Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles
Abstract
:1. Introduction
2. Materials and Methods
2.1. C. elegans Strains
2.2. Primary Cultures of Embryonic Nematode Cells
2.3. Microarray Experiments and Analysis
2.3.1. Amplification, Labeling and Chip Hybridization
2.3.2. Microarray Data Analysis
2.4. Quantitative Real-Time Polymerase Chain Reaction
2.5. Whole C. elegans HR-MAS NMR Spectroscopy
2.5.1. Sample Preparation
2.5.2. HR-MAS NMR Spectroscopy
2.5.3. HR-MAS Data Processing and Statistical Analysis
2.6. Chemicals
2.7. Plasmid Construction
2.8. Cell Culture
2.9. Transfection and 48 Wells Plate Screening Protocol
2.10. Cell Viability Assays
- Eoxi570 = molar extinction coefficient (E) of oxidized alamarBlue Reagent at 570 nm = 80,586
- Eoxi600 = E of oxidized alamarBlue Reagent at 600 nm = 117,216
- A570 = absorbance of test wells at 570 nm
- A600 = absorbance of test wells at 600 nm
- Ered570 = E of reduced alamarBlue at 570 nm = 155,677
- Ered600 = E of reduced alamarBlue at 600 nm = 14,652
- C570 = absorbance of negative control well (media, AlamarBlue Reagent, no cells, no treatment) at 570 nm
- C600 = absorbance of negative control well (media, AlamarBlue Reagent, no cells, no treatment) at 600 nm
2.11. Firefly Luciferase Inhibition Assays
2.12. Statistics
3. Results
3.1. Transcriptomic and Metabolomic Profiling of C. elegans WT vs. ahr-1(ia03) Mutant
3.1.1. Whole C. elegans HR-MAS NMR Spectroscopy: The Inactivation of the AHR-1 Receptor Impacts Metabolism
3.1.2. Global Gene Expression Profiles Are Altered in C. elegans ahr-1-Expressing Cells
3.2. Identification of AHR-1 Activity and Modulators In Vitro in Cos-7 Cells
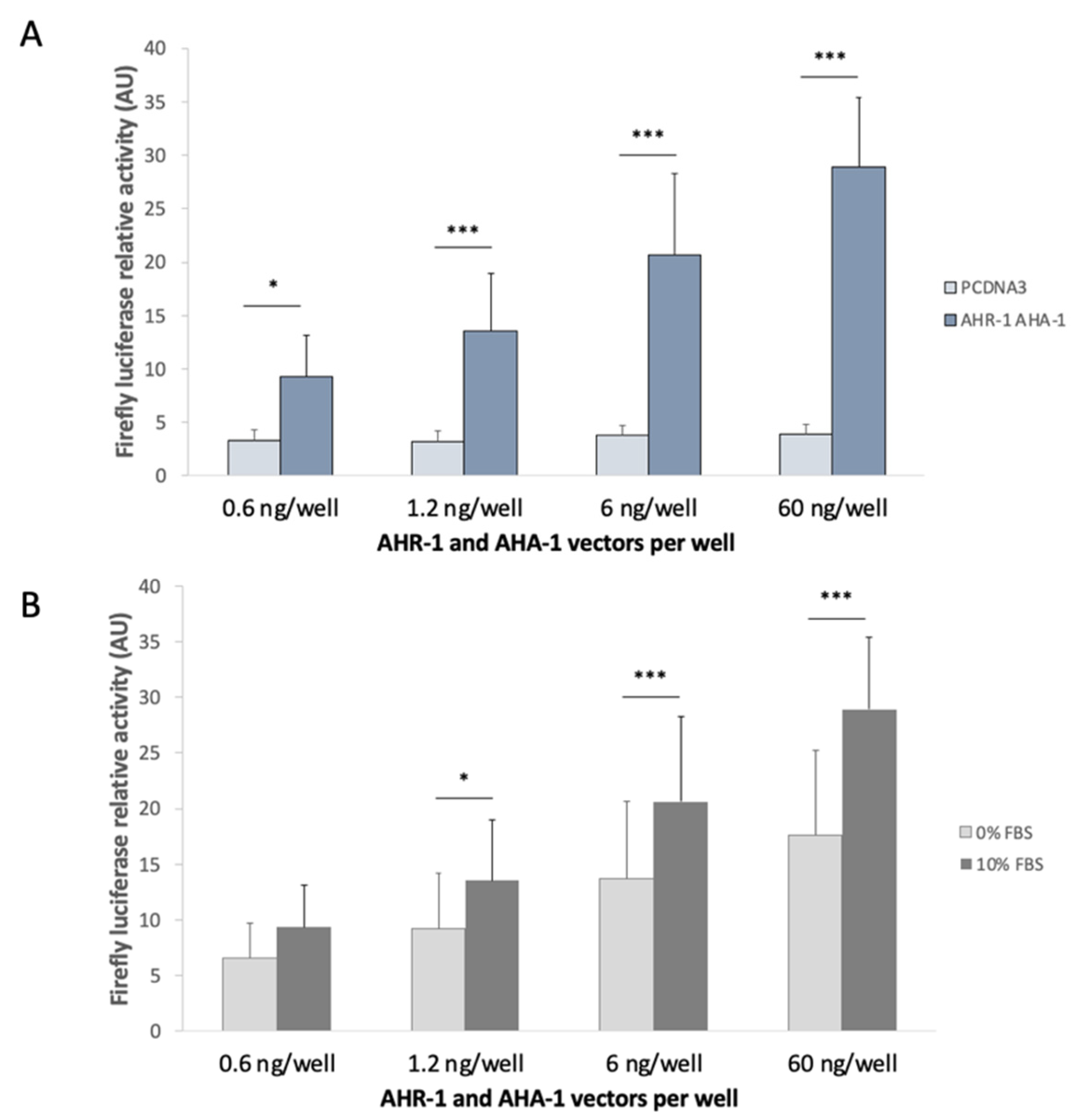
3.2.1. Optimization of the in vitro System
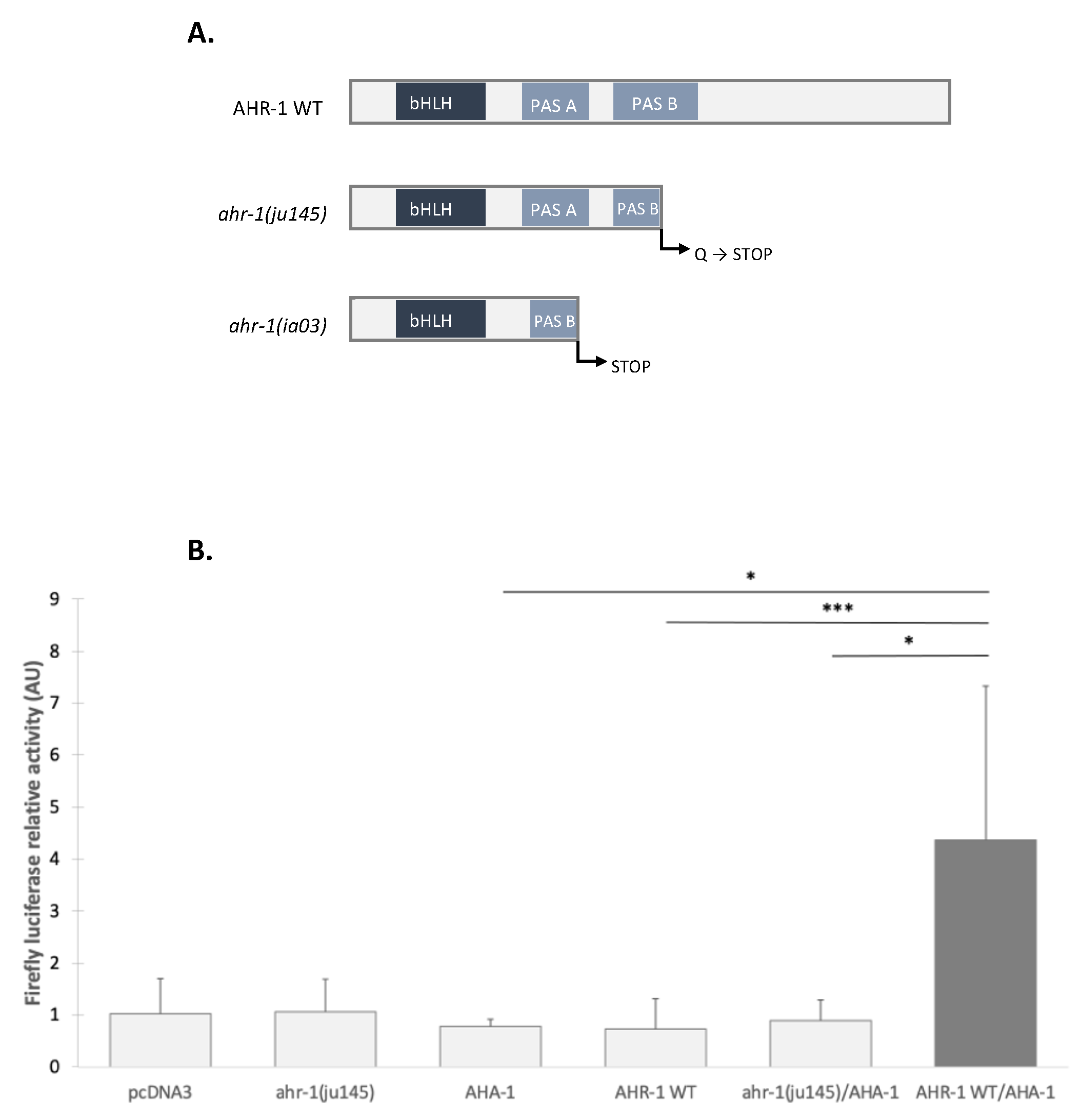
3.2.2. The Transcriptional Activation Domain of AHR-1 Is Required for Gene Expression
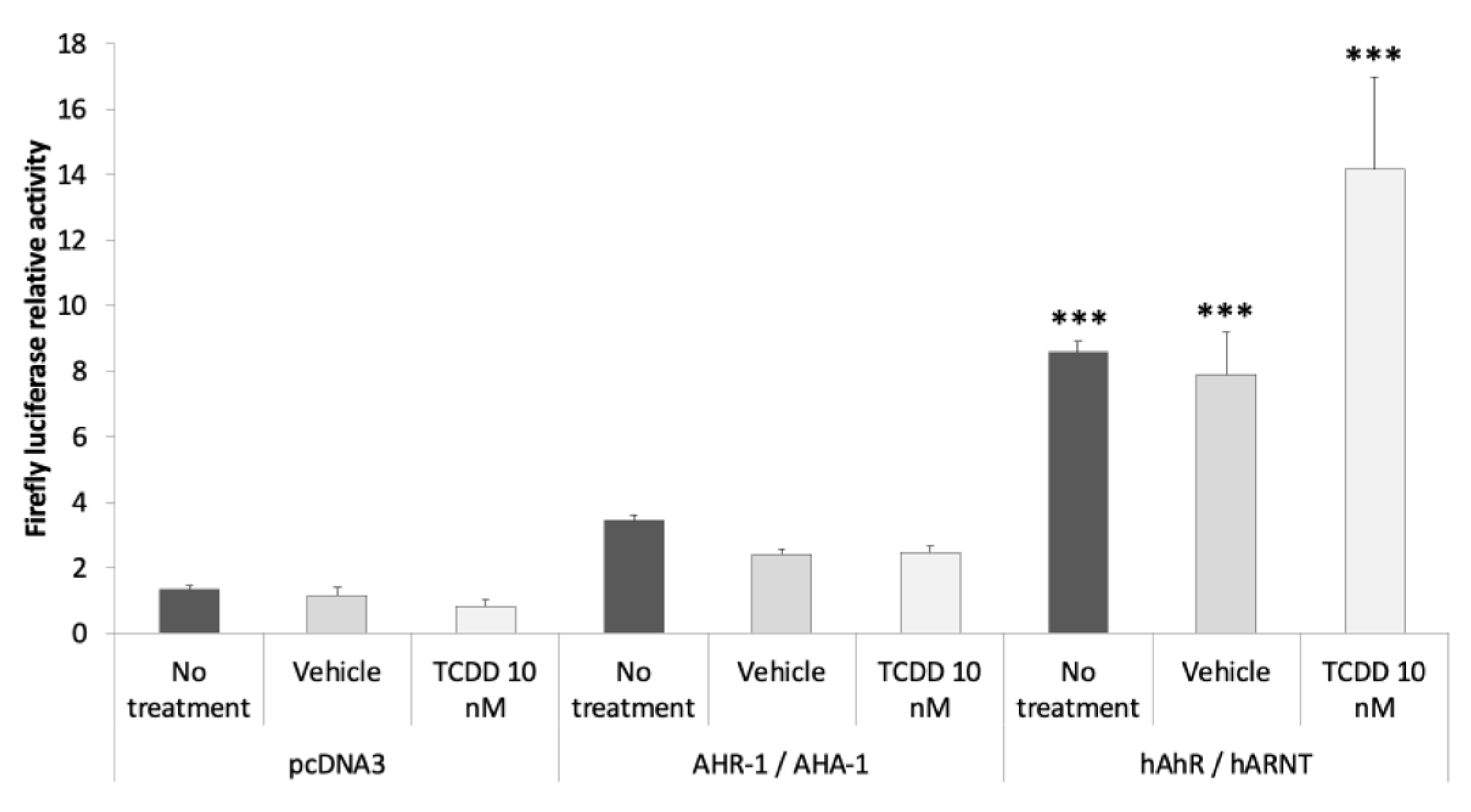
3.2.3. AHR-1 Basal Activity in Cos-7 Cells
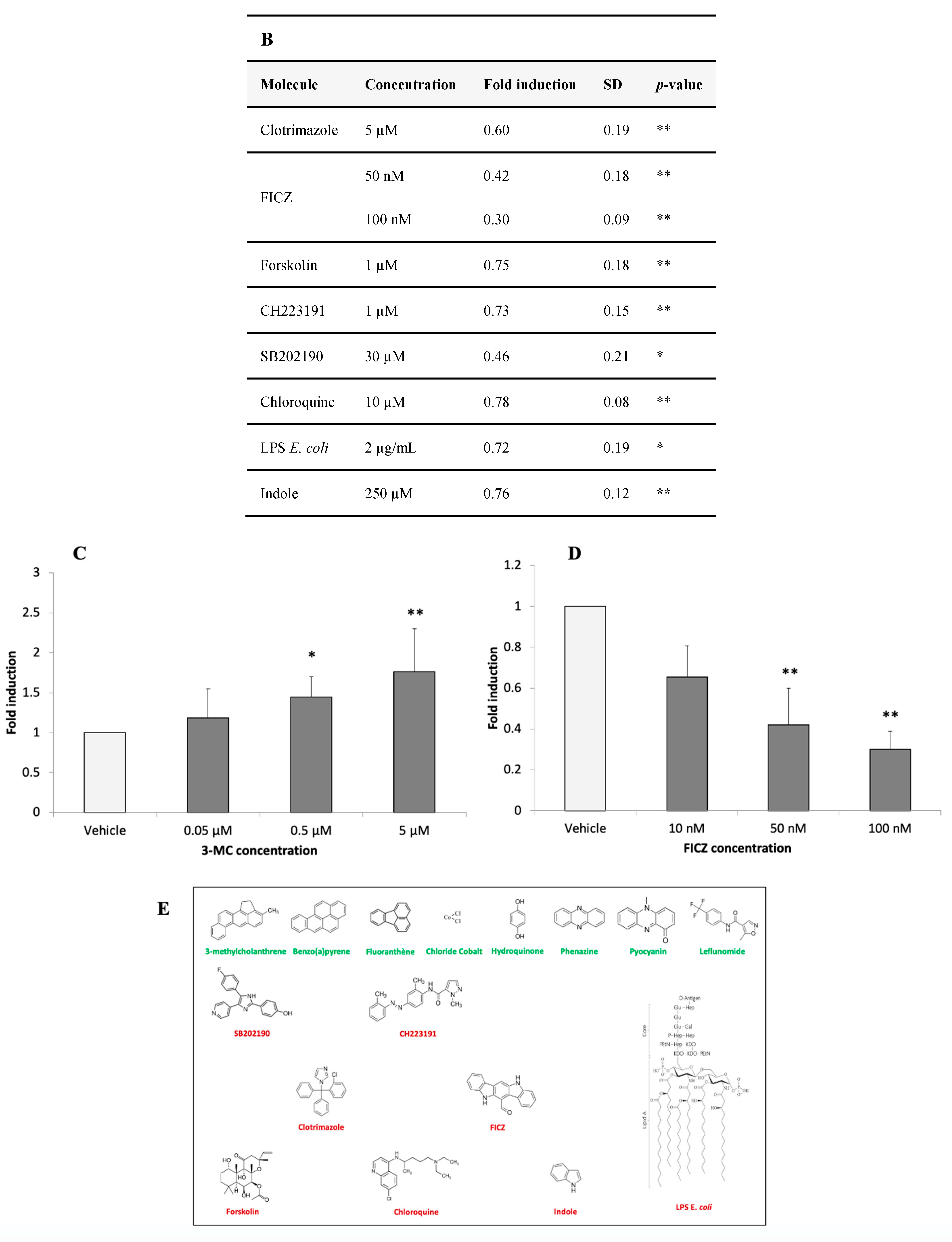
3.2.4. Identification of AHR-1 Positive and Negative Modulators
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poland, A.; Clover, E.; Kende, A.S.; DeCamp, M.; Giandomenico, C.M. 3,4,3′,4′-Tetrachloro Azoxybenzene and Azobenzene: Potent Inducers of Aryl Hydrocarbon Hydroxylase|Science. Science 1976, 194, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Poland, A.; Palen, D.; Glover, E. Tumour Promotion by TCDD in Skin of HRS/J Hairless Mice. Nature 1982, 300, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, J.P. Genetic and Molecular Aspects of 2,3,7,8-Tetra-Chlorodibenzo-P-Dioxin Action. Annu. Rev. Pharmacol. Toxicol. 1990, 30, 251–277. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.P.; Bradfield, C.A. The Search for Endogenous Activators of the Aryl Hydrocarbon Receptor. Chem. Res. Toxicol. 2008, 21, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, E.; Reyes, H.; Chu, F.; Sander, F.; Conley, L.; Brooks, B.; Hankinson, O. Cloning of a Factor Required for Activity of the Ah (Dioxin) Receptor. Science 1991, 252, 954–958. [Google Scholar] [CrossRef]
