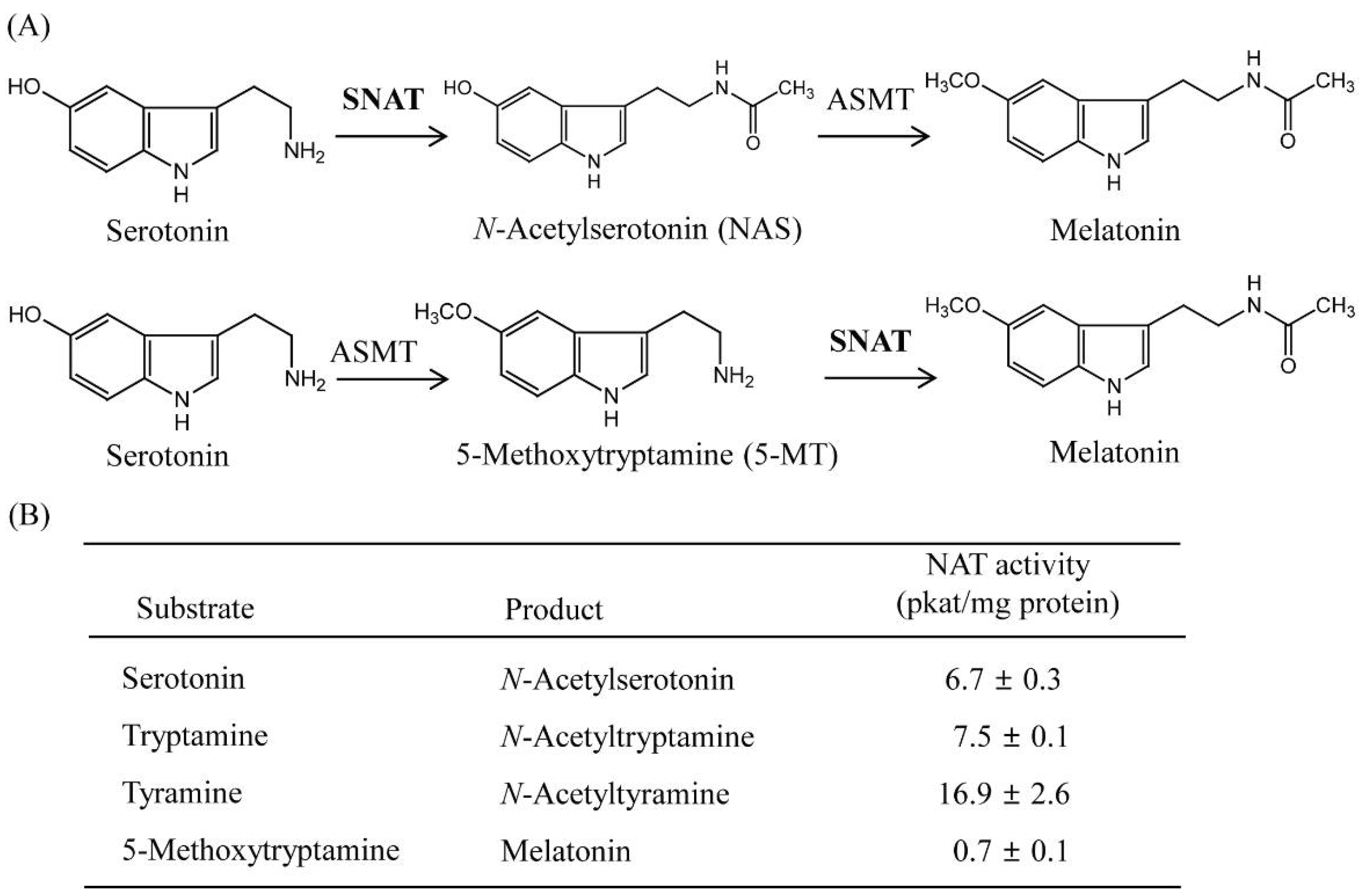
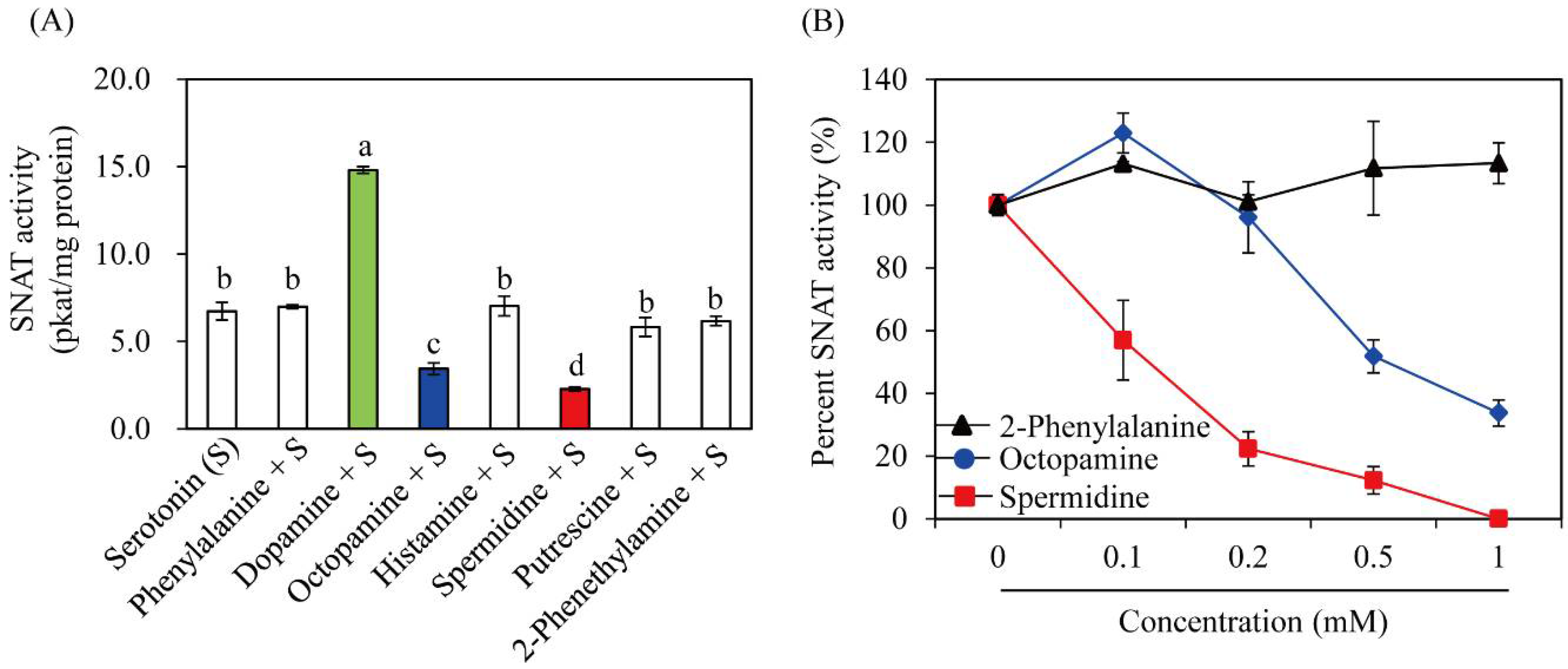
3.3. Substrate Specificity
3.4. Phylogenetic Analysis
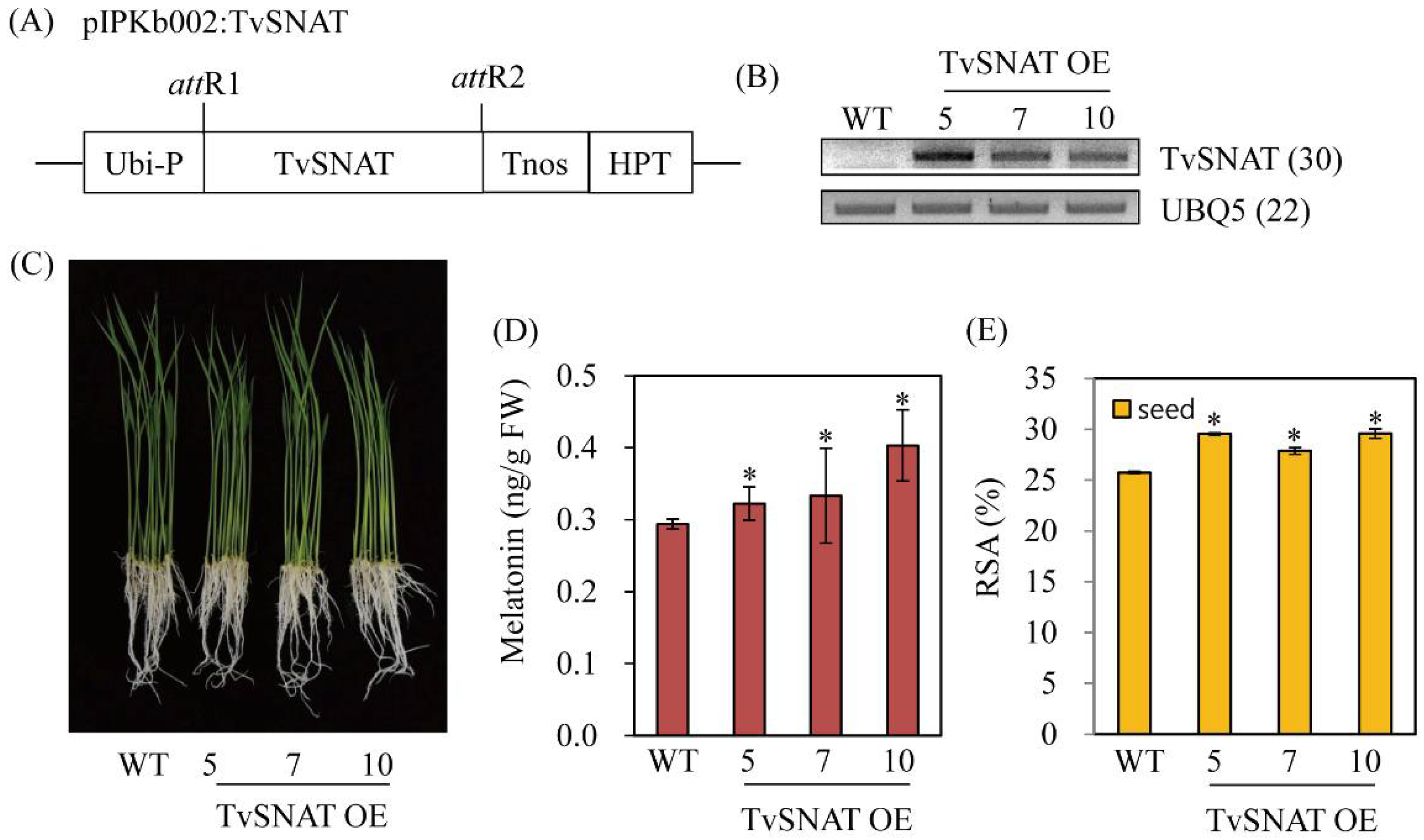
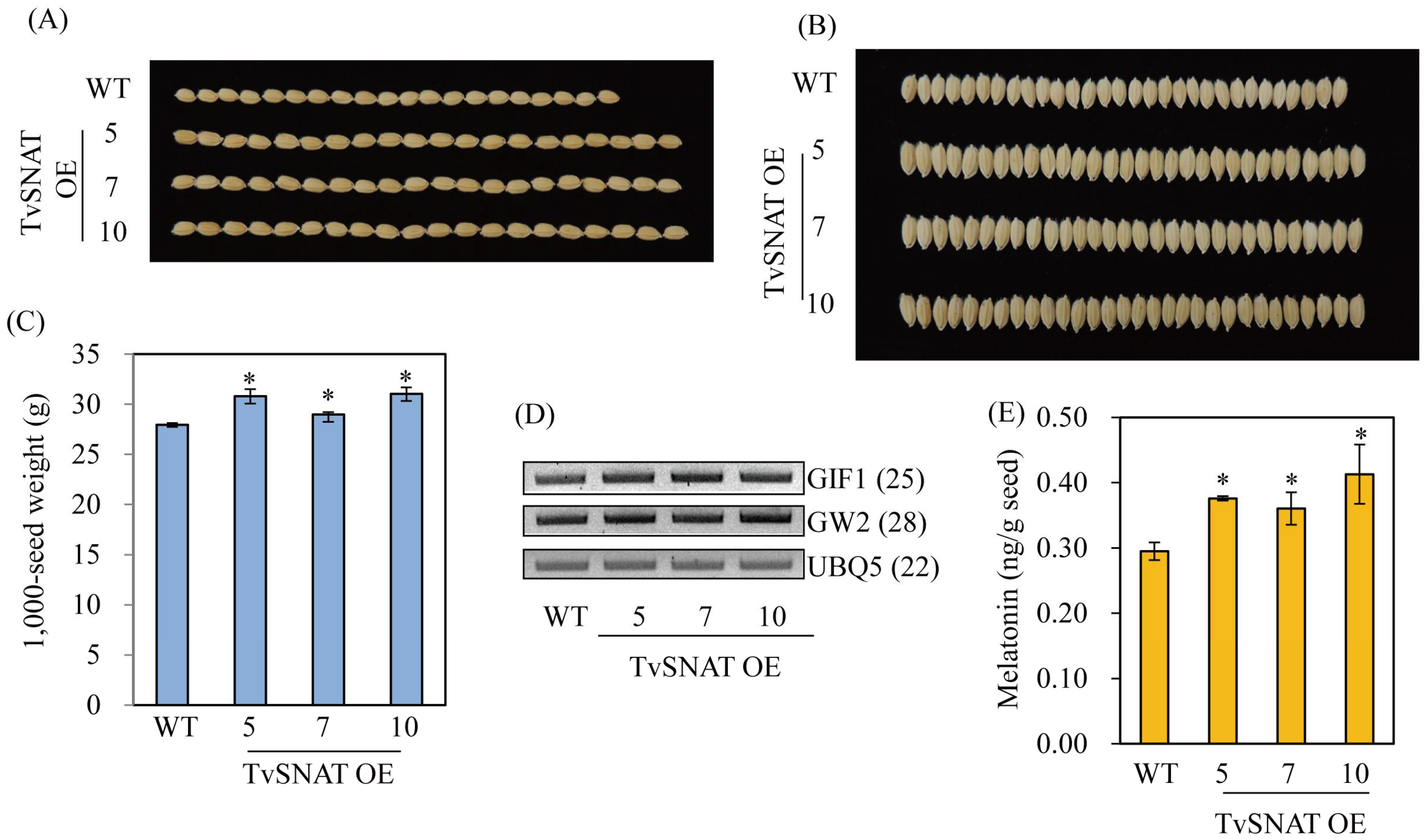
3.5. Characterization of Transgenic Rice Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin synthesis and function: Evolutionary history in animals and plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019, 2, 10–36. [Google Scholar] [CrossRef]
- Reiter, R.J.; Tan, D.X.; Sharma, R. Historical perspective and evaluation of the mechanisms by which melatonin mediates seasonal reproduction in mammals. Melatonin Res. 2018, 1, 59–77. [Google Scholar] [CrossRef]
- Yu, Y.; Lv, Y.; Shi, Y.; Li, T.; Chen, Y.; Zhao, D.; Zhao, Z. The role of phyto-melatonin and related metabolites in response to stress. Molecules 2018, 23, 1887. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Back, K. Melatonin plays a pivotal role in conferring tolerance against endoplasmic reticulum stress via mitogen-activated protein kinases and bZIP60 in Arabidopsis thaliana. Melatonin Res. 2018, 1, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Hwang, O.J.; Back, K. Phytomelatonin as a signaling molecule for protein quality control via chaperone, autophagy, and ubiquitin–proteasome systems in plants. J. Exp. Bot. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.H.; Back, K. Choi GH, Back K. Cyclic 3-hydroxymelatonin exhibits diurnal rhythm and cyclic 3-hydroxymelatonin overproduction increases secondary tillers in rice by upregulating MOC1 expression. Melatonin Res. 2019, 2, 120–138. [Google Scholar] [CrossRef]
