Investigation of DHA-Induced Regulation of Redox Homeostasis in Retinal Pigment Epithelium Cells through the Combination of Metabolic Imaging and Molecular Biology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells Culture and Treatments
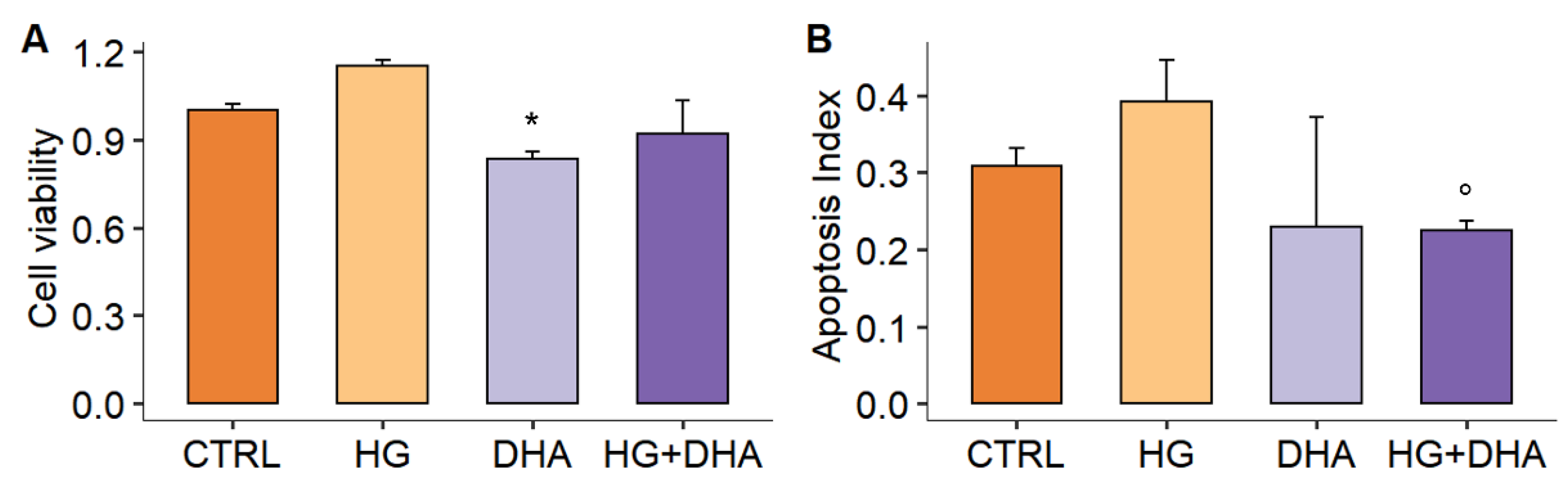
2.2. Cell Viability
