A Genotype–Phenotype Analysis of Glutathione Peroxidase 4 in Human Atrial Myocardium and Its Association with Postoperative Atrial Fibrillation
Abstract
:1. Introduction
2. Materials and Methods
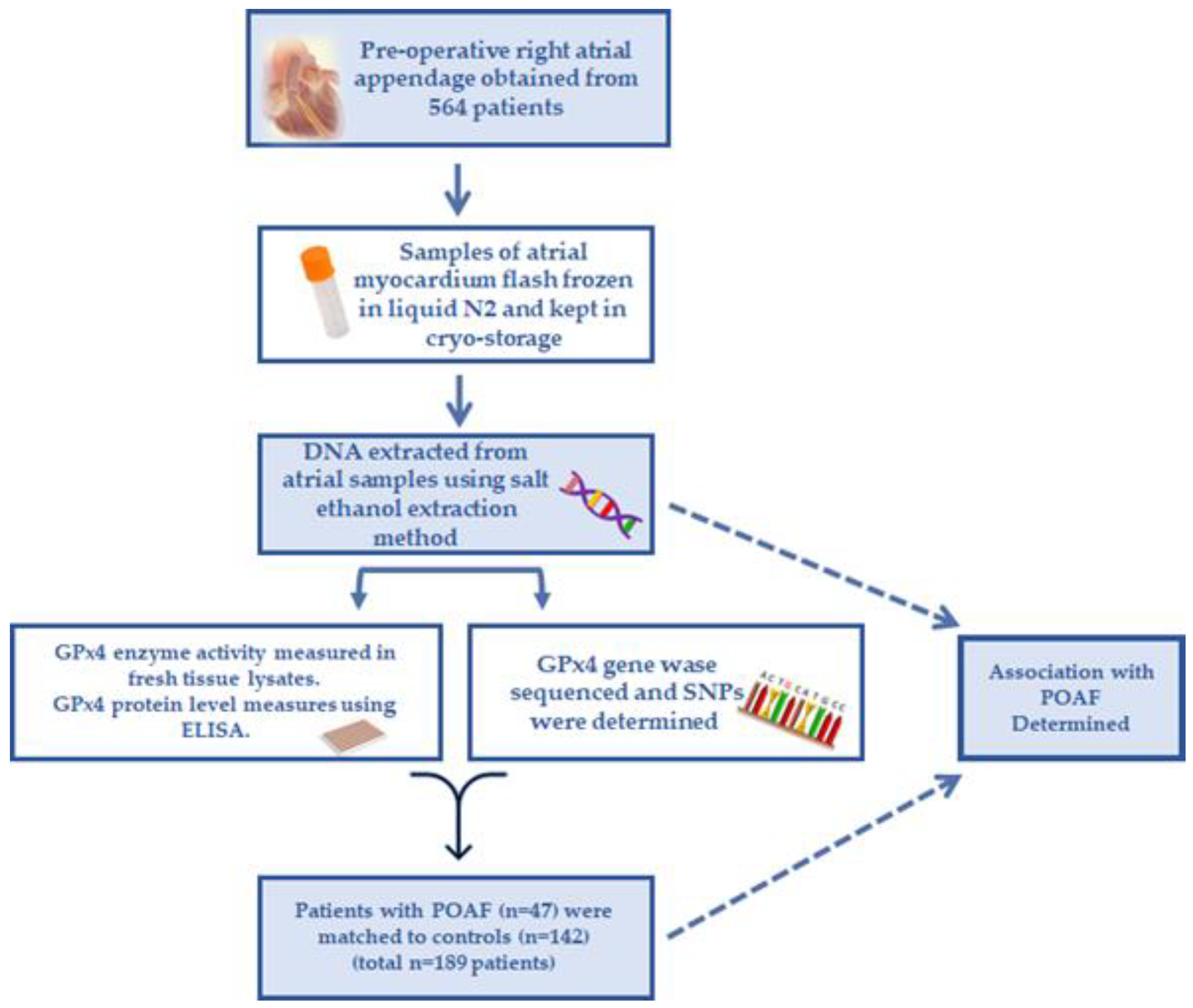
2.1. Study Design and Tissues Used for Analysis
2.2. Analyzing Genotype–Phenotype and POAF Risk
2.3. DNA Extraction and Storage
2.4. Myocardial Protein Preparation
2.5. Myocardial GPx4 Activity Analysis
2.6. Myocardial GPx4 Enzyme Quantitation
2.7. DNA Sequencing
2.8. Statistical Analysis
3. Results
3.1. Patient Characteristics and Incidence of POAF
3.2. Variants of GPX4 and POAF Risk
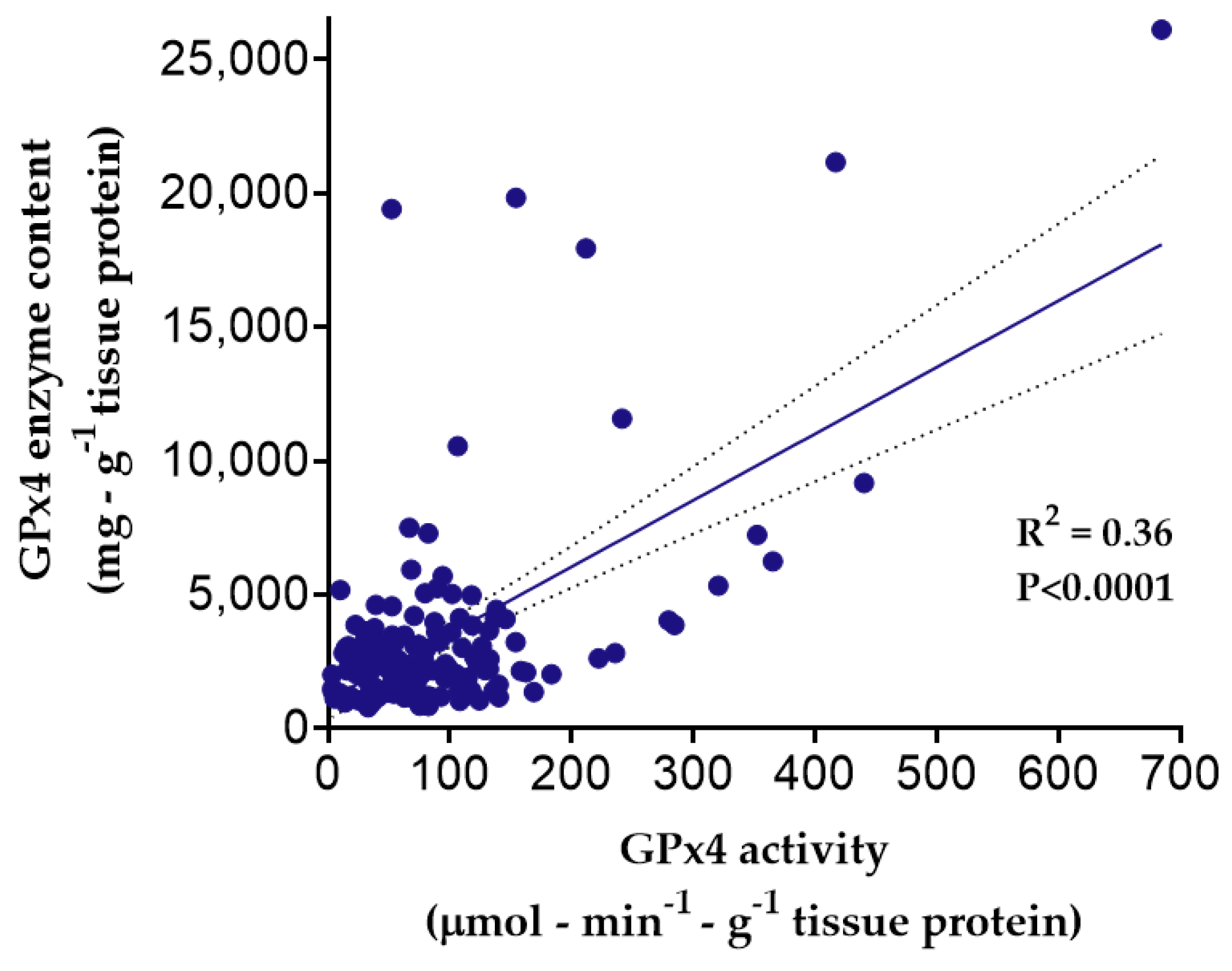
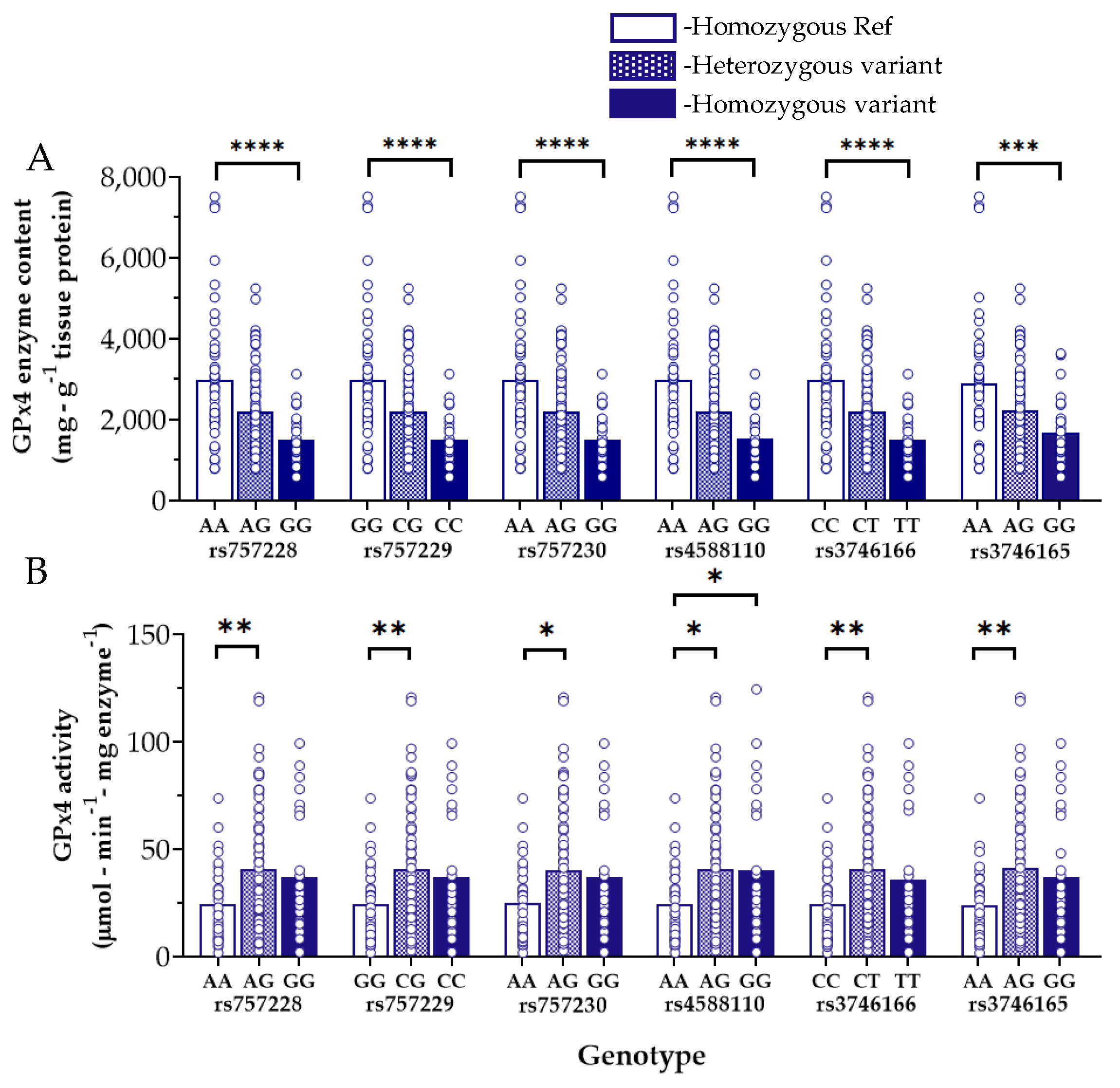
3.3. Myocardial GPx4 Content and Activity and Influence of GPX4 Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kornej, J.; Börschel, C.S.; Benjamin, E.J.; Schnabel, R.B. Epidemiology of Atrial Fibrillation in the 21st Century. Circ. Res. 2020, 127, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Zakkar, M.; Ascione, R.; James, A.F.; Angelini, G.D.; Suleiman, M.S. Inflammation, oxidative stress and postoperative atrial fibrillation in cardiac surgery. Pharmacol. Ther. 2015, 154, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmoneim, S.S.; Rosenberg, E.; Meykler, M.; Patel, B.; Reddy, B.; Ho, J.; Klem, I.; Singh, J.; Worku, B.; Tranbaugh, R.F.; et al. The Incidence and Natural Progression of New-Onset Postoperative Atrial Fibrillation. JACC Clin. Electrophysiol. 2021, 7, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Higgs, M.; Sim, J.; Traynor, V. Incidence and risk factors for new-onset atrial fibrillation following coronary artery bypass grafting: A systematic review and meta-analysis. Intensive Crit. Care Nurs. 2020, 60, 102897. [Google Scholar] [CrossRef]
- Qureshi, M.; Ahmed, A.; Massie, V.; Marshall, E.; Harky, A. Determinants of atrial fibrillation after cardiac surgery. Rev. Cardiovasc. Med. 2021, 22, 329–341. [Google Scholar] [CrossRef]
- Dobrev, D.; Aguilar, M.; Heijman, J.; Guichard, J.-B.; Nattel, S. Postoperative atrial fibrillation: Mechanisms, manifestations and management. Nat. Rev. Cardiol. 2019, 16, 1. [Google Scholar] [CrossRef]
- Karam, B.S.; Chavez-Moreno, A.; Koh, W.; Akar, J.G.; Akar, F.G. Oxidative stress and inflammation as central mediators of atrial fibrillation in obesity and diabetes. Cardiovasc. Diabetol. 2017, 16, 120. [Google Scholar] [CrossRef]