- Choi, G.H.; Back, K. Suppression of melatonin 2-hydroxylase increases melatonin production leading to the enhanced abiotic stress tolerance against cadmium, senescence, salt, and tunicamycin in rice plants. Biomolecules 2019, 9, 589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Back, K. 2-Hydroxymelatonin, rather than melatonin, is responsible for RBOH-dependent reactive oxygen species production leading to premature senescence in plants. Antioxidant 2021, 10, 1782. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. The phytomelatonin receptor (PMRT1) Arabidopsis Cand2 is not a bona fide G protein-coupled melatonin receptor. Melatonin Res. 2020, 3, 177–186. [Google Scholar] [CrossRef]
- Tan, D.-X.; Reiter, R.J. Mitochondria: The birth place, battle ground and site of melatonin metabolism in cells. Melatonin Res. 2019, 2, 44–66. [Google Scholar] [CrossRef]
- Back, K. Melatonin metabolism, signaling and possible roles in plants. Plant J. 2021, 105, 376–391. [Google Scholar] [CrossRef]
- Coon, S.L.; Klein, D.C. Evolution of arylalkylamine N-acetyltransferase: Emergence and divergence. Mol. Cell. Endocrinol. 2006, 252, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.X.; Zheng, X.; Kong, J.; Manchester, L.C.; Hardeland, R.; Kim, S.J.; Xu, X.; Reiter, R.J. Fundamental issues related to the origin of melatonin and melatonin isomers during evolution: Relation to their biological functions. Int. J. Mol. Sci. 2014, 15, 15858–15890. [Google Scholar] [CrossRef] [Green Version]
- Dyda, F.; Klein, D.C.; Hickman, A.B. GCN5-related N-acetyltransferases: A structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 81–103. [Google Scholar] [CrossRef]
- Pan, Q.; Zhao, F.-L.; Ye, B.-C. Eis, a novel family of arylalkylamine N-acetyltransferase (EC 2.3.1.87). Sci. Rep. 2018, 8, 2435. [Google Scholar] [CrossRef] [Green Version]
