Mechanism of Cadmium Exposure Induced Hepatotoxicity in the Mud Crab (Scylla paramamosain): Activation of Oxidative Stress and Nrf2 Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mud Crabs
2.2. Cadmium Exposure
2.3. Measurement of Oxidative Stress Parameters
2.4. Comprehensive Toxicity Assessment
2.5. Histopathologic Investigation
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.7. Nrf2 Silencing in Mud Crabs
2.8. Cadmium Exposure in Nrf2-Silenced Mud Crabs
2.9. Statistical Analysis
3. Results
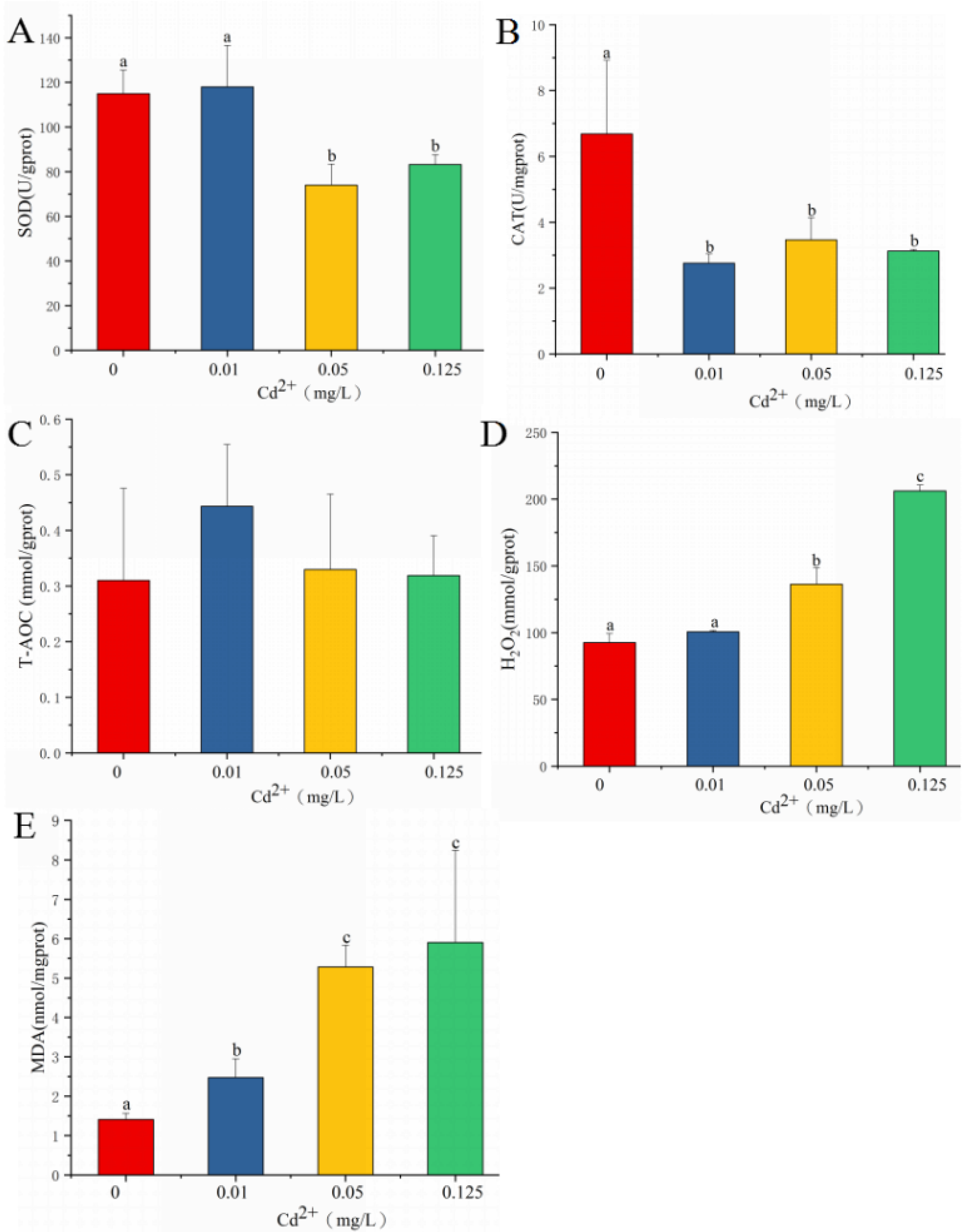
3.1. Oxidative Stress and IBRv2 after Cadmium Exposure
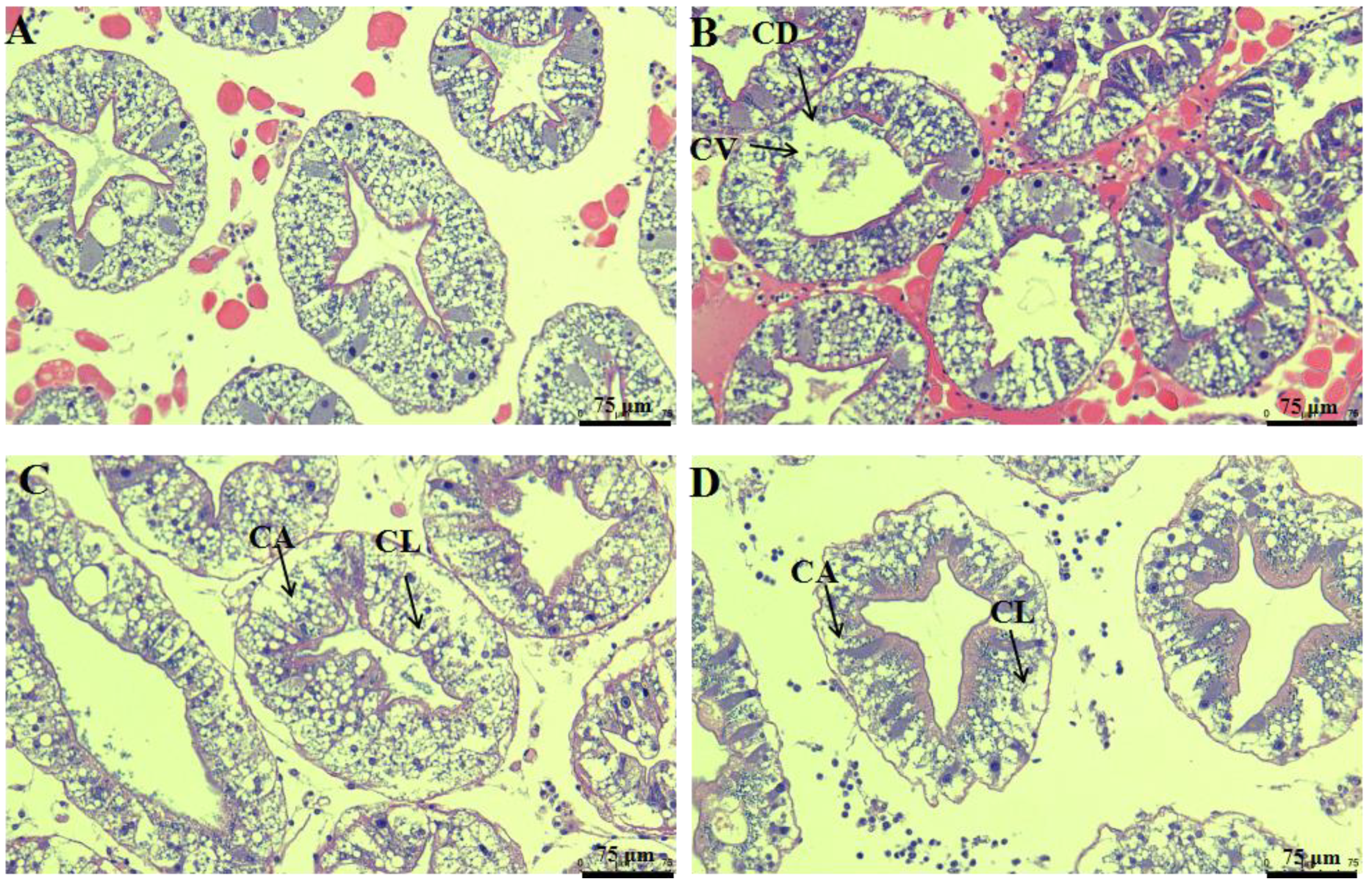
3.2. Histopathological Changes after Cadmium Exposure
3.3. Effects of Cadmium Exposure on the Expression of Nrf2, CYP2, and MT
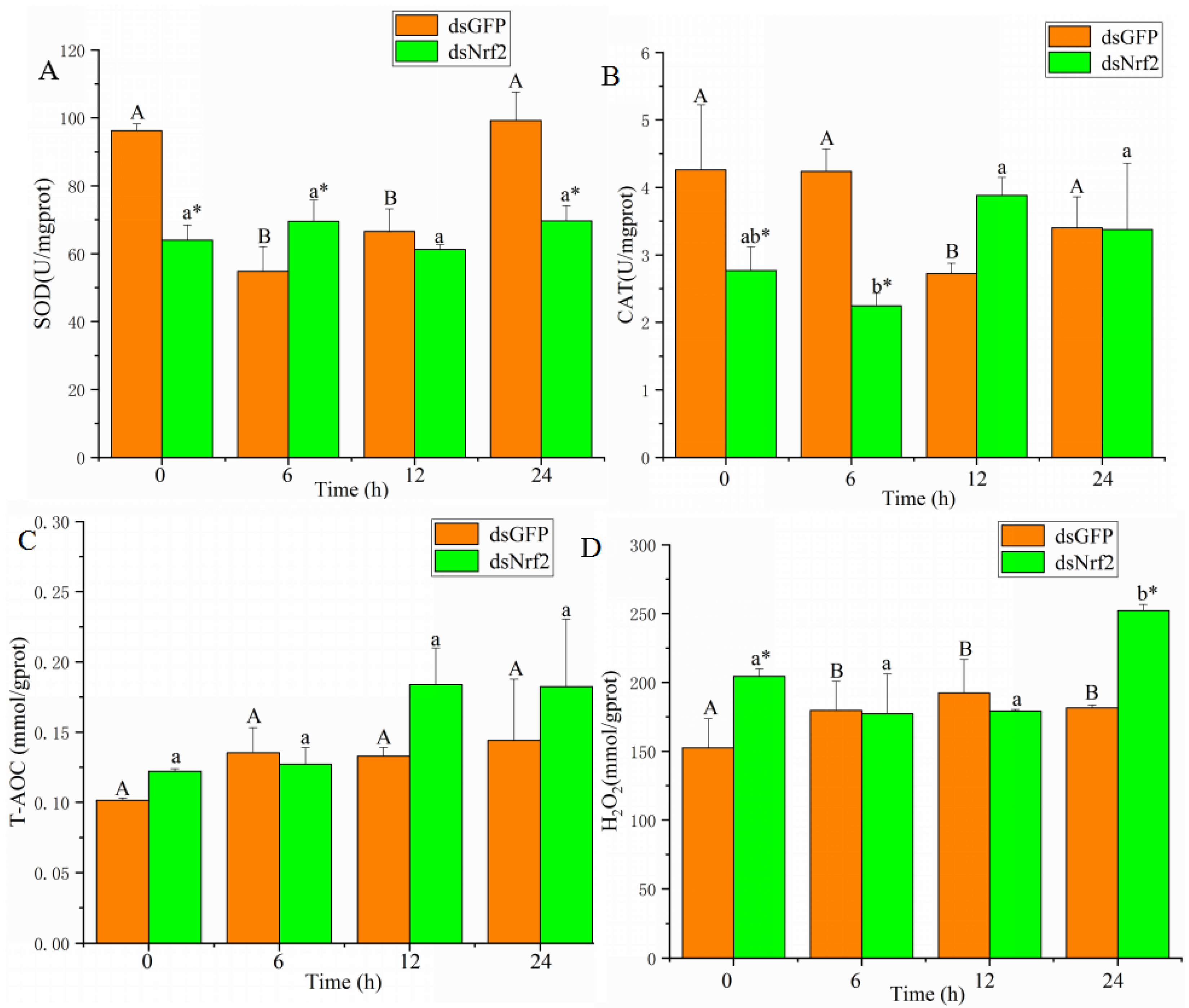
3.4. The Expression of Genes Related to Antioxidants after Knockdown of Nrf2
3.5. Effects of Nrf2-Interfered on Oxidative Stress Biomarkers after Cadmium Exposure
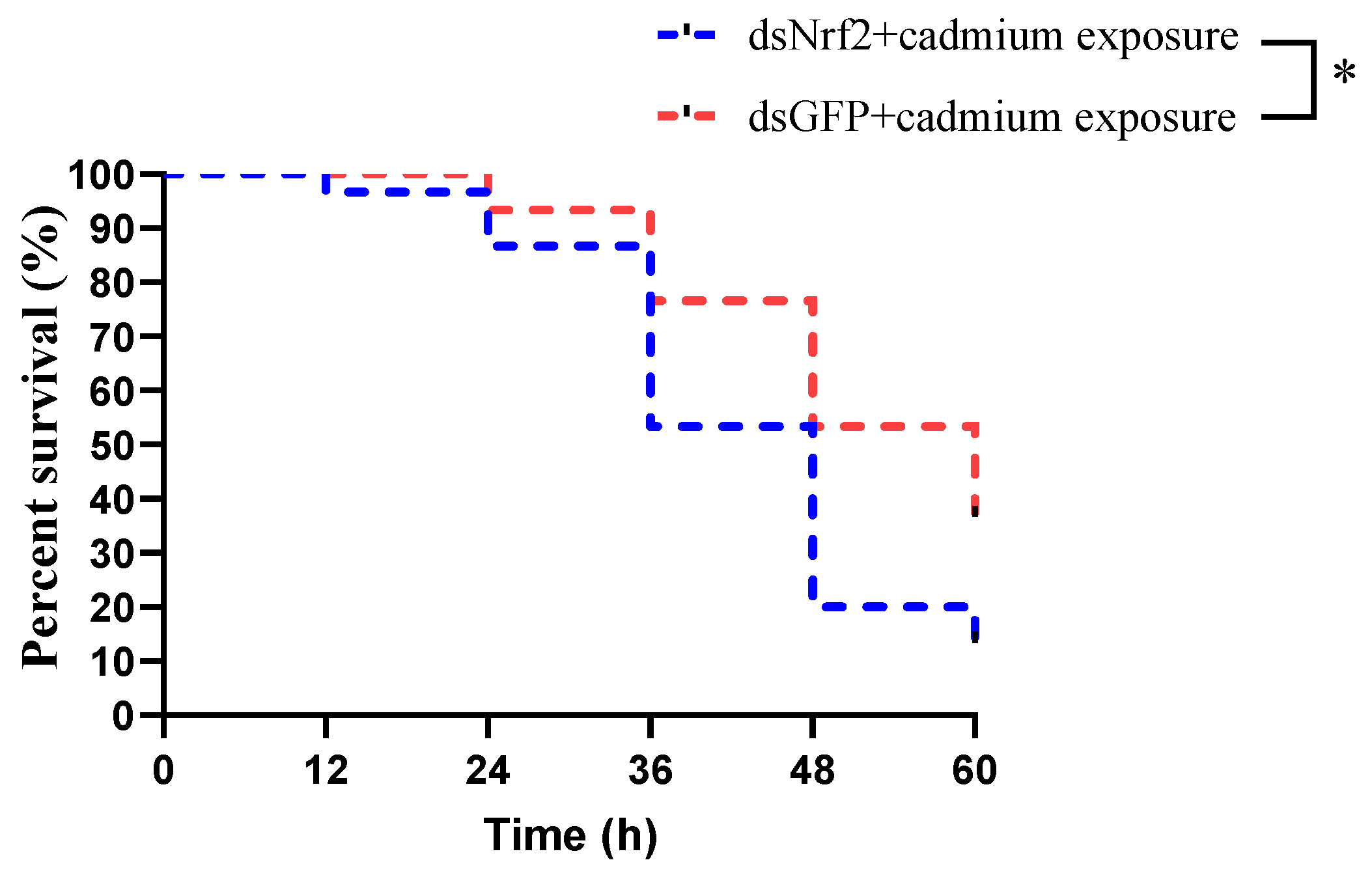
3.6. The Mortality of Mud Crabs after Cadmium Exposure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yi, Y.; Yang, Z.; Zhang, S. Ecological risk assessment of heavy metals in sediment and human health risk assessment of heavy metals in fishes in the middle and lower reaches of the Yangtze River basin. Environ. Pollut. 2011, 159, 2575–2585. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Li, M.; Li, H.; Yin, L.Y.; Li, W. Exposure to cadmium causes declines in growth and photosynthesis in the endangered aquatic fern (Ceratopteris pteridoides). Aquat. Bot. 2014, 112, 23–32. [Google Scholar] [CrossRef]