- Park, T.-J.; Park, J.H.; Lee, G.S.; Lee, J.-Y.; Shin, J.H.; Kim, M.W.; Kim, Y.S.; Kim, J.-Y.; Oh, K.-J.; Han, B.-S.; et al. Quantitative proteomic analyses reveal that GPX4 downregulation during myocardial infarction contributes to ferroptosis in cardiomyocytes. Cell Death Dis. 2019, 10, 835. [Google Scholar] [CrossRef] [Green Version]
- Jeganathan, J.; Saraf, R.; Mahmood, F.; Pal, A.; Bhasin, M.K.; Huang, T.; Mittel, A.; Knio, Z.; Simons, R.; Khabbaz, K.; et al. Mitochondrial Dysfunction in Atrial Tissue of Patients Developing Postoperative Atrial Fibrillation. Ann. Thorac. Surg. 2017, 104, 1547–1555. [Google Scholar] [CrossRef] [Green Version]
- Emelyanova, L.; Ashary, Z.; Cosic, M.; Negmadjanov, U.; Ross, G.; Rizvi, F.; Olet, S.; Kress, D.; Sra, J.; Tajik, A.J.; et al. Selective downregulation of mitochondrial electron transport chain activity and increased oxidative stress in human atrial fibrillation. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, H54–H63. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Kattach, H.; Ratnatunga, C.; Pillai, R.; Channon, K.M.; Casadei, B. Association of Atrial Nicotinamide Adenine Dinucleotide Phosphate Oxidase Activity with the Development of Atrial Fibrillation after Cardiac Surgery. J. Am. Coll. Cardiol. 2008, 51, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, E.J.; Efird, J.T.; Davies, S.W.; O’Neal, W.T.; Darden, T.M.; Thayne, K.A.; Katunga, L.A.; Kindell, L.C.; Ferguson, T.B.; Anderson, C.A.; et al. Monoamine Oxidase is a Major Determinant of Redox Balance in Human Atrial Myocardium and Is Associated with Postoperative Atrial Fibrillation. J. Am. Heart Assoc. 2014, 3, e000713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshwal, S.; Di Sante, M.; Di Lisa, F.; Kaludercic, N. Emerging role of monoamine oxidase as a therapeutic target for cardiovascular disease. Curr. Opin. Pharmacol. 2017, 33, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.P.; Geiger, P.G.; Maiorino, M.; Ursini, F.; Girotti, A.W. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim. Biophys. Acta 1990, 1045, 252–260. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.-L.; Huang, Z.-J.; Lin, Z.-T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef]
- Seiler, A.; Schneider, M.; Förster, H.; Roth, S.; Wirth, E.K.; Culmsee, C.; Plesnila, N.; Kremmer, E.; Rådmark, O.; Wurst, W.; et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008, 8, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Porto Freitas, F.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422.e21. [Google Scholar] [CrossRef] [Green Version]