- Byeon, Y.; Lee, K.; Park, Y.I.; Park, S.; Back, K. Molecular cloning and functional analysis of serotonin N-acetyltransferase from the cyanobacterium Synechocystis sp. PCC 6803. J. Pineal Res. 2013, 55, 371–376. [Google Scholar] [CrossRef]
- Ganguly, S.; Mummaneni, P.; Steinbach, P.J.; Klein, D.C.; Coon, S.L. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 2001, 276, 47239–47247. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, J.; Gastel, J.; Klein, D.C.; Cole, P.A. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem. 1998, 273, 3045–3050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferry, G.; Loynel, A.; Kucharczyk, N.; Bertin, S.; Rodriguez, M.; Delagrange, P.; Galizzi, J.-P.; Jacoby, E.; Volland, J.-P.; Lesieur, D.; et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem. 2000, 275, 8794–8805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Lee, K.; Back, K. Knockout of Arabidopsis serotonin N-acetyltransferase-2 reduces melatonin levels and delays flowering. Biomolecules 2019, 9, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Hwang, O.J.; Back, K. Functional characterization of tobacco (Nicotiana benthamiana) serotonin N-acetyltransferases (NbSNAT1 and NbSNAT2). Melatonin Res. 2021, 4, 507–521. [Google Scholar]
- Biarrotte-Sorin, S.; Mayer, C. Cloning, purification, crystallization and preliminary crystallographic analysis of a hypothetical acetyltransferase from Pyrococcus furious. Acta Cryst. 2005, F61, 269–270. [Google Scholar]