- Sellin, M.K.; Kolok, A.S. Cadmium exposures during early development: DO they lead to reproductive impairment in fathead minnows? Environ. Toxicol. Chem. 2006, 25, 2957–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mebane, C.A.; Hennessy, D.P.; Dillon, F.S. Developing acute-to-chronic toxicity ratios for lead, cadmium, and zinc using rainbow trout, a mayfly, and a midge. Water Air Soil Pollut. 2008, 188, 41–66. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Kholodkevich, S.; Sharov, A.; Chen, C.; Feng, Y. Effects of cadmium on intestinal histology and microbiota in freshwater crayfish (Procambarus clarkii). Chemosphere 2020, 242, 125105. [Google Scholar] [CrossRef]
- Cheng, C.H.; Ma, H.L.; Deng, Y.Q.; Feng, J.; Jie, Y.K.; Guo, Z.X. Oxidative stress, cell cycle arrest, DNA damage and apoptosis in the mud crab (Scylla paramamosain) induced by cadmium exposure. Chemosphere 2021, 263, 128277. [Google Scholar] [CrossRef]
- Duan, Y.F.; Wang, Y.; Huang, J.H.; Li, H.; Dong, H.B.; Zhang, J.S. Toxic effects of cadmium and lead exposure on intestinal histology, oxidative stress response, and microbial community of Pacific white shrimp Litopenaeus vannamei. Mar. Pollut. Bull. 2021, 167, 112220. [Google Scholar] [CrossRef]
- Cho, H.Y.; Reddy, S.P.; Kleeberger, S.R. Nrf2 defends the lung from oxidative stress. Antioxid. Redox Sign. 2006, 8, 76–87. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [Green Version]