- Denison, M.S.; Fisher, J.M.; Whitlock, J.P. Protein-DNA Interactions at Recognition Sites for the Dioxin-Ah Receptor Complex. J. Biol. Chem. 1989, 264, 16478–16482. [Google Scholar] [CrossRef]
- Rannug, A.; Rannug, U.; Rosenkranz, H.S.; Winqvist, L.; Westerholm, R.; Agurell, E.; Grafström, A.K. Certain Photooxidized Derivatives of Tryptophan Bind with Very High Affinity to the Ah Receptor and Are Likely to Be Endogenous Signal Substances. J. Biol. Chem. 1987, 262, 15422–15427. [Google Scholar] [CrossRef]
- Helferich, W.G.; Denison, M.S. Ultraviolet Photoproducts of Tryptophan Can Act as Dioxin Agonists. Mol. Pharmacol. 1991, 40, 674–678. [Google Scholar]
- Lahvis, G.P.; Lindell, S.L.; Thomas, R.S.; McCuskey, R.S.; Murphy, C.; Glover, E.; Bentz, M.; Southard, J.; Bradfield, C.A. Portosystemic Shunting and Persistent Fetal Vascular Structures in Aryl Hydrocarbon Receptor-Deficient Mice. Proc. Natl. Acad. Sci. USA 2000, 97, 10442–10447. [Google Scholar] [CrossRef] [Green Version]
- Abbott, B.D.; Schmid, J.E.; Brown, J.G.; Wood, C.R.; White, R.D.; Buckalew, A.R.; Held, G.A. RT-PCR Quantification of AHR, ARNT, GR, and CYP1A1 MRNA in Craniofacial Tissues of Embryonic Mice Exposed to 2,3,7,8-Tetrachlorodibenzo-p- Dioxin and Hydrocortisone. Toxicol. Sci. 1999, 47, 76–85. [Google Scholar] [CrossRef]
- Fernandez-Salguero, P.; Pineau, T.; Hilbert, D.M.; McPhail, T.; Lee, S.S.; Kimura, S.; Nebert, D.W.; Rudikoff, S.; Ward, J.M.; Gonzalez, F.J. Immune System Impairment and Hepatic Fibrosis in Mice Lacking the Dioxin-Binding Ah Receptor. Science 1995, 268, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.V.; Su, G.H.; Reddy, J.K.; Simon, M.C.; Bradfield, C.A. Characterization of a Murine Ahr Null Allele: Involvement of the Ah Receptor in Hepatic Growth and Development. Proc. Natl. Acad. Sci. USA 1996, 93, 6731–6736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juricek, L.; Carcaud, J.; Pelhaitre, A.; Riday, T.T.; Chevallier, A.; Lanzini, J.; Auzeil, N.; Laprévote, O.; Dumont, F.; Jacques, S.; et al. AhR-Deficiency as a Cause of Demyelinating Disease and Inflammation. Sci. Rep. 2017, 7, 9794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevallier, A.; Mialot, A.; Petit, J.-M.; Fernandez-Salguero, P.; Barouki, R.; Coumoul, X.; Beraneck, M. Oculomotor Deficits in Aryl Hydrocarbon Receptor Null Mouse. PLoS ONE 2013, 8, e53520. [Google Scholar] [CrossRef] [Green Version]
- Shackleford, G.; Sampathkumar, N.K.; Hichor, M.; Weill, L.; Meffre, D.; Juricek, L.; Laurendeau, I.; Chevallier, A.; Ortonne, N.; Larousserie, F.; et al. Involvement of Aryl Hydrocarbon Receptor in Myelination and in Human Nerve Sheath Tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E1319–E1328. [Google Scholar] [CrossRef] [Green Version]
- Juricek, L.; Coumoul, X. The Aryl Hydrocarbon Receptor and the Nervous System. Int. J. Mol. Sci. 2018, 19, 2504. [Google Scholar] [CrossRef] [Green Version]
