Over-Expression of Dehydroascorbate Reductase Improves Salt Tolerance, Environmental Adaptability and Productivity in Oryza sativa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vector Construction and Agrobacterium-Mediated Rice Transformation
2.2. Plant Materials and Growing Conditions
2.3. Copy Number and Location of the OsDHAR1 Transgene
2.4. Measurement of the Ascorbate Content
2.5. Enzyme Activity Assay
2.6. Redox State Analysis
2.7. Ion Leakage and Chlorophyll Fluorescence Analysis under Conditions of MV Treatment
2.8. Microarray-Based Gene Expression Profiling
2.9. Estimation of Agronomic Traits in Paddy Fields
2.10. Data Analysis
3. Results
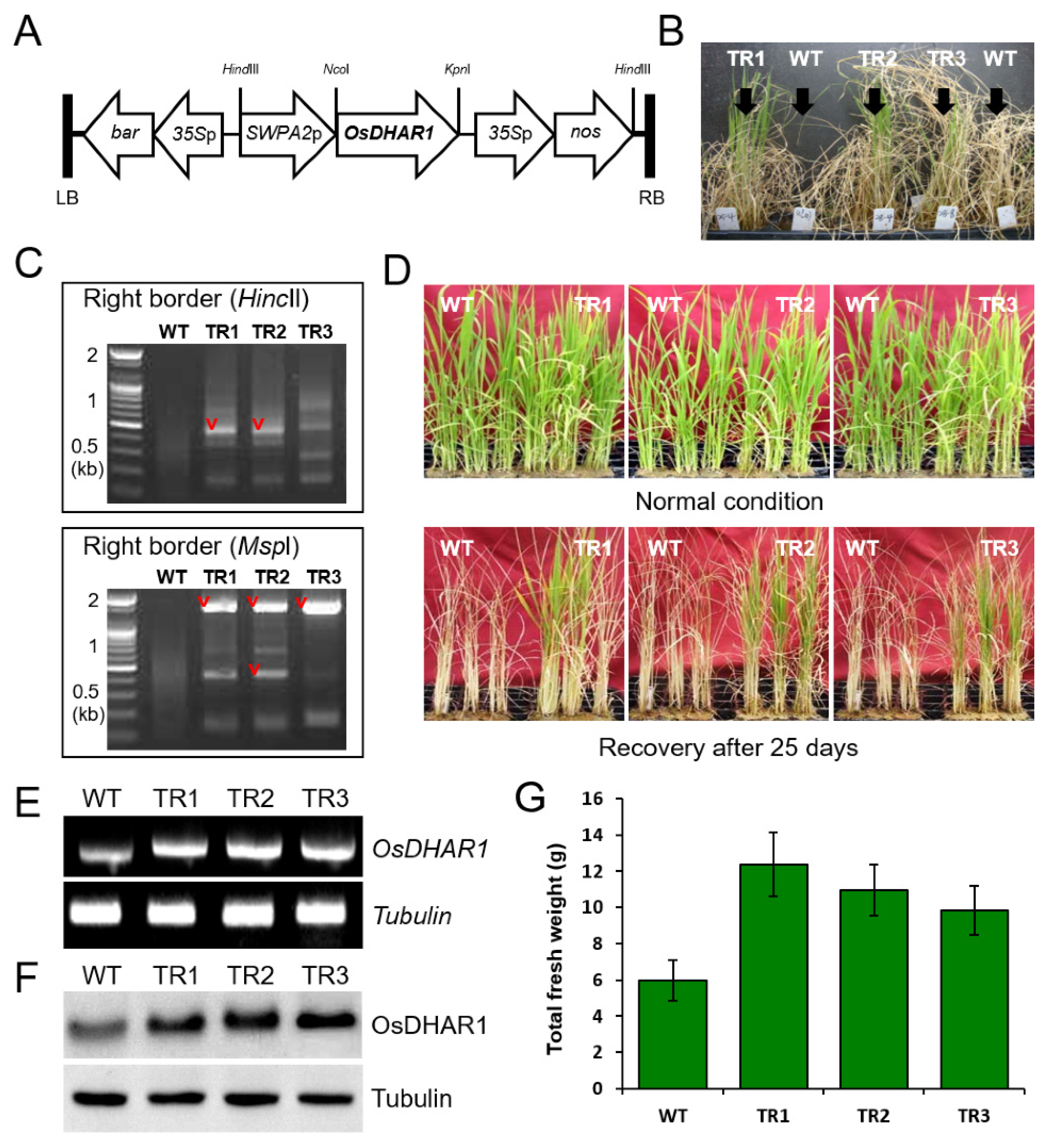
3.1. Development of SWPA2::OsDHAR1 Transgenic Rice Plants
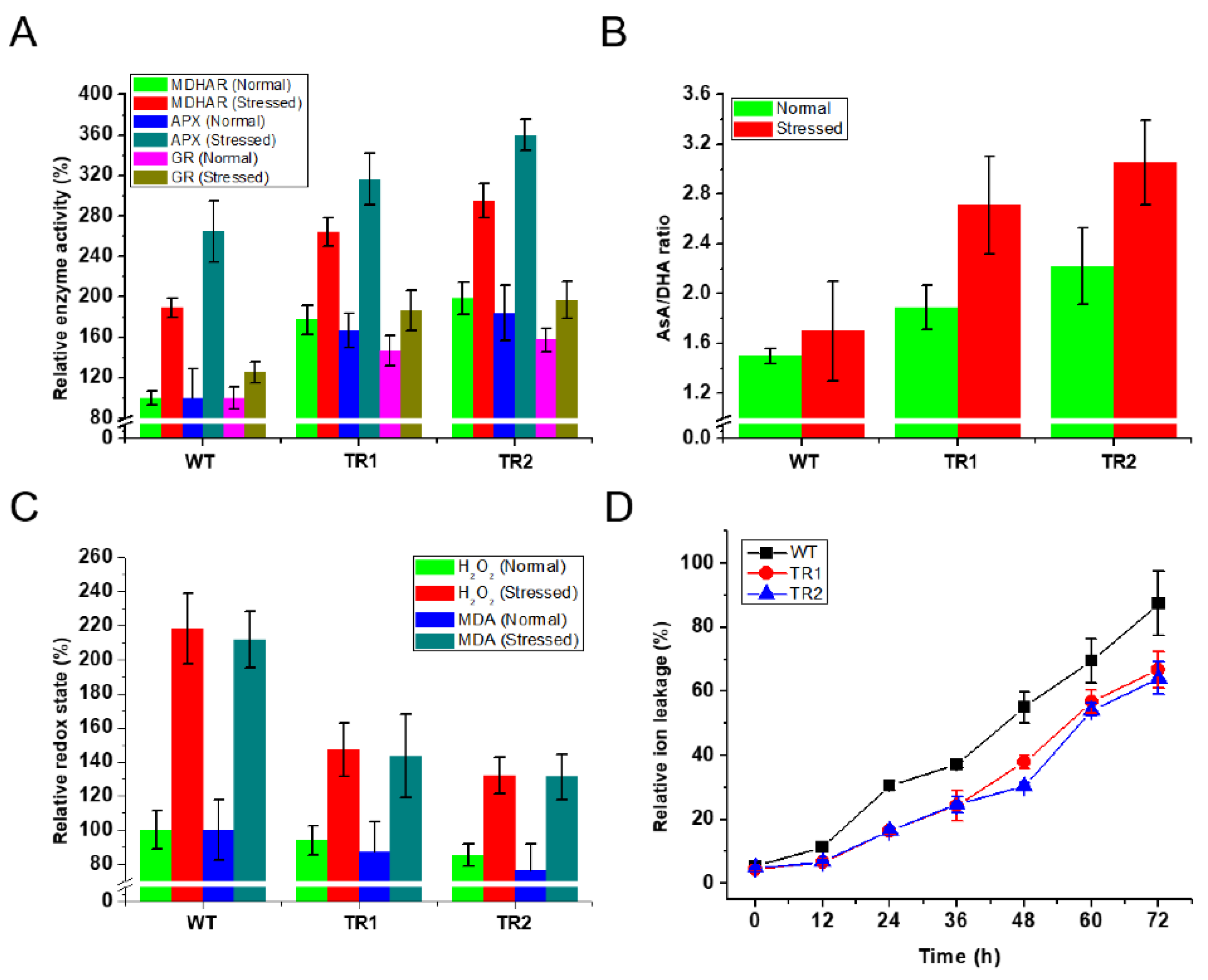
3.2. Performance of OsDHAR1-Expressing Transgenic Plants under Salt Stress