- Osburn, W.O.; Kensler, T.W. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat. Res. 2008, 659, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Duan, Y.F.; Huang, J.H.; Wang, J.; Zhou, C.P.; Jiang, S.G.; Lin, H.Z.; Zhang, Z. Characterization and functional study of nuclear factor erythroid 2-related factor 2 (Nrf2) in black tiger shrimp (Penaeus monodon). Fish Shellfish Immun. 2021, 119, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gallagher, E.P. Role of Nrf2 antioxidant defense in mitigating cadmium-induced oxidative stress in the olfactory system of zebrafish. Toxicol. Appl. Pharmacol. 2013, 266, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.D.; Liu, Y.; Jiang, J.; Wu, P.; Feng, L.; Zhou, X.Q. Copper exposure induces toxicity to the antioxidant system via the destruction of Nrf2/ARE signaling and caspase-3-regulated DNA damage in fish muscle: Amelioration by myo-inositol. Aquat. Toxicol. 2015, 159, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Uno, T.; Ishizuka, M.; Itakur, T. Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 2012, 34, 1–13. [Google Scholar] [CrossRef]
- Sun, J.X.; Wang, S.C.; Cao, Y.R.; Wang, S.T.; Li, S. Cadmium exposure induces apoptosis, inflammation and immunosuppression through CYPs activation and antioxidant dysfunction in common carp neutrophils. Fish Shellfish Immun. 2020, 99, 284–290. [Google Scholar]
- Cao, X.; Bi, R.; Song, Y. Toxic responses of cytochrome P450 sub-enzyme activities to heavy metals exposure in soil and correlation with their bioaccumulation in Eisenia fetida. Ecotoxicol. Environ. Saf. 2017, 144, 158–165. [Google Scholar] [CrossRef]