- Ma, C.; Pathak, C.; Jang, S.; Lee, S.J.; Nam, M.; Kim, S.J.; Im, H.; Lee, B.J. Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A. Biochim. Biophys. Acta 2014, 1844, 1790–1797. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Himmelbach, A.; Zierold, U.; Hensel, G.; Riechen, J.; Douchkov, D.; Schweizer, P.; Kumlehn, J. A set of modular binary vectors for transformation of cereals. Plant Physiol. 2007, 145, 1192–1200. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Lee, S.B.; Chung, J.S.; Han, S.U.; Han, O.; Guh, J.O.; Jeon, J.S.; An, G.; Back, K. Transgenic rice plants expressing a Bacillus subtilis protoporphyrinogen oxidase gene are resistant to diphenyl ether herbicide oxyfluorfen. Plant Cell Physiol. 2000, 41, 743–749. [Google Scholar] [CrossRef]
- Kang, K.; Kim, Y.S.; Park, S.; Back, K. Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 2009, 150, 1380–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawashima, T.; Amano, N.; Koike, H.; Makino, S.I.; Higuchi, S.; Kawashima-Ohya, T.; Watanabe, K.; Yamazaki, M.; Kanehori, K.; Kawamoto, T.; et al. Archaeal adaptation to higher temperatures reveals by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 2000, 97, 14257–14262. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef]
- Yu, Y.; Bian, L.; Jiao, Z.; Keke, Y.; Wan, Y.; Zhang, G.; Guo, D. Molecular cloning and characterization of a grapevine (Vitis vinifera L.) serotonin N-acetyltransferase (VvSNAT2) gene involved in plant defense. BMC Genom. 2019, 20, 880. [Google Scholar] [CrossRef] [PubMed]
- Falcón, J.; Coon, S.L.; Besseau, L.; Cazamea-Catalan, D.; Fuentes, M.; Magnanou, E.; Paulin, C.H.; Boeuf, G.; Sauzet, S.; Jorgensen, E.H.; et al. Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya. Proc. Natl. Acad. Sci. USA 2014, 111, 314–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Celano, P.; Mank, A.R.; Pegg, A.E.; Casero, R.A., Jr. Characterization of a full-length cDNA which codes for the human spermidine/spermine N1-acetyltransferase. Biochem. Biophys. Res. Commun. 1991, 179, 407–415. [Google Scholar] [CrossRef]