- Powell-Coffman, J.A.; Bradfield, C.A.; Wood, W.B. Caenorhabditis Elegans Orthologs of the Aryl Hydrocarbon Receptor and Its Heterodimerization Partner the Aryl Hydrocarbon Receptor Nuclear Translocator. Proc. Natl. Acad. Sci. USA 1998, 95, 2844–2849. [Google Scholar] [CrossRef] [Green Version]
- Butler, R.A.; Kelley, M.L.; Powell, W.H.; Hahn, M.E.; Van Beneden, R.J. An Aryl Hydrocarbon Receptor (AHR) Homologue from the Soft-Shell Clam, Mya Arenaria: Evidence That Invertebrate AHR Homologues Lack 2,3,7,8-Tetrachlorodibenzo-p-Dioxin and Beta-Naphthoflavone Binding. Gene 2001, 278, 223–234. [Google Scholar] [CrossRef]
- Huang, X.; Powell-Coffman, J.A.; Jin, Y. The AHR-1 Aryl Hydrocarbon Receptor and Its Co-Factor the AHA-1 Aryl Hydrocarbon Receptor Nuclear Translocator Specify GABAergic Neuron Cell Fate in C. Elegans. Development 2004, 131, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.J.; O’Brien, T.; Chatzigeorgiou, M.; Spencer, W.C.; Feingold-Link, E.; Husson, S.J.; Hori, S.; Mitani, S.; Gottschalk, A.; Schafer, W.R.; et al. Sensory Neuron Fates Are Distinguished by a Transcriptional Switch That Regulates Dendrite Branch Stabilization. Neuron 2013, 79, 266–280. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Powell-Coffman, J.A. The Caenorhabditis Elegans Aryl Hydrocarbon Receptor, AHR-1, Regulates Neuronal Development. Dev. Biol. 2004, 270, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.D.; Jan, L.Y.; Jan, Y.N. The BHLH-PAS Protein Spineless Is Necessary for the Diversification of Dendrite Morphology of Drosophila Dendritic Arborization Neurons. Genes Dev. 2006, 20, 2806–2819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernet, M.F.; Mazzoni, E.O.; Çelik, A.; Duncan, D.M.; Duncan, I.; Desplan, C. Stochastic Spineless Expression Creates the Retinal Mosaic for Colour Vision. Nature 2006, 440, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aarnio, V.; Storvik, M.; Lehtonen, M.; Asikainen, S.; Reisner, K.; Callaway, J.; Rudgalvyte, M.; Lakso, M.; Wong, G. Fatty Acid Composition and Gene Expression Profiles Are Altered in Aryl Hydrocarbon Receptor-1 Mutant Caenorhabditis Elegans. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010, 151, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Aarnio, V.; Heikkinen, L.; Peltonen, J.; Goldsteins, G.; Lakso, M.; Wong, G. Transcriptional Profiling Reveals Differential Expression of a Neuropeptide-like Protein and Pseudogenes in Aryl Hydrocarbon Receptor-1 Mutant Caenorhabditis Elegans. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 9, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tourette, C.; Farina, F.; Vazquez-Manrique, R.P.; Orfila, A.-M.; Voisin, J.; Hernandez, S.; Offner, N.; Parker, J.A.; Menet, S.; Kim, J.; et al. The Wnt Receptor Ryk Reduces Neuronal and Cell Survival Capacity by Repressing FOXO Activity During the Early Phases of Mutant Huntingtin Pathogenicity. PLoS Biol. 2014, 12, e1001895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marisa, L.; Ichante, J.-L.; Reymond, N.; Aggerbeck, L.; Delacroix, H.; Mucchielli-Giorgi, M.-H. MAnGO: An Interactive R-Based Tool for Two-Colour Microarray Analysis. Bioinformatics 2007, 23, 2339–2341. [Google Scholar] [CrossRef] [Green Version]