2.3. Detection of Apoptosis and Intracellular Levels of Nrf2
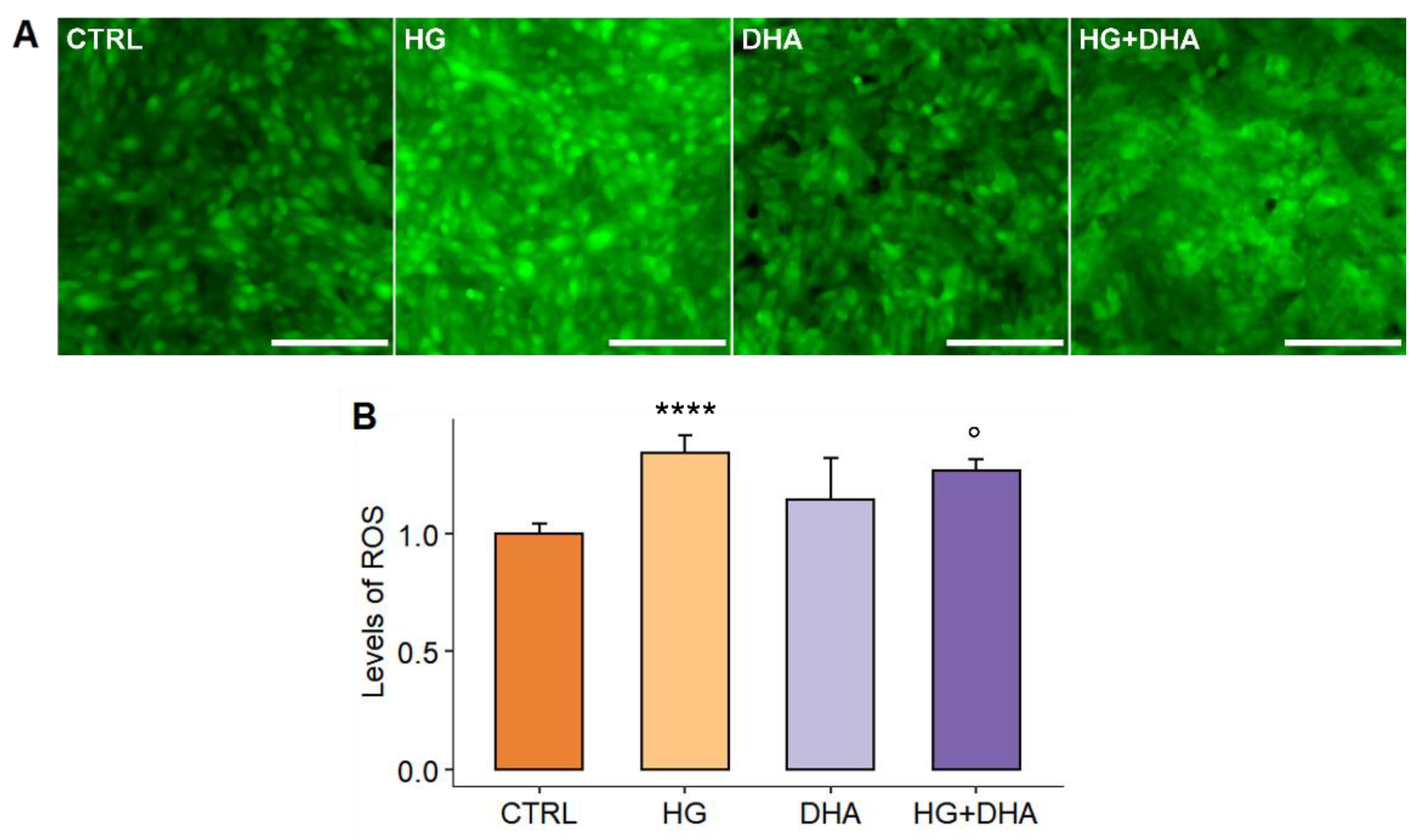
2.4. Quantification of Intracellular Levels of ROS
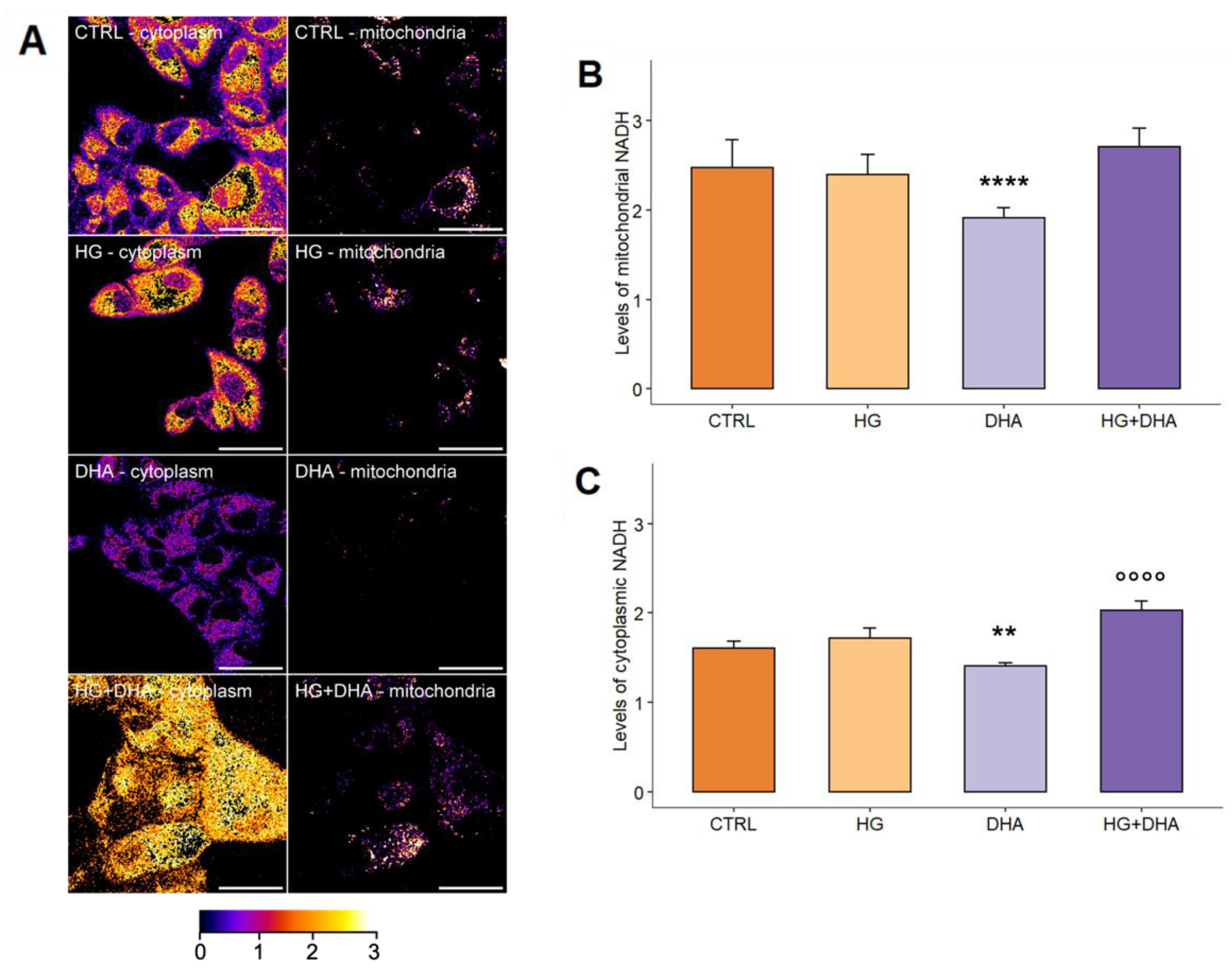
2.5. Metabolic Imaging for the Evaluation of Intracellular Levels of NADH
2.6. Molecular Characterization: RNA Isolation and RT-PCR
- Stage 1 (1 cycle) at 50 °C, for 2 min, followed by 95 °C for 10 min;
- Stage 2 (40 cycles) at 95 °C for 15 s, followed by 60 °C for 1 min; the melt standard curve was at 95 °C for 15 s followed by 60 °C for 1 min, 95 °C for 15 s and finally 60 °C for 15 s.