- Sunde, R.A. Selenium regulation of selenoprotein enzyme activity and transcripts in a pilot study with Founder strains from the Collaborative Cross. PLoS ONE 2018, 13, e0191449. [Google Scholar] [CrossRef]
- Sneddon, A.A.; Wu, H.C.; Farquharson, A.; Grant, I.; Arthur, J.R.; Rotondo, D.; Choe, S.N.; Wahle, K.W. Regulation of selenoprotein GPx4 expression and activity in human endothelial cells by fatty acids, cytokines and antioxidants. Atherosclerosis 2003, 171, 57–65. [Google Scholar] [CrossRef]
- Xie, B.; Guo, Y. Molecular mechanism of cell ferroptosis and research progress in regulation of ferroptosis by noncoding RNAs in tumor cells. Cell Death Discov. 2021, 7, 101. [Google Scholar] [CrossRef]
- Wei, X.; Yi, X.; Zhu, X.-H.; Jiang, D.-S. Posttranslational Modifications in Ferroptosis. Oxidative Med. Cell. Longev. 2020, 2020, 8832043. [Google Scholar] [CrossRef] [PubMed]
- Reinke, E.N.; Ekoue, D.N.; Bera, S.; Mahmud, N.; Diamond, A.M. Translational regulation of GPx-1 and GPx-4 by the mTOR pathway. PLoS ONE 2014, 9, e93472. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, X.; Yang, Q.; Chen, J.; Huang, Q.; Yao, L.; Yan, D.; Wu, J.; Zhang, P.; Tang, D.; et al. Broad Spectrum Deubiquitinase Inhibition Induces Both Apoptosis and Ferroptosis in Cancer Cells. Front. Oncol. 2020, 10, 949. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Friedmann Angeli, J.P. Glutathione peroxidase 4 (Gpx4) and ferroptosis: What’s so special about it? Mol. Cell. Oncol. 2015, 2, e995047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockwell, B.R.; Jiang, X.; Gu, W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol. 2020, 30, 478–490. [Google Scholar] [CrossRef]
- Ursini, F.; Maiorino, M. Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic. Biol. Med. 2020, 152, 175–185. [Google Scholar] [CrossRef]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef]
- Crosley, L.K.; Bashir, S.; Nicol, F.; Arthur, J.R.; Hesketh, J.E.; Sneddon, A.A. The single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene influences endothelial cell function: Interaction with selenium and fatty acids. Mol. Nutr. Food Res. 2013, 57, 2185–2194. [Google Scholar] [CrossRef] [Green Version]
- Polonikov, A.V.; Vialykh, E.K.; Churnosov, M.I.; Illig, T.; Freidin, M.B.; Vasil’eva, O.V.; Bushueva, O.Y.; Ryzhaeva, V.N.; Bulgakova, I.V.; Solodilova, M.A. The C718T polymorphism in the 3′-untranslated region of glutathione peroxidase-4 gene is a predictor of cerebral stroke in patients with essential hypertension. Hypertens. Res. 2012, 35, 507–512. [Google Scholar] [CrossRef]