- Isani, G.; Carpen, E. Metallothioneins, unconventional proteins from unconventional animals: A long journey from nematodes to mammals. Biomolecules 2014, 4, 435–457. [Google Scholar] [CrossRef] [Green Version]
- Babula, P.; Masarik, M.; Adam, V.; Eckschlager, T.; Stiborova, M.; Trnkova, L.; Skutkova, H.; Provaznik, I.; Hubalek, J.; Kizek, R. Mammalian metallothioneins: Properties and functions. Metallomics 2012, 4, 739–750. [Google Scholar] [CrossRef]
- Yen Le, T.T.; Zimmermann, S.; Sures, B. How does the metallothionein induction in bivalves meet the criteria for biomarkers of metal exposure? Environ. Pollut. 2016, 212, 257–268. [Google Scholar]
- Zhang, Y.; Li, Z.; Kholodkevich, S.; Sharov, A.; Feng, Y.; Ren, N.; Sun, K. Cadmium-induced oxidative stress, histopathology, and transcriptome changes in the hepatopancreas of freshwater crayfish (Procambarus clarkii). Sci. Total Environ. 2019, 666, 944–955. [Google Scholar] [CrossRef]
- Jia, R.; Han, C.; Lei, J.L.; Liu, B.L.; Huang, B.; Huo, H.H.; Yin, S.T. Effects of nitrite exposure on haematological parameters, oxidative stress and apoptosis in juvenile turbot (Scophthalmus maximus). Aquat. Toxicol. 2015, 169, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Huang, J.J.; Dahms, H.U.; Zhen, J.J.; Ying, X.P. Cell damage and apoptosis in the hepatopancreas of Eriocheir sinensis induced by cadmium. Aquat. Toxicol. 2017, 190, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Fraga, C.G.; Oteiza, P.I.; Galleano, M. In vitro measurements and interpretation of total antioxidant capacity. Biochim. Biophys. Acta-Gen. Subj. 2014, 2, 931–934. [Google Scholar] [CrossRef]