- Blaise, B.J.; Giacomotto, J.; Elena, B.; Dumas, M.-E.; Toulhoat, P.; Ségalat, L.; Emsley, L. Metabotyping of Caenorhabditis Elegans Reveals Latent Phenotypes. Proc. Natl. Acad. Sci. USA 2007, 104, 19808–19812. [Google Scholar] [CrossRef] [Green Version]
- Morel, Y.; Barouki, R. Down-Regulation of Cytochrome P450 1A1 Gene Promoter by Oxidative Stress. Critical Contribution of Nuclear Factor 1. J. Biol. Chem. 1998, 273, 26969–26976. [Google Scholar] [CrossRef] [Green Version]
- Poutiainen, P.K.; Palvimo, J.J.; Hinkkanen, A.E.; Valkonen, A.; Väisänen, T.K.; Laatikainen, R.; Pulkkinen, J.T. Discovery of 5-Benzyl-3-Phenyl-4,5-Dihydroisoxazoles and 5-Benzyl-3-Phenyl-1,4,2-Dioxazoles as Potent Firefly Luciferase Inhibitors. J. Med. Chem. 2013, 56, 1064–1073. [Google Scholar] [CrossRef]
- Pontoizeau, C.; Mouchiroud, L.; Molin, L.; Mergoud-dit-Lamarche, A.; Dallière, N.; Toulhoat, P.; Elena-Herrmann, B.; Solari, F. Metabolomics Analysis Uncovers That Dietary Restriction Buffers Metabolic Changes Associated with Aging in Caenorhabditis Elegans. J. Proteome Res. 2014, 13, 2910–2919. [Google Scholar] [CrossRef] [PubMed]
- Reyes, H.; Reisz-Porszasz, S.; Hankinson, O. Identification of the Ah Receptor Nuclear Translocator Protein (Arnt) as a Component of the DNA Binding Form of the Ah Receptor. Science 1992, 256, 1193–1195. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, B.N.; Probst, M.R.; Reisz-Porszasz, S.; Hankinson, O. Identification of Functional Domains of the Aryl Hydrocarbon Receptor. J. Biol. Chem. 1995, 270, 29270–29278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-Y.; Puga, A. Constitutive Activation of the Aromatic Hydrocarbon Receptor. Mol. Cell. Biol. 1998, 18, 525–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffer, A.; Chang, C.Y.; Puga, A. Dioxin Induces Transcription of Fos and Jun Genes by Ah Receptor-Dependent and -Independent Pathways. Toxicol. Appl. Pharmacol. 1996, 141, 238–247. [Google Scholar] [CrossRef]
- Bak, S.-M.; Nakata, H.; Koh, D.-H.; Yoo, J.; Iwata, H.; Kim, E.-Y. In Vitro and in Silico AHR Assays for Assessing the Risk of Heavy Oil-Derived Polycyclic Aromatic Hydrocarbons in Fish. Ecotoxicol. Environ. Saf. 2019, 181, 214–223. [Google Scholar] [CrossRef]
- Kim, E.-Y.; Suda, T.; Tanabe, S.; Batoev, V.B.; Petrov, E.A.; Iwata, H. Evaluation of Relative Potencies for in Vitro Transactivation of the Baikal Seal Aryl Hydrocarbon Receptor by Dioxin-Like Compounds. Environ. Sci. Technol. 2011, 45, 1652–1658. [Google Scholar] [CrossRef]