- Zilberman-Peled, B.; Bransburg-Zabary, S.; Klein, D.C.; Gothilf, Y. Molecular evolution of multiple arylalkylamine N-acetyltransferase (AANAT) in fish. Mar. Drugs 2011, 9, 906–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, O.J.; Back, K. Melatonin deficiency confers tolerance to multiple abiotic stresses in rice via decreased brassinosteroid levels. Int. J. Mol. Sci. 2019, 20, 5173. [Google Scholar] [CrossRef] [Green Version]
- Arnesen, T.; Anderson, D.; Torsvik, J.; Halseth, H.B.; Varhaug, J.E.; Lillehaug, J.R. Cloning and characterization of hNAT5/hSAN: An evolutionarily conserved component of the NatA protein N-α-acetyltransferase complex. Gene 2006, 371, 291–295. [Google Scholar] [CrossRef]
- Linster, E.; Wirtz, M. N-terminal acetylation: An essential protein modification emerges as an important regulator of stress responses. J. Exp. Bot. 2018, 69, 4555–4568. [Google Scholar] [CrossRef]
- McGarry, R.C.; Barron, Y.D.; Carvalho, M.F.; Hill, J.E.; Gold, D.; Cheung, E.; Kraus, W.L.; Lazarowitz, S.G. A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell 2003, 15, 1605–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koskela, M.M.; Brünje, A.; Ivanauskaite, A.; Grabsztunowicz, M.; Lassowskat, I.; Neumann, U.; Dinh, T.V.; Sindlinger, J.; Schwarzer, D.; Wirtz, M.; et al. Chloroplast acetyltransferase NSI is required for state transition in Arabidopsis thaliana. Plant Cell 2018, 30, 1695–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Back, K. Melatonin regulates chloroplast protein quality control via a mitogen-activated protein kinase signaling