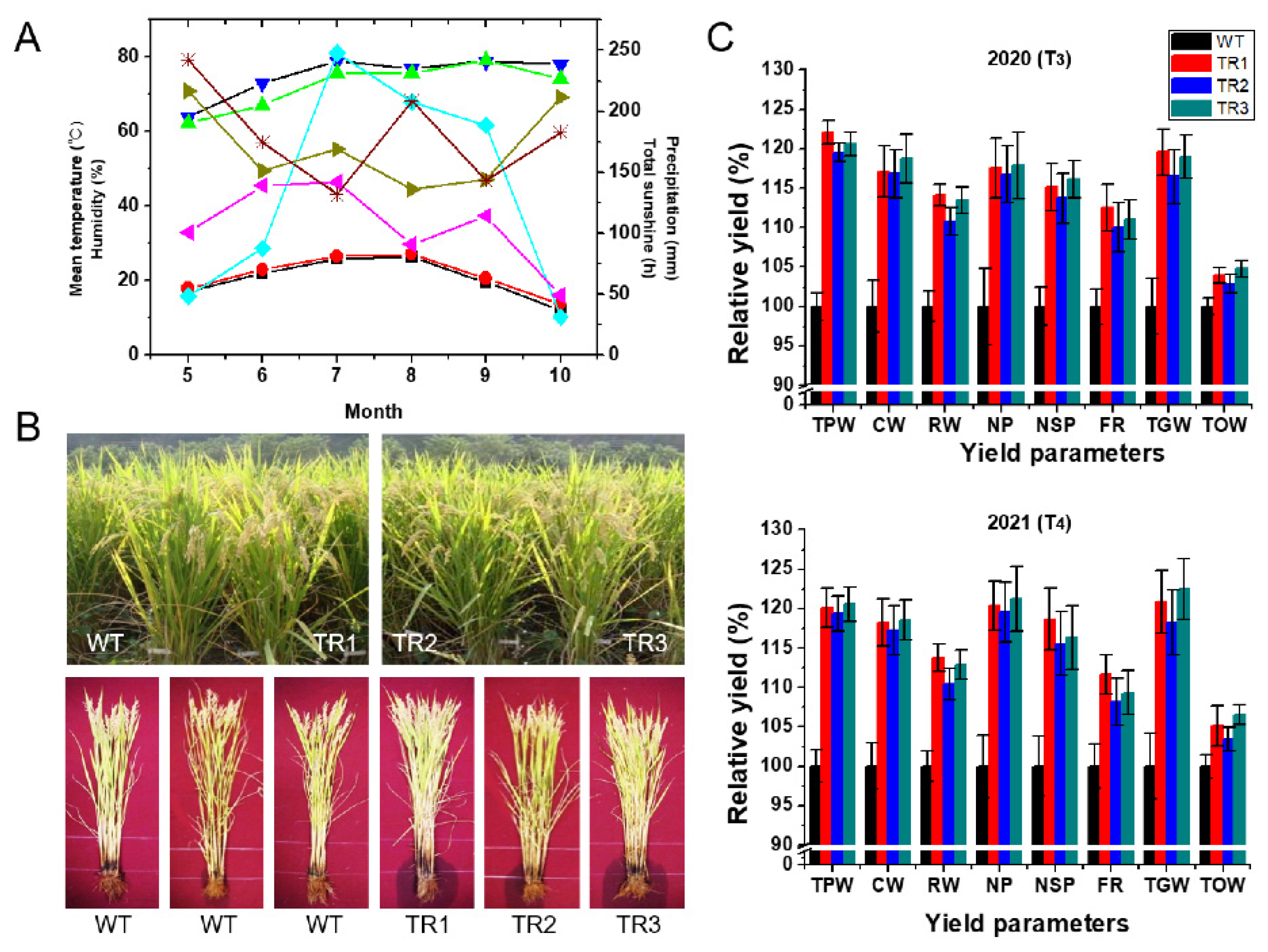
3.3. Agronomic Traits of OsDHAR1-Expressing Transgenic Plants
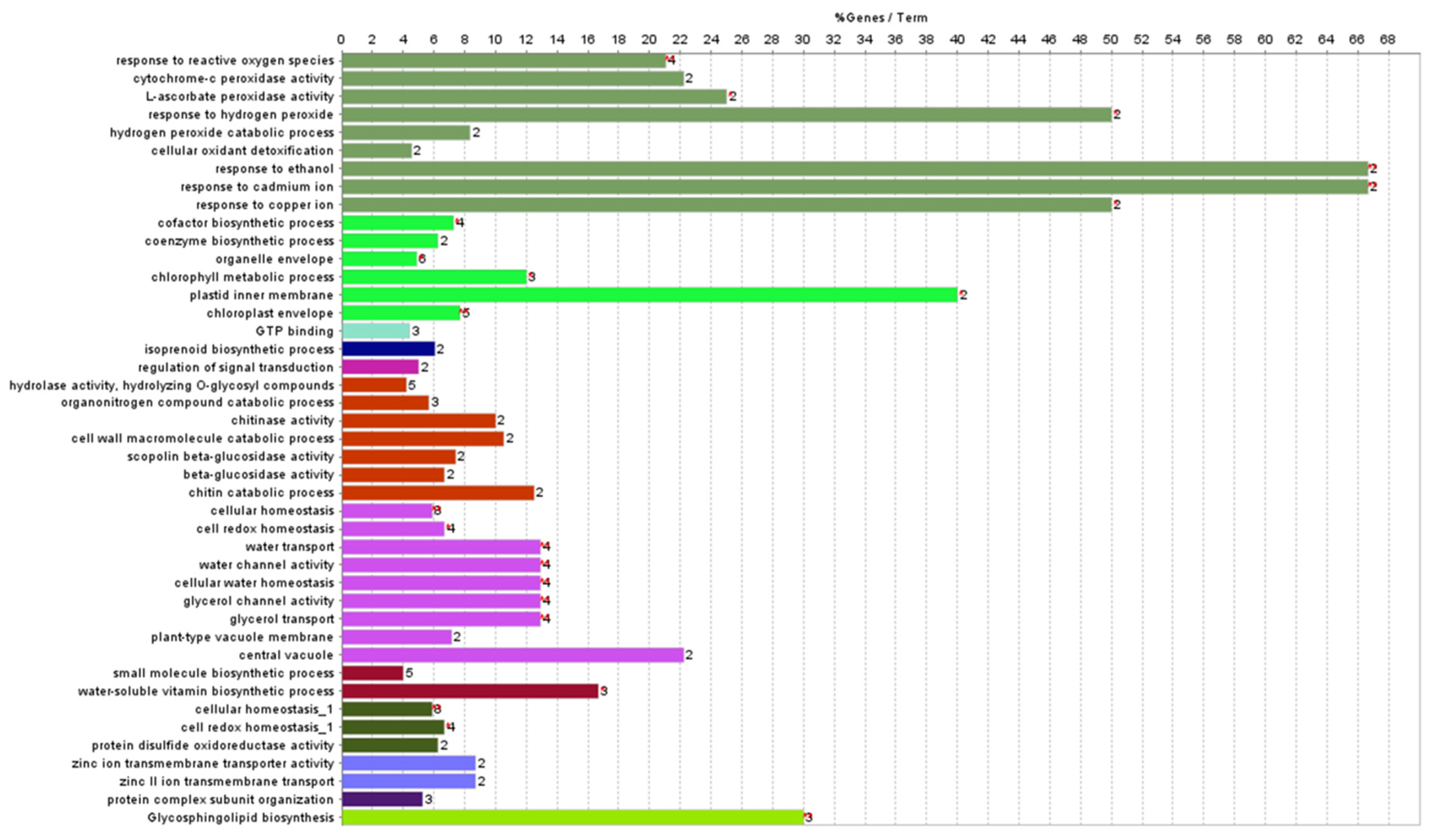
3.4. Effect of OsDHAR1 Overexpression on Overall Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hasegawa, T.; Yin, X.; Zhu, Y.; Boote, K.; Adam, M.; Bregaglio, S.; Buis, S.; Confalonieri, R.; Fumoto, T.; et al. Uncertainties in predicting rice yield by current crop models under a wide range of climatic conditions. Glob. Chang. Biol. 2015, 21, 1328–1341. [Google Scholar] [CrossRef] [PubMed]
- Pittelkow, C.M.; Adviento-Borbe, M.A.; van Kessel, C.; Hill, J.E.; Linquist, B.A. Optimizing rice yields while minimizing yield-scaled global warming potential. Glob. Chang. Biol. 2014, 20, 1382–1393. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.J.; Shao, H.B.; Teixeira da Silva, J.A. A critical review on the improvement of photosynthetic carbon assimilation in C3 plants using genetic engineering. Crit. Rev. Biotechnol. 