- Fujisawa, N.; Darwish, W.S.; Ikenaka, Y.; Kim, E.; Lee, J.; Iwata, H.; Nakayama, S.; Ishizuka, M. Molecular Evaluation of a New Highly Sensitive Aryl Hydrocarbon Receptor in Ostriches. Poult. Sci. 2013, 92, 1921–1929. [Google Scholar] [CrossRef]
- Karchner, S.I.; Jenny, M.J.; Tarrant, A.M.; Evans, B.R.; Kang, H.J.; Bae, I.; Sherr, D.H.; Hahn, M.E. The Active Form of Human Aryl Hydrocarbon Receptor (AHR) Repressor Lacks Exon 8, and Its Pro185 and Ala185 Variants Repress Both AHR and Hypoxia-Inducible Factor. Mol. Cell. Biol. 2009, 29, 3465–3477. [Google Scholar] [CrossRef] [Green Version]
- Lanham, K.A.; Prasch, A.L.; Weina, K.M.; Peterson, R.E.; Heideman, W. A Dominant Negative Zebrafish Ahr2 Partially Protects Developing Zebrafish from Dioxin Toxicity. PLoS ONE 2011, 6, e28020. [Google Scholar] [CrossRef] [Green Version]
- Farmahin, R.; Crump, D.; Kennedy, S.W. Sensitivity of Avian Species to the Aryl Hydrocarbon Receptor Ligand 6-Formylindolo [3,2-b] Carbazole (FICZ). Chemico-Biol. Interact. 2014, 221, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.E. Aryl Hydrocarbon Receptors: Diversity and Evolution. Chemico-Biological Interactions 2002, 141, 131–160. [Google Scholar] [CrossRef]
- Kim, J.H.; Stallcup, M.R. Role of the Coiled-Coil Coactivator (CoCoA) in Aryl Hydrocarbon Receptor-Mediated Transcription. J. Biol. Chem. 2004, 279, 49842–49848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.T.Y. β-Naphthoflavone, an Inducer of Xenobiotic Metabolizing Enzymes, Inhibits Firefly Luciferase Activity. Analytical Biochemistry 2002, 304, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiarova, A.; Taslimi, P.; Elliman, S.J.; Kosinski, P.A.; Hubbard, B.; Kavana, M.; Kemp, D.M. Resveratrol Inhibits Firefly Luciferase. Biochem. Biophys. Res. Commun. 2006, 351, 481–484. [Google Scholar] [CrossRef]
- Braeuning, A. Firefly Luciferase Inhibition: A Widely Neglected Problem. Arch. Toxicol. 2015, 89, 141–142. [Google Scholar] [CrossRef] [Green Version]
- Reitzel, A.M.; Passamaneck, Y.J.; Karchner, S.I.; Franks, D.G.; Martindale, M.Q.; Tarrant, A.M.; Hahn, M.E. Aryl Hydrocarbon Receptor (AHR) in the Cnidarian Nematostella Vectensis: Comparative Expression, Protein Interactions, and Ligand Binding. Dev. Genes Evol. 2014, 224, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Hahn, M.E.; Karchner, S.I.; Merson, R.R. Diversity as Opportunity: Insights from 600 Million Years of AHR Evolution. Curr. Opin. Toxicol. 2017, 2, 58–71. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, E.F.; Saili, K.S.; Koch, D.C.; Kopparapu, P.R.; Farrer, D.; Bisson, W.H.; Mathew, L.K.; Sengupta, S.; Kerkvliet, N.I.; Tanguay, R.L.; et al. The Anti-Inflammatory Drug Leflunomide Is an Agonist of the Aryl Hydrocarbon Receptor. PLoS ONE 2010, 5, e13128. [Google Scholar] [CrossRef]
- Eckers, A.; Jakob, S.; Heiss, C.; Haarmann-Stemmann, T.; Goy, C.; Brinkmann, V.; Cortese-Krott, M.M.; Sansone, R.; Esser, C.; Ale-Agha, N.; et al. The Aryl Hydrocarbon Receptor Promotes Aging Phenotypes across Species. Sci. Rep. 2016, 6, 19618. [Google Scholar] [CrossRef] [Green Version]