2.7. Statistical Analysis
3. Results
3.1. Effect of High Glucose on Cell Viability and Apoptosis
3.2. Effect of High-Glucose and DHA on ARPE Redox Homeostasis
3.3. DHA Modulates the High Glucose-Induced Oxidative Stress by Activating the Nrf2-Nqo1-HO Signaling Pathway
3.4. DHA Activates the Production of Intracellular NADH to Promote the Formation of Reductive Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aronson, D.; Edelman, E.R. Coronary Artery Disease and Diabetes Mellitus. Cardiol. Clin. 2014, 32, 439–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiruvoipati, T.; Kielhorn, C.E.; Armstrong, E.J. Peripheral Artery Disease in Patients with Diabetes: Epidemiology, Mechanisms, and Outcomes. World J. Diabetes 2015, 6, 961–969. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2009, 32, S62–S67. [CrossRef] [PubMed] [Green Version]
- Athyros, V.G.; Doumas, M.; Imprialos, K.P.; Stavropoulos, K.; Georgianou, E.; Katsimardou, A.; Karagiannis, A. Diabetes and Lipid Metabolism. Hormones 2018, 17, 61–67. [Google Scholar] [CrossRef] [Green Version]
- King, G.L.; Loeken, M.R. Hyperglycemia-Induced Oxidative Stress in Diabetic Complications. Histochem. Cell Biol. 2004, 122, 333–338. [Google Scholar] [CrossRef]
- Fiorentino, T.V.; Prioletta, A.; Zuo, P.; Folli, F. Hyperglycemia-Induced Oxidative Stress and Its Role in Diabetes Mellitus Related Cardiovascular Diseases. Curr. Pharm. Des. 2013, 19, 5695–5703. [Google Scholar] [CrossRef]
- Ramasamy, R.; Yan, S.F.; Herold, K.; Clynes, R.; Schmidt, A.M. Receptor for Advanced Glycation End Products: Fundamental Roles in the Inflammatory Response: Winding the Way to the Pathogenesis of Endothelial Dysfunction and Atherosclerosis. Ann. N. Y. Acad. Sci. 2008, 1126, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.P.; Bali, A.; Singh, N.; Jaggi, A.S. Advanced Glycation End Products and Diabetic Complications. Korean J. Physiol. Pharmacol. 2014, 18, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.Y.; Kim, Y.S. The Role of Advanced Glycation End Products in Diabetic Vascular Complications. Diabetes Metab. J. 2018, 42, 188–195. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and Classification of Diabetes Mellitus. Diabetes Care 2014, 37 (Suppl. S1), S81–S90. [Google Scholar] [CrossRef] [Green Version]
- Cade, W.T. Diabetes-Related Microvascular and Macrovascular Diseases in the Physical Therapy Setting. Phys. Ther. 2008, 88, 1322–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhruddin, S.; Alanazi, W.; Jackson, K.E. Diabetes-Induced Reactive Oxygen Species: Mechanism of Their Generation and Role in Renal Injury. J. Diabetes Res. 2017, 2017, e8379327. [Google Scholar] [CrossRef] [PubMed]
- Kida, T.; Oku, H.; Osuka, S.; Horie, T.; Ikeda, T. Hyperglycemia-Induced VEGF and ROS Production in Retinal Cells Is Inhibited by the MTOR Inhibitor, Rapamycin. Sci. Rep. 2021, 11, 1885. [Google Scholar] [CrossRef] [PubMed]
- Swanson, M. Retinopathy Screening in Individuals with Type 2 Diabetes: Who, How, How Often, and at What Cost—An Epidemiologic Review. Optometry 2005, 76, 636–646. [Google Scholar] [CrossRef]
- Yang, X.; Yu, X.-W.; Zhang, D.-D.; Fan, Z.-G. Blood-Retinal Barrier as a Converging Pivot in Understanding the Initiation and Development of Retinal Diseases. Chin. Med. J. 2020, 133, 2586–2594. [Google Scholar] [CrossRef]
- Boulton, M.; Dayhaw-Barker, P. The Role of the Retinal Pigment Epithelium: Topographical Variation and Ageing Changes. Eye 2001, 15, 384–389. [Google Scholar] [CrossRef] [Green Version]