- Strauss, E.; Tomczak, J.; Staniszewski, R.; Oszkinis, G. Associations and interactions between variants in selenoprotein genes, selenoprotein levels and the development of abdominal aortic aneurysm, peripheral arterial disease, and heart failure. PLoS ONE 2018, 13, e0203350. [Google Scholar] [CrossRef]
- Admoni, S.N.; Santos-Bezerra, D.P.; Perez, R.V.; Patente, T.A.; Monteiro, M.B.; Cavaleiro, A.M.; Parisi, M.C.; Moura Neto, A.; Pavin, E.J.; Queiroz, M.S.; et al. Glutathione peroxidase 4 functional variant rs713041 modulates the risk for cardiovascular autonomic neuropathy in individuals with type 1 diabetes. Diabetes Vasc. Dis. Res. 2019, 16, 297–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosby, A.J.; Wahle, K.W.; Duthie, G.G. Modulation of glutathione peroxidase activity in human vascular endothelial cells by fatty acids and the cytokine interleukin-1 beta. Biochim. Biophys. Acta 1996, 1303, 187–192. [Google Scholar] [CrossRef]
- Anderson, E.J.; Kypson, A.P.; Rodriguez, E.; Anderson, C.A.; Lehr, E.J.; Neufer, P.D. Substrate-specific derangements in mitochondrial metabolism and redox balance in the atrium of the type 2 diabetic human heart. J. Am. Coll. Cardiol. 2009, 54, 1891–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, E.J.; Rodriguez, E.; Anderson, C.A.; Thayne, K.; Chitwood, W.R.; Kypson, A.P. Increased propensity for cell death in diabetic human heart is mediated by mitochondrial-dependent pathways. Am. J. Physiol. Heart Circ. Physiol. 2011, 300, H118–H124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamchandani, K.; Khanna, A.K.; Bose, S.; Fernando, R.J.; Walkey, A.J. Atrial Fibrillation: Current Evidence and Management Strategies during the Perioperative Period. Anesth. Analg. 2020, 130, 2–13. [Google Scholar] [CrossRef]
- Nelson, M.M.; Builta, Z.J.; Monroe, T.B.; Doorn, J.A.; Anderson, E.J. Biochemical characterization of the catecholaldehyde reactivity of L-carnosine and its therapeutic potential in human myocardium. Amino Acids 2019, 51, 97–102. [Google Scholar] [CrossRef]
- Nelson, M.M.; Efird, J.T.; Kew, K.A.; Katunga, L.A.; Monroe, T.B.; Doorn, J.A.; Beatty, C.N.; Shi, Q.; Akhter, S.A.; Alwair, H.; et al. Enhanced Catecholamine Flux and Impaired Carbonyl Metabolism Disrupt Cardiac Mitochondrial Oxidative Phosphorylation in Diabetes Patients. Antioxid. Redox Signal. 2021, 35, 235–251. [Google Scholar] [CrossRef]