- Sanchez, W.; Burgeot, T.; Porcher, J.M. A novel “Integrated Biomarker Response” calculation based on reference deviation concept. Environ. Sci. Pollut. Res. 2013, 20, 2721–2725. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xia, L.P.; Chen, S.H.; Dahms, H.U.; Ying, X.P.; Peng, X. Cadmium induced oxidative damage and apoptosis in the hepatopancreas of Meretrix meretrix. Ecotoxicology 2016, 25, 959–969. [Google Scholar] [CrossRef]
- Zhou, L.; Li, M.; Zhong, Z.S.; Chen, H.; Wang, X.C.; Wang, M.X.; Xu, Z.; Cao, L.; Lian, C.; Zhang, H.; et al. Biochemical and metabolic responses of the deep-sea mussel Bathymodiolus platifrons to cadmium and copper exposure. Aquat. Toxicol. 2021, 236, 105845. [Google Scholar] [CrossRef]
- Copple, I.M.; Goldring, C.E.; Jenkins, R.E. The hepatotoxic metabolite of acetaminophen directly activates the Keap1-Nrf2 cell defense system. Hepatology 2008, 48, 1292–1301. [Google Scholar] [CrossRef]
- Sara, B.; Cullinan, J.D.; Jin, J.P.; Wade, J.; Alan, D. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 2004, 24, 8477–8486. [Google Scholar]
- Dong, W.X.; Liu, G.; Zhang, K.L.; Tan, Y.; Zou, H.; Yuan, Y.; Gu, J.H.; Song, R.L.; Zhu, J.; Liu, Z.P. Cadmium exposure induces rat proximal tubular cells injury via p62-dependent Nrf2 nucleus translocation mediated activation of AMPK/AKT/mTOR pathway. Ecotoxicol. Environ. Saf. 2021, 214, 112058. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.C.; Liu, J.J.; Klaassen, C.C. Nrf2 activation prevents cadmium-induced acute liver injury. Toxicol. Appl. Pharmacol. 