- Ablonczy, Z.; Dahrouj, M.; Tang, P.H.; Liu, Y.; Sambamurti, K.; Marmorstein, A.D.; Crosson, C.E. Human Retinal Pigment Epithelium Cells as Functional Models for the RPE In Vivo. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8614–8620. [Google Scholar] [CrossRef]
- Dornan, T.L.; Carter, R.D.; Bron, A.J.; Turner, R.C.; Mann, J.I. Low Density Lipoprotein Cholesterol: An Association with the Severity of Diabetic Retinopathy. Diabetologia 1982, 22, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Su, D.H.W.; Yeo, K.T. Diabetic Retinopathy and Serum Lipids. Singap. Med. J. 2000, 41, 295–297. [Google Scholar]
- Sasongko, M.B.; Wong, T.Y.; Nguyen, T.T.; Kawasaki, R.; Jenkins, A.; Shaw, J.; Wang, J.J. Serum Apolipoprotein AI and B Are Stronger Biomarkers of Diabetic Retinopathy Than Traditional Lipids. Diabetes Care 2011, 34, 474–479. [Google Scholar] [CrossRef] [Green Version]
- Sala-Vila, A.; Díaz-López, A.; Valls-Pedret, C.; Cofán, M.; García-Layana, A.; Lamuela-Raventós, R.-M.; Castañer, O.; Zanon-Moreno, V.; Martinez-Gonzalez, M.A.; Toledo, E.; et al. Dietary Marine ω-3 Fatty Acids and Incident Sight-Threatening Retinopathy in Middle-Aged and Older Individuals With Type 2 Diabetes: Prospective Investigation From the PREDIMED Trial. JAMA Ophthalmol. 2016, 134, 1142–1149. [Google Scholar] [CrossRef] [PubMed]
- Opreanu, M.; Tikhonenko, M.; Bozack, S.; Lydic, T.A.; Reid, G.E.; McSorley, K.M.; Sochacki, A.; Perez, G.I.; Esselman, W.J.; Kern, T.; et al. The Unconventional Role of Acid Sphingomyelinase in Regulation of Retinal Microangiopathy in Diabetic Human and Animal Models. Diabetes 2011, 60, 2370–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Jump, D.B.; Esselman, W.J.; Busik, J.V. Inhibition of Cytokine Signaling in Human Retinal Endothelial Cells through Modification of Caveolae/Lipid Rafts by Docosahexaenoic Acid. Investig. Ophthalmol. Vis. Sci. 2007, 48, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Connor, K.M.; SanGiovanni, J.P.; Lofqvist, C.; Aderman, C.M.; Chen, J.; Higuchi, A.; Hong, S.; Pravda, E.A.; Majchrzak, S.; Carper, D.; et al. Increased Dietary Intake of Omega-3-Polyunsaturated Fatty Acids Reduces Pathological Retinal Angiogenesis. Nat. Med. 2007, 13, 868–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tikhonenko, M.; Lydic, T.A.; Wang, Y.; Chen, W.; Opreanu, M.; Sochacki, A.; McSorley, K.M.; Renis, R.L.; Kern, T.; Jump, D.B.; et al. Remodeling of Retinal Fatty Acids in an Animal Model of Diabetes: A Decrease in Long-Chain Polyunsaturated Fatty Acids Is Associated with a Decrease in Fatty Acid Elongases Elovl2 and Elovl4. Diabetes 2010, 59, 219–227. [Google Scholar] [CrossRef] [Green Version]
- Hassinen, I.; Chance, B. Oxidation-Reduction Properties of the Mitochondrial Flavoprotein Chain. Biochem. Biophys. Res. Commun. 1968, 31, 895–900. [Google Scholar] [CrossRef]
- Bianchetti, G.; Ciccarone, F.; Ciriolo, M.R.; De Spirito, M.; Pani, G.; Maulucci, G. Label-Free Metabolic Clustering through Unsupervised Pixel Classification of Multiparametric Fluorescent Images. Anal. Chim. Acta 2021, 1148, 238173. [Google Scholar] [CrossRef]
- Maulucci, G.; Labate, V.; Mele, M.; Panieri, E.; Arcovito, G.; Galeotti, T.; Østergaard, H.; Winther, J.R.; De Spirito, M.; Pani, G. High-Resolution Imaging of Redox Signaling in Live Cells through an Oxidation-Sensitive Yellow Fluorescent Protein. Sci. Signal. 2008, 1, pl3. [Google Scholar] [CrossRef]