pathway. Antioxidants 2021, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Byeon, Y.; Tan, D.X.; Reiter, R.J.; Back, K. Arabidopsis serotonin N-acetyltransferase knockout plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 2015, 58, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Back, K. Melatonin induction and its role in high light stress tolerance in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12504. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Back, K. An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. J. Pineal Res. 2014, 56, 408–414. [Google Scholar] [CrossRef]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Janas, K.M.; Posmyk, M.M. Melatonin, an underestimated natural substance with great potential for agricultural application. Acta Physiol. Plant. 2013, 35, 3285–3292. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Kamran, M.; Zhou, X.; Ahmad, I.; Meng, X.; Javed, T.; Iqbal, A.; Wang, G.; Su, W.; Wu, X.; et al. 2021. Melatonin improves the seed filling rate and endogenous hormonal mechanism in grains of summer maize. Physiol. Plant. 2021, 172, 1059–1072. [Google Scholar] [CrossRef]
- Yan, S.; Zou, G.; Li, S.; Wang, H.; Liu, H.; Zhai, G.; Guo, P.; Song, H.; Yan, C.; Tao, Y. Seed size is determined by the combinations of the genes controlling different seed characteristics in rice. Theor. Appl. Genet. 2011, 123, 1173–1181. [Google Scholar] [CrossRef]
- Huang, R.; Jiang, L.; Zheng, J.; Wang, T.; Wang, H.; Huang, Y.; Hong, Z. Genetic bases of rice grain shape: So many genes, so little known. Trends Plant Sci. 2013, 18, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623630. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Cheng, M.L.; Hsing, Y.C.; Chen, Y.S.; Lee, K.W.; Hong, Y.F.; Hsiao, Y.; Hsiao, A.S.; Chen, P.J.; Wong, L.I.; et al. Rice Big Grain 1 promotes cell division to enhance organ development, stress tolerance and grain yield. Plant Biotechnol. J. 2020, 18, 1969–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Xu, L.; Su, T.; Jiang, Y.; Hu, L.; Ma, F. Melatonin regulates carbohydrate metabolism and defenses against Pseudomonas syringae pv. tomato DC3000 infection in Arabidopsis thaliana. J. Pineal Res. 2015, 59, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef]
- Bai, Y.; Xiao, S.; Zhang, Z.; Zhang, Y.; Sun, H.; Zhang, K.; Wang, X.; Bai, Z.; Li, C.; Liu, L. Melatonin improves the germination rate of cotton seeds under drought stress by opening pores in the seed coat. Peer J. 2020, 8, e9450. [Google Scholar] [CrossRef]
- Li, D.; Batchelor, W.D.; Zhang, D.; Miao, H.; Li, H.; Song, S.; Li, R. Analysis of melatonin regulation of germination and antioxidant metabolism in different wheat cultivars under polyethylene glycol stress. PLoS ONE 2020, 15, e0237536. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Choi, G.-H.; Back, K. Functional Characterization of Serotonin N-Acetyltransferase in Archaeon Thermoplasma volcanium. Antioxidants 2022, 11, 596. https://doi.org/10.3390/antiox11030596
Lee K, Choi G-H, Back K. Functional Characterization of Serotonin N-Acetyltransferase in Archaeon Thermoplasma volcanium. Antioxidants. 2022; 11(3):596. https://doi.org/10.3390/antiox11030596
Chicago/Turabian StyleLee, Kyungjin, Geun-Hee Choi, and Kyoungwhan Back. 2022. "Functional Characterization of Serotonin N-Acetyltransferase in Archaeon Thermoplasma volcanium" Antioxidants 11, no. 3: 596. https://doi.org/10.3390/antiox11030596