- Sonowal, R.; Swimm, A.; Sahoo, A.; Luo, L.; Matsunaga, Y.; Wu, Z.; Bhingarde, J.A.; Ejzak, E.A.; Ranawade, A.; Qadota, H.; et al. Indoles from Commensal Bacteria Extend Healthspan. Proc. Natl. Acad. Sci. USA 2017, 114, E7506–E7515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satsu, H.; Yoshida, K.; Mikubo, A.; Ogiwara, H.; Inakuma, T.; Shimizu, M. Establishment of a Stable Aryl Hydrocarbon Receptor-Responsive HepG2 Cell Line. Cytotechnology 2015, 67, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coates, J.; de Bono, M. Antagonistic Pathways in Neurons Exposed to Body Fluid Regulate Social Feeding in Caenorhabditis Elegans. Nature 2002, 419, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Carreira, V.S.; Fan, Y.; Wang, Q.; Zhang, X.; Kurita, H.; Ko, C.-I.; Naticchioni, M.; Jiang, M.; Koch, S.; Medvedovic, M.; et al. Ah Receptor Signaling Controls the Expression of Cardiac Development and Homeostasis Genes. Toxicol. Sci. 2015, 147, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Marín, N.; Merino, J.M.; Alvarez-Barrientos, A.; Patel, D.P.; Takahashi, S.; González-Sancho, J.M.; Gandolfo, P.; Rios, R.M.; Muñoz, A.; Gonzalez, F.J.; et al. Aryl Hydrocarbon Receptor Promotes Liver Polyploidization and Inhibits PI3K, ERK, and Wnt/β-Catenin Signaling. iScience 2018, 4, 44–63. [Google Scholar] [CrossRef]
- Kawano, Y.; Nishiumi, S.; Tanaka, S.; Nobutani, K.; Miki, A.; Yano, Y.; Seo, Y.; Kutsumi, H.; Ashida, H.; Azuma, T.; et al. Activation of the Aryl Hydrocarbon Receptor Induces Hepatic Steatosis via the Upregulation of Fatty Acid Transport. Arch. Biochem. Biophys. 2010, 504, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-C.; Chen, H.-J.; Tseng, W.-C.; Hsu, J.-M.; Huang, T.-T.; Chen, C.-H.; Pan, C.-L.A.C. Elegans Thermosensory Circuit Regulates Longevity through Crh-1 /CREB-Dependent Flp-6 Neuropeptide Signaling. Dev. Cell 2016, 39, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Kudo, K.; Takeuchi, T.; Murakami, Y.; Ebina, M.; Kikuchi, H. Characterization of the Region of the Aryl Hydrocarbon Receptor Required for Ligand Dependency of Transactivation Using Chimeric Receptor between Drosophila and Mus Musculus. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2009, 1789, 477–486. [Google Scholar] [CrossRef]
- Wang, X.; Li, S.; Liu, L.; Jian, Z.; Cui, T.; Yang, Y.; Guo, S.; Yi, X.; Wang, G.; Li, C.; et al. Role of the Aryl Hydrocarbon Receptor Signaling Pathway in Promoting Mitochondrial Biogenesis against Oxidative Damage in Human Melanocytes. J. Dermatol. Sci. 2019, 96, 33–41. [Google Scholar] [CrossRef]
- Ikuta, T.; Kobayashi, Y.; Kawajiri, K. Phosphorylation of Nuclear Localization Signal Inhibits the Ligand-Dependent Nuclear Import of Aryl Hydrocarbon Receptor. Biochem. Biophys. Res. Commun. 2004, 317, 545–550. [Google Scholar] [CrossRef]