- Maulucci, G.; Bačić, G.; Bridal, L.; Schmidt, H.H.; Tavitian, B.; Viel, T.; Utsumi, H.; Yalçın, A.S.; De Spirito, M. Imaging Reactive Oxygen Species-Induced Modifications in Living Systems. Antioxid. Redox Signal. 2016, 24, 939–958. [Google Scholar] [CrossRef]
- Koch, O.R.; Fusco, S.; Ranieri, S.C.; Maulucci, G.; Palozza, P.; Larocca, L.M.; Cravero, A.A.M.; Farre’, S.M.; De Spirito, M.; Galeotti, T.; et al. Role of the Life Span Determinant P66shcA in Ethanol-Induced Liver Damage. Lab. Investig. 2008, 88, 750–760. [Google Scholar] [CrossRef] [Green Version]
- Chia, T.H.; Williamson, A.; Spencer, D.D.; Levene, M.J. Multiphoton Fluorescence Lifetime Imaging of Intrinsic Fluorescence in Human and Rat Brain Tissue Reveals Spatially Distinct NADH Binding. Opt. Express 2008, 16, 4237–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maulucci, G.; Troiani, D.; Eramo, S.L.M.; Paciello, F.; Podda, M.V.; Paludetti, G.; Papi, M.; Maiorana, A.; Palmieri, V.; De Spirito, M.; et al. Time Evolution of Noise Induced Oxidation in Outer Hair Cells: Role of NAD(P)H and Plasma Membrane Fluidity. Biochim. Biophys. Acta 2014, 1840, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Balu, M.; Mazhar, A.; Hayakawa, C.K.; Mittal, R.; Krasieva, T.B.; König, K.; Venugopalan, V.; Tromberg, B.J. In Vivo Multiphoton NADH Fluorescence Reveals Depth-Dependent Keratinocyte Metabolism in Human Skin. Biophys. J. 2013, 104, 258–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clementi, M.E.; Pani, G.; Sampaolese, B.; Tringali, G. Punicalagin Reduces H2O2-Induced Cytotoxicity and Apoptosis in PC12 Cells by Modulating the Levels of Reactive Oxygen Species. Nutr. Neurosci. 2018, 21, 447–454. [Google Scholar] [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Bianchetti, G.; Di Giacinto, F.; De Spirito, M.; Maulucci, G. Machine-Learning Assisted Confocal Imaging of Intracellular Sites of Triglycerides and Cholesteryl Esters Formation and Storage. Anal. Chim. Acta 2020, 1121, 57–66. [Google Scholar] [CrossRef]
- Bianchetti, G.; Viti, L.; Scupola, A.; Di Leo, M.; Tartaglione, L.; Flex, A.; De Spirito, M.; Pitocco, D.; Maulucci, G. Erythrocyte Membrane Fluidity as a Marker of Diabetic Retinopathy in Type 1 Diabetes Mellitus. Eur. J. Clin. Investig. 2021, 51, e13455. [Google Scholar] [CrossRef]
- Bianchetti, G.; Taralli, S.; Vaccaro, M.; Indovina, L.; Mattoli, M.V.; Capotosti, A.; Scolozzi, V.; Calcagni, M.L.; Giordano, A.; De Spirito, M.; et al. Automated Detection and Classification of Tumor Histotypes on Dynamic PET Imaging Data through Machine-Learning Driven Voxel Classification. Comput. Biol. Med. 2022, 145, 105423. [Google Scholar] [CrossRef]
- Patterson, G.H.; Knobel, S.M.; Arkhammar, P.; Thastrup, O.; Piston, D.W. Separation of the Glucose-Stimulated Cytoplasmic and Mitochondrial NAD(P)H Responses in Pancreatic Islet Beta Cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5203–5207. [Google Scholar] [CrossRef] [Green Version]
- Clementi, M.E.; Pizzoferrato, M.; Bianchetti, G.; Brancato, A.; Sampaolese, B.; Maulucci, G.; Tringali, G. Cytoprotective Effect of Idebenone through Modulation of the Intrinsic Mitochondrial Pathway of Apoptosis in Human Retinal Pigment Epithelial Cells Exposed to Oxidative Stress Induced by Hydrogen Peroxide. Biomedicines 2022, 10, 503. [Google Scholar] [CrossRef]