- Stolwijk, J.M.; Falls-Hubert, K.C.; Searby, C.C.; Wagner, B.A.; Buettner, G.R. Simultaneous detection of the enzyme activities of GPx1 and GPx4 guide optimization of selenium in cell biological experiments. Redox Biol. 2020, 32, 101518. [Google Scholar] [CrossRef]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef] [Green Version]
- Roselli, C.; Chaffin, M.D.; Weng, L.C.; Aeschbacher, S.; Ahlberg, G.; Albert, C.M.; Almgren, P.; Alonso, A.; Anderson, C.D.; Aragam, K.G.; et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018, 50, 1225–1233. [Google Scholar] [CrossRef]
- Shiroshita-Takeshita, A.; Schram, G.; Lavoie, J.; Nattel, S. Effect of simvastatin and antioxidant vitamins on atrial fibrillation promotion by atrial-tachycardia remodeling in dogs. Circulation 2004, 110, 2313–2319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korantzopoulos, P.; Letsas, K.; Fragakis, N.; Tse, G.; Liu, T. Oxidative stress and atrial fibrillation: An update. Free Radic. Res. 2018, 52, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Shiroshita-Takeshita, A.; Brundel, B.J.J.M.; Burstein, B.; Leung, T.-K.; Mitamura, H.; Ogawa, S.; Nattel, S. Effects of simvastatin on the development of the atrial fibrillation substrate in dogs with congestive heart failure. Cardiovasc. Res. 2007, 74, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguet, F.; Brown, A.A.; Castel, S.E.; Davis, J.R.; He, Y.; Jo, B.; Mohammadi, P.; Park, Y.; Parsana, P.; Segrè, A.V.; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Kanagaratnam, P.; Kojodjojo, P.; Peters, N.S. Electrophysiological abnormalities occur prior to the development of clinical episodes of atrial fibrillation: Observations from human epicardial mapping. Pacing Clin. Electrophysiol. 2008, 31, 443–453. [Google Scholar] [CrossRef]
- Yadava, M.; Hughey, A.B.; Crawford, T.C. Postoperative atrial fibrillation: Incidence, mechanisms, and clinical correlates. Cardiol. Clin. 2014, 32, 627–636. [Google Scholar] [CrossRef]
- Turagam, M.K.; Mirza, M.; Werner, P.H.; Sra, J.; Kress, D.C.; Tajik, A.J.; Jahangir, A. Circulating Biomarkers Predictive of Postoperative Atrial Fibrillation. Cardiol. Rev. 2016, 24, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Gawałko, M.; Dobrev, D. Surgery-related cardiac stress: A susceptibility test of late atrial fibrillation recurrence? Int. J. Cardiol. Heart Vasc. 2020, 32, 100693. [Google Scholar] [CrossRef]
- Maesen, B.; Nijs, J.; Maessen, J.; Allessie, M.; Schotten, U. Post-operative atrial fibrillation: A maze of mechanisms. Europace 2012, 14, 159–174. [Google Scholar] [CrossRef]