- Henklová, P.; Vrzal, R.; Ulrichová, J.; Dvorák, Z. Role of Mitogen-Activated Protein Kinases in Aryl Hydrocarbon Receptor Signaling. Chem. Biol. Interact. 2008, 172, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Park, J.C.; Hagiwara, A.; Park, H.G.; Lee, J.-S. Identification of the Full 26 Cytochrome P450 (CYP) Genes and Analysis of Their Expression in Response to Benzo[α]Pyrene in the Marine Rotifer Brachionus Rotundiformis. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Pan, L.; Miao, J. Molecular Evidence for the Existence of an Aryl Hydrocarbon Receptor Pathway in Scallops Chlamys Farreri. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 196–197, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Jenny, M.J.; Walton, W.C.; Payton, S.L.; Powers, J.M.; Findlay, R.H.; O’Shields, B.; Diggins, K.; Pinkerton, M.; Porter, D.; Crane, D.M.; et al. Transcriptomic Evaluation of the American Oyster, Crassostrea Virginica, Deployed during the Deepwater Horizon Oil Spill: Evidence of an Active Hydrocarbon Response Pathway. Mar. Environ. Res. 2016, 120, 166–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.P.; Mishra, M.; Sharma, A.; Shukla, A.K.; Mudiam, M.K.R.; Patel, D.K.; Ram, K.R.; Chowdhuri, D.K. Genotoxicity and Apoptosis in Drosophila Melanogaster Exposed to Benzene, Toluene and Xylene: Attenuation by Quercetin and Curcumin. Toxicol. Appl. Pharmacol. 2011, 253, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Pan, L.; Wang, L. Metabolic Enzyme Activities, Metabolism-Related Genes Expression and Bioaccumulation in Juvenile White Shrimp Litopenaeus Vannamei Exposed to Benzo[a]Pyrene. Ecotoxicol. Environ. Saf. 2014, 104, 79–86. [Google Scholar] [CrossRef]


| Gene Symbol | Gene Description | Fold Change KO/WT |
|---|---|---|
| gcy-35 npl-20 B0207.7 flp-8 gcy-36 K09E4.4 gcy-34 gcy-32 T09B9.5 cyp-13A10 agr-1 F18G5.1 C18C4.1 C05D9.4 kin-1 sra-21 C17F4.8 srw-129 hlh-26 Y57G11C.20 gcy-33 T05A7.6 hlh-14 T27C5.6 F14F9.4 gcy-37 F14F9.3 che-11 srh-261 clec-18 | Guanylate CYclase Neuropeptide-Like Protein Predicted to have serine/threonine kinase activity FMRF-Like peptide Guanylate CYclase Orthoilog of human NAGLU (N-acetyl-alpha-glucosaminidase) Guanylate CYclase Guanylate CYclase T09B9.5 CYtochrome P450 family AGRin (synaptic protein) homolog F18G5.1 glb-5 (GLoBin related) C05D9.4 Protein kinase Serpentin receptor, class A (alpha) Predicted to encode a protein with BTB/POZ domain Serpentin receptor, class W Helix Loop Helix Uncharacterized protein Guanylate CYclase Predicted to have serine/threonine kinase activity Helix Loop Helix F14F9.4 Guanylate CYclase F14F9.3 Abnormal CHEmotaxis Serpentin Receptor, class H C-type LECtin | 0.07 0.11 0.15 0.20 0.28 0.29 0.29 0.30 0.35 0.37 0.37 0.41 0.42 0.43 0.47 0.48 0.49 0.50 0.50 0.52 0.52 0.52 0.53 0.53 0.54 0.54 0.55 0.55 0.55 0.55 |