- Villarroel, M.; García-Ramírez, M.; Corraliza, L.; Hernández, C.; Simó, R. Effects of High Glucose Concentration on the Barrier Function and the Expression of Tight Junction Proteins in Human Retinal Pigment Epithelial Cells. Exp. Eye Res. 2009, 89, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Clementi, M.; Lazzarino, G.; Sampaolese, B.; Brancato, A.; Tringali, G. DHA Protects PC12 Cells against Oxidative Stress and Apoptotic Signals through the Activation of the NFE2L2/HO-1 Axis. Int. J. Mol. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Molz, P.; Schröder, N. Potential Therapeutic Effects of Lipoic Acid on Memory Deficits Related to Aging and Neurodegeneration. Front. Pharm. 2017, 8, 849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, R.K.; Singh, M.; Roy, S.; Ansari, M.N.; Saeedan, A.S.; Kaithwas, G. Modulation of Oxidative Stress Response by Flaxseed Oil: Role of Lipid Peroxidation and Underlying Mechanisms. Prostaglandins Other Lipid Mediat. 2018, 135, 21–26. [Google Scholar] [CrossRef]
- Guo, R.; Li, W.; Liu, B.; Li, S.; Zhang, B.; Xu, Y. Resveratrol Protects Vascular Smooth Muscle Cells against High Glucose-Induced Oxidative Stress and Cell Proliferation in Vitro. Med. Sci. Monit. Basic Res. 2014, 20, 82–92. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.-M.; Ran, F.; Ni, H.-Z.; Sun, L.-L.; Xiao, L.; Li, X.-Q.; Li, W.-D. Metformin Inhibits High Glucose-Induced Smooth Muscle Cell Proliferation and Migration. Aging 2020, 12, 5352–5361. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. The Bcl-2-Regulated Apoptosis Switch: Mechanism and Therapeutic Potential. Curr. Opin. Immunol. 2007, 19, 488–496. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of Apoptosis Signalling Pathways by Reactive Oxygen Species. Biochim. Biophys. Acta 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Jiang, S.; Deng, C.; Lv, J.; Fan, C.; Hu, W.; Di, S.; Yan, X.; Ma, Z.; Liang, Z.; Yang, Y. Nrf2 Weaves an Elaborate Network of Neuroprotection Against Stroke. Mol. Neurobiol. 2017, 54, 1440–1455. [Google Scholar] [CrossRef]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.-L. The Keap1-Nrf2 Pathway: Mechanisms of Activation and Dysregulation in Cancer. Redox Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasik, U.; Milkiewicz, M.; Kempinska-Podhorodecka, A.; Milkiewicz, P. Protection against Oxidative Stress Mediated by the Nrf2/Keap1 Axis Is Impaired in Primary Biliary Cholangitis. Sci. Rep. 2017, 7, 44769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prochaska, H.J.; Santamaria, A.B.; Talalay, P. Rapid Detection of Inducers of Enzymes That Protect against Carcinogens. Proc. Natl. Acad. Sci. USA 1992, 89, 2394–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, P.; Xie, T.; Xu, Y.; Jaiswal, A.K. NAD(P)H:Quinone Oxidoreductase1 (DT-Diaphorase): Expression, Regulation, and Role in Cancer. Oncol. Res. 1994, 6, 525–532. [Google Scholar]
- Vasiliou, V.; Theurer, M.J.; Puga, A.; Reuter, S.F.; Nebert, D.W. Mouse Dioxin-Inducible NAD(P)H: Menadione Oxidoreductase: NMO1 CDNA Sequence and Genetic Differences in MRNA Levels. Pharmacogenetics 1994, 4, 341–348. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Regulation of Genes Encoding NAD(P)H:Quinone Oxidoreductases. Free Radic Biol. Med. 2000, 29, 254–262. [Google Scholar] [CrossRef]
- Thor, H.; Smith, M.T.; Hartzell, P.; Bellomo, G.; Jewell, S.A.; Orrenius, S. The Metabolism of Menadione (2-Methyl-1,4-Naphthoquinone) by Isolated Hepatocytes. A Study of the Implications of Oxidative Stress in Intact Cells. J. Biol. Chem. 1982, 257, 12419–12425. [Google Scholar] [CrossRef]