- Nattel, S.; Harada, M. Atrial remodeling and atrial fibrillation: Recent advances and translational perspectives. J. Am. Coll. Cardiol. 2014, 63, 2335–2345. [Google Scholar] [CrossRef] [Green Version]
- Pinho-Gomes, A.C.; Reilly, S.; Brandes, R.P.; Casadei, B. Targeting Inflammation and Oxidative Stress in Atrial Fibrillation: Role of 3-Hydroxy-3-Methylglutaryl-Coenzyme A Reductase Inhibition with Statins. Antioxid. Redox Signal. 2013, 20, 1268–1285. [Google Scholar] [CrossRef] [PubMed]
- Granger, D.N.; Vowinkel, T.; Petnehazy, T. Modulation of the Inflammatory Response in Cardiovascular Disease. Hypertension 2004, 43, 924–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Montero, J.; Brito, R.; Gajardo, A.I.; Rodrigo, R. Myocardial reperfusion injury and oxidative stress: Therapeutic opportunities. World J. Cardiol. 2018, 10, 74–86. [Google Scholar] [CrossRef]
- Semchyshyn, H.M. Reactive Carbonyl SpeciesIn Vivo: Generation and Dual Biological Effects. Sci. World J. 2014, 2014, 417842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, T.; Ding, W.; Ji, X.; Ao, X.; Liu, Y.; Yu, W.; Wang, J. Oxidative Stress in Cell Death and Cardiovascular Diseases. Oxidative Med. Cell. Longev. 2019, 2019, 9030563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.H.Y.; Marchioli, R.; Silletta, M.G.; Masson, S.; Sellke, F.W.; Libby, P.; Milne, G.L.; Brown, N.J.; Lombardi, F.; Damiano, R.J.; et al. Oxidative Stress Biomarkers and Incidence of Postoperative Atrial Fibrillation in the Omega-3 Fatty Acids for Prevention of Postoperative Atrial Fibrillation (OPERA) Trial. J. Am. Heart Assoc. 2015, 4, e001886. [Google Scholar] [CrossRef] [Green Version]
- Rodrigo, R.; Gutiérrez, R.; Fernández, R.; Guzmán, P. Ageing improves the antioxidant response against postoperative atrial fibrillation: A randomized controlled trial. Interact. Cardiovasc. Thorac. Surg. 2012, 15, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Ran, Q.; Roberts, L.J., 2nd; Zhou, L.; Richardson, A.; Sharan, C.; Wu, D.; Yang, H. Suppression of atherogenesis by overexpression of glutathione peroxidase-4 in apolipoprotein E-deficient mice. Free Radic. Biol. Med. 2008, 44, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Baseler, W.A.; Dabkowski, E.R.; Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Croston, T.L.; Powell, M.; Razunguzwa, T.T.; Lewis, S.E.; et al. Reversal of mitochondrial proteomic loss in Type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 304, R553–R565. [Google Scholar] [CrossRef] [Green Version]