| Ribonuclease T27D12.1 C12D5.3 nspc-12 Y62H9A.2 nscp-10 C13A2.5 bus-12 fbxb-54 irg-5 clec-34 F22E5.8 T21C12.8 W06D4.3 str-208 oac-36 T21C12.8 R07E3.2 dnc-6 clec-116 pho-10 F07E5.7 C13A2.10 col-41 dpy-13 R13H4.8 T22B2.6 sqt-1 pgs-1 cnk-1 | K10C9.3 T27S12.1 C12D5.3 Nematode Specific Peptide family, group C Y62H9A.2 Nematode Specific Peptide family, group C Predicted to have transferase activity, transferring glycosyl groups Ortholog of human SLC35D3 (solute carrier family 35 membre D3) F-box B protein Infection Response Gene C-type LECtin F22E5.8 T21C12.8 W06D4.3 Seven TM Receptor Predicted to have transferase activity R07E3.2 DyNactine Complex component Is predicted to have carbohydrate binding activity intestinal acid PHOsphatase Predicted to encode a protein with TRA-1 regulated domain Predicted to encode a protein with the methyltransferase FkbM domain Predicted tob e a structural constituent of cuticle DumPY:shorter than wild-type R13H4.8 T22B2.6 Ortholog oh human METTL24 and SCARA3 PhosphatidylGlycerophosphate Synthase Connector/eNhancer of KSR | 1.75 1.76 1.76 1.77 1.78 1.80 1.88 1.89 1.94 1.94 1.99 2.06 2.06 2.12 2.16 2.21 2.27 2.32 2.51 2.52 2.60 2.70 2.80 2.83 3.67 4.13 4.32 4.44 4.81 10.64 |
| Gene Set Name (C. elegans Gene Only) | NES | p-Value | AhR KO Phenotype |
|---|---|---|---|
| Regulation of nervous system development | 0.73 | 0.002 | Depleted |
| Mecanosensory behavior | 0.63 | 0.031 | |
| Neuron migration | 0.58 | 0.084 | |
| Synaptic transmission | 0.52 | 0.010 | |
| Neurogenesis | 0.51 | 0.035 | |
| Learning and memory | 0.48 | 0.115 | |
| Fatty acid biosynthetic process | −0.78 | 0.008 | Enriched |
| Glycolysis | −0.67 | 0.054 | |
| Fatty acid metabolic process | −0.66 | 0.058 | |
| Oxidative phisphorylation | −0.64 | 0.004 | |
| Ribosome | −0.52 | 0.065 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larigot, L.; Bui, L.-C.; de Bouvier, M.; Pierre, O.; Pinon, G.; Fiocca, J.; Ozeir, M.; Tourette, C.; Ottolenghi, C.; Imbeaud, S.; et al. Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles. Antioxidants 2022, 11, 1030. https://doi.org/10.3390/antiox11051030
Larigot L, Bui L-C, de Bouvier M, Pierre O, Pinon G, Fiocca J, Ozeir M, Tourette C, Ottolenghi C, Imbeaud S, et al. Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles. Antioxidants. 2022; 11(5):1030. https://doi.org/10.3390/antiox11051030
Chicago/Turabian StyleLarigot, Lucie, Linh-Chi Bui, Marine de Bouvier, Ophélie Pierre, Grégory Pinon, Justine Fiocca, Mohammad Ozeir, Cendrine Tourette, Chris Ottolenghi, Sandrine Imbeaud, and et al. 2022. "Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles" Antioxidants 11, no. 5: 1030. https://doi.org/10.3390/antiox11051030
APA StyleLarigot, L., Bui, L. -C., de Bouvier, M., Pierre, O., Pinon, G., Fiocca, J., Ozeir, M., Tourette, C., Ottolenghi, C., Imbeaud, S., Pontoizeau, C., Blaise, B. J., Chevallier, A., Tomkiewicz, C., Legrand, B., Elena-Herrmann, B., Néri, C., Brinkmann, V., Nioche, P., ... Coumoul, X. (2022). Identification of Modulators of the C. elegans Aryl Hydrocarbon Receptor and Characterization of Transcriptomic and Metabolic AhR-1 Profiles. Antioxidants, 11(5), 1030. https://doi.org/10.3390/antiox11051030