- Lee, E.-J.; Ko, H.-M.; Jeong, Y.-H.; Park, E.-M.; Kim, H.-S. β-Lapachone Suppresses Neuroinflammation by Modulating the Expression of Cytokines and Matrix Metalloproteinases in Activated Microglia. J. Neuroinflammation 2015, 12, 133. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-L.; Varner, S.E.; Rao, A.S.; Grey, J.Y.; Thomas, S.; Cook, C.K.; Wasserman, M.A.; Medford, R.M.; Jaiswal, A.K.; Kunsch, C. Laminar Flow Induction of Antioxidant Response Element-Mediated Genes in Endothelial Cells. A Novel Anti-Inflammatory Mechanism. J. Biol. Chem. 2003, 278, 703–711. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-O.; Chang, Y.-C.; Whang, K.; Kim, C.-H.; Lee, I.-S. Role of NAD(P)H:Quinone Oxidoreductase 1 on Tumor Necrosis Factor-Alpha-Induced Migration of Human Vascular Smooth Muscle Cells. Cardiovasc. Res. 2007, 76, 331–339. [Google Scholar] [CrossRef] [Green Version]
- Hur, K.Y.; Kim, S.H.; Choi, M.-A.; Williams, D.R.; Lee, Y.; Kang, S.W.; Yadav, U.C.S.; Srivastava, S.K.; Jung, M.; Cho, J.W.; et al. Protective Effects of Magnesium Lithospermate B against Diabetic Atherosclerosis via Nrf2-ARE-NQO1 Transcriptional Pathway. Atherosclerosis 2010, 211, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.J.; Collier, A.C.; Bowen, L.D.; Pritsos, K.L.; Goodrich, G.G.; Arger, K.; Cutter, G.; Pritsos, C.A. Polymorphisms in the NQO1, GSTT and GSTM Genes Are Associated with Coronary Heart Disease and Biomarkers of Oxidative Stress. Mutat. Res. 2009, 674, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Kensler, T.W. New Player on An Old Field; the Keap1/Nrf2 Pathway as a Target for Treatment of Type 2 Diabetes and Metabolic Syndrome. Curr. Diabetes Rev. 2013, 9, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Li, Y. NAD(P)H: Quinone Oxidoreductase 1 and Its Potential Protective Role in Cardiovascular Diseases and Related Conditions. Cardiovasc. Toxicol. 2012, 12, 39–45. [Google Scholar] [CrossRef] [PubMed]

| Gene Target | Accession Code | Primer Sequence Forward (5′ to 3′) | Primer Sequence Reverse (5′ to 3′) |
|---|---|---|---|
| Β-Actin | NM_001101.5 | AAACTGGAACGGTGAAGGTG | GTGGCTTTTAGGATGGCAAG |
| Nrf2 | NM_006164.4 | GTCACATCGAGAGCCCAGTC | ACCATGGTAGTCTCAACCAGC |
| Nqo1 | X06985.1 | GGTTTGAGCGAGTGTTCATAGG | CAGAGAGTACATGGAGCCAC |
| HO-1 | J03934.1 | CTGGAGGAGGAGATTGAGCG | ATGGCTGGTGTGTAGGGGAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchetti, G.; Clementi, M.E.; Sampaolese, B.; Serantoni, C.; Abeltino, A.; De Spirito, M.; Sasson, S.; Maulucci, G. Investigation of DHA-Induced Regulation of Redox Homeostasis in Retinal Pigment Epithelium Cells through the Combination of Metabolic Imaging and Molecular Biology. Antioxidants 2022, 11, 1072. https://doi.org/10.3390/antiox11061072
Bianchetti G, Clementi ME, Sampaolese B, Serantoni C, Abeltino A, De Spirito M, Sasson S, Maulucci G. Investigation of DHA-Induced Regulation of Redox Homeostasis in Retinal Pigment Epithelium Cells through the Combination of Metabolic Imaging and Molecular Biology. Antioxidants. 2022; 11(6):1072. https://doi.org/10.3390/antiox11061072
Chicago/Turabian StyleBianchetti, Giada, Maria Elisabetta Clementi, Beatrice Sampaolese, Cassandra Serantoni, Alessio Abeltino, Marco De Spirito, Shlomo Sasson, and Giuseppe Maulucci. 2022. "Investigation of DHA-Induced Regulation of Redox Homeostasis in Retinal Pigment Epithelium Cells through the Combination of Metabolic Imaging and Molecular Biology" Antioxidants 11, no. 6: 1072. https://doi.org/10.3390/antiox11061072