- Dabkowski, E.R.; Williamson, C.L.; Hollander, J.M. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic. Biol. Med. 2008, 45, 855–865. [Google Scholar] [CrossRef]
- Méplan, C.; Crosley, L.K.; Nicol, F.; Horgan, G.W.; Mathers, J.C.; Arthur, J.R.; Hesketh, J.E. Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: Interaction with sex. Am. J. Clin. Nutr. 2008, 87, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Maia, A.-T.; Spiteri, I.; Lee, A.J.X.; O’Reilly, M.; Jones, L.; Caldas, C.; Ponder, B.A.J. Extent of differential allelic expression of candidate breast cancer genes is similar in blood and breast. Breast Cancer Res. 2009, 11, R88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermano, G.; Pagmantidis, V.; Holloway, N.; Kadri, S.; Mowat, N.A.; Shiel, R.S.; Arthur, J.R.; Mathers, J.C.; Daly, A.K.; Broom, J.; et al. Evidence that a polymorphism within the 3′UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutr. 2007, 2, 225–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christophersen, I.E.; Rienstra, M.; Roselli, C.; Yin, X.; Geelhoed, B.; Barnard, J.; Lin, H.; Arking, D.E.; Smith, A.V.; Albert, C.M.; et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 2017, 49, 946–952. [Google Scholar] [CrossRef] [Green Version]
- Weng, L.C.; Preis, S.R.; Hulme, O.L.; Larson, M.G.; Choi, S.H.; Wang, B.; Trinquart, L.; McManus, D.D.; Staerk, L.; Lin, H.; et al. Genetic Predisposition, Clinical Risk Factor Burden, and Lifetime Risk of Atrial Fibrillation. Circulation 2018, 137, 1027–1038. [Google Scholar] [CrossRef]

| Variables | POAF N (%) | Sinus Rhythm N (%) | p-Value |
|---|---|---|---|
| Overall | 47 (25) | 142 (75) | |
| Demographics | |||
| Age > 65 years | 33 (70.21) | 76 (53.52) | 0.0447 |
| White race | 42 (89.36) | 113 (79.58) | 0.1301 |
| Obese | 21 (44.68) | 60 (42.25) | 0.7707 |
| Male | 37 (78.72) | 111 (78.17) | 0.9363 |
| Smoking Status | 12 (25.53) | 40 (28.17) | 0.7257 |
| Comorbidities | |||
| History of AF | 17 (36.17) | 9 (6.34) | <0.0001 |
| COPD | 5 (10.64) | 14 (9.86) | 0.8776 |
| DM | 28 (59.57) | 63 (44.37) | 0.0705 |
| HF | 15 (31.91) | 21 (14.79) | 0.0095 |
| HTN | 41 (87.23) | 119 (83.80) | 0.5716 |
| Prior MI | 19 (40.43) | 52 (36.62) | 0.6405 |
| Medications | |||
| ACEi/ARB | 21 (44.68) | 56 (39.44) | 0.5259 |
| β- blockers | 40 (85.11) | 109 (76.76) | 0.2247 |
| CCBs | 10 (21.28) | 32 (22.54) | 0.8572 |
| Diuretics | 24 (51.06) | 59 (41.55) | 0.2546 |