2012, 263, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.D.; Pan, L.Q.; Xu, R.Y.; Si, L.G.; Zhang, X. The molecular mechanism of Nrf2-Keap1 signaling pathway in the antioxidant defense response induced by BaP in the scallop Chlamys farreri. Fish Shellfish Immun. 2019, 92, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Wang, W.N.; Gu, M.M.; Xie, C.Y.; Xiao, Y.C.; Liu, Y.; Wang, L. Essential roles of Cdc42 and MAPK in cadmium-induced apoptosis in Litopenaeus vannamei. Aquat. Toxicol. 2015, 163, 89–96. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; Ma, H.; Liu, G.; Fan, S.; Guo, Z. Mechanism of Cadmium Exposure Induced Hepatotoxicity in the Mud Crab (Scylla paramamosain): Activation of Oxidative Stress and Nrf2 Signaling Pathway. Antioxidants 2022, 11, 978. https://doi.org/10.3390/antiox11050978
Cheng C, Ma H, Liu G, Fan S, Guo Z. Mechanism of Cadmium Exposure Induced Hepatotoxicity in the Mud Crab (Scylla paramamosain): Activation of Oxidative Stress and Nrf2 Signaling Pathway. Antioxidants. 2022; 11(5):978. https://doi.org/10.3390/antiox11050978
Chicago/Turabian StyleCheng, Changhong, Hongling Ma, Guangxin Liu, Sigang Fan, and Zhixun Guo. 2022. "Mechanism of Cadmium Exposure Induced Hepatotoxicity in the Mud Crab (Scylla paramamosain): Activation of Oxidative Stress and Nrf2 Signaling Pathway" Antioxidants 11, no. 5: 978. https://doi.org/10.3390/antiox11050978
APA StyleCheng, C., Ma, H., Liu, G., Fan, S., & Guo, Z. (2022). Mechanism of Cadmium Exposure Induced Hepatotoxicity in the Mud Crab (Scylla paramamosain): Activation of Oxidative Stress and Nrf2 Signaling Pathway. Antioxidants, 11(5), 978. https://doi.org/10.3390/antiox11050978