| DM Meds | 25 (53.19) | 59 (41.55) | 0.1638 |
| Nitrates | 28 (59.57) | 92 (64.79) | 0.5198 |
| Statins | 36 (76.60) | 118 (83.10) | 0.3198 |
| Cardiac Function | |||
| EF > 35 | 35 (74.47) | 127 (90.07) | 0.0073 |
| HR ≥ 70 bpm | 19 (40.43) | 69 (48.59) | 0.3306 |
| LAD ≥ 5 cm a | 2 (4.26) | 2 (1.41) | 0.2442 |
| GPx4 values | Mean ± SD | ||
| GPX4 activity a | 129.7 ± 154.4 | 113.5 ± 134.7 | 0.5405 |
| GPX4 protein level a | 2293.2 ± 1119.4 | 2507.5 ± 1542.3 | 0.3165 |
| SNP | Minor Allele | Consequence | OR a | p-Value b | Permutated p-Value | Number of Permutations | FDR_BH c | FDR_BY d |
|---|---|---|---|---|---|---|---|---|
| rs2075710 | T | Regulatory region | 1.869 | 0.0206 | 0.0199 | 1006 | 0.0965 | 0.3137 |
| rs8178977 | C | Intron variant | 1.737 | 0.0546 | 0.0531 | 376 | 0.1531 | 0.4978 |
| rs2074452 | T | TF binding site e | 0.5028 | 0.0393 | 0.0465 | 429 | 0.1377 | 0.4478 |
| rs3826961 | T | Intron variant | 0.3317 | 0.0049 | 0.0037 | 5721 | 0.0339 | 0.1103 |
| rs3746162 | T | Intron variant | 0.3169 | 0.0035 | 0.0024 | 8716 | 0.0339 | 0.1103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berdaweel, I.A.; Hart, A.A.; Jatis, A.J.; Karlan, N.; Akhter, S.A.; Gaine, M.E.; Smith, R.M.; Anderson, E.J. A Genotype–Phenotype Analysis of Glutathione Peroxidase 4 in Human Atrial Myocardium and Its Association with Postoperative Atrial Fibrillation. Antioxidants 2022, 11, 721. https://doi.org/10.3390/antiox11040721
Berdaweel IA, Hart AA, Jatis AJ, Karlan N, Akhter SA, Gaine ME, Smith RM, Anderson EJ. A Genotype–Phenotype Analysis of Glutathione Peroxidase 4 in Human Atrial Myocardium and Its Association with Postoperative Atrial Fibrillation. Antioxidants. 2022; 11(4):721. https://doi.org/10.3390/antiox11040721
Chicago/Turabian StyleBerdaweel, Islam A., Alexander A. Hart, Andrew J. Jatis, Nathan Karlan, Shahab A. Akhter, Marie E. Gaine, Ryan M. Smith, and Ethan J. Anderson. 2022. "A Genotype–Phenotype Analysis of Glutathione Peroxidase 4 in Human Atrial Myocardium and Its Association with Postoperative Atrial Fibrillation" Antioxidants 11, no. 4: 721. https://doi.org/10.3390/antiox11040721
APA StyleBerdaweel, I. A., Hart, A. A., Jatis, A. J., Karlan, N., Akhter, S. A., Gaine, M. E., Smith, R. M., & Anderson, E. J. (2022). A Genotype–Phenotype Analysis of Glutathione Peroxidase 4 in Human Atrial Myocardium and Its Association with Postoperative Atrial Fibrillation. Antioxidants, 11(4), 721. https://doi.org/10.3390/antiox11040721