2012, 32, 1–21. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, N.A.; Reddy, P.S.; Masood, A.; Hasanuzzaman, M.; Khan, M.I.; Anjum, N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: Phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017, 115, 126–140. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Cheung, C.Y.; Mohanty, B.; Lee, D.Y. Modeling rice metabolism: From elucidating environmental effects on cellular phenotype to guiding crop improvement. Front. Plant Sci. 2016, 7, 1795. [Google Scholar] [CrossRef] [Green Version]
- Asada, K. The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1419–1431. [Google Scholar] [CrossRef] [Green Version]
- Hossain, M.A.; Asada, K. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J. Biol. Chem. 1985, 260, 12920–12926. [Google Scholar] [CrossRef]
- Yoon, H.S.; Lee, H.; Lee, I.A.; Kim, K.Y.; Jo, J. Molecular cloning of the monodehydroascorbate reductase gene from Brassica campestris and analysis of its mRNA level in response to oxidative stress. Biochim. Biophys. Acta 2004, 1658, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Deutsch, J.C. Dehydroascorbic acid. J. Chromatogr. A 2000, 881, 299–307. [Google Scholar] [CrossRef]
- Shimaoka, T.; Yokota, A.; Miyake, C. Purification and characterization of chloroplast dehydroascorbate reductase from spinach leaves. Plant Cell Physiol. 2000, 41, 1110–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urano, J.; Nakagawa, T.; Maki, Y.; Masumura, T.; Tanaka, K.; Murata, N.; Ushimaru, T. Molecular cloning and characterization of a rice dehydroascorbate reductase. FEBS Lett. 2000, 466, 107–111. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Tamaoki, M.; Shikano, T.; Nakajima, N.; Ogawa, D.; Ioki, M.; Aono, M.; Kubo, A.; Kamada, H.; Inoue, Y.; et al. Cytosolic dehydroascorbate reductase is important for ozone tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.C.; Huang, C.Y.; Wen, L.; Lin, C.T. Dehydroascorbate reductase cDNA from sweet potato (Ipomoea batatas [L.] Lam): Expression, enzyme properties, and kinetic studies. J. Agric. Food Chem. 2008, 56, 3623–3627. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gallie, D.R. Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 2006, 142, 775–787. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.Y.; Choi, S.M.; Ahn, Y.O.; Lee, H.S.; Lee, H.B.; Park, Y.M.; Kwak, S.S. Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J. Plant Physiol. 2003, 160, 347–353. [Google Scholar] [CrossRef]
- Chew, O.; Whelan, J.; Millar, A.H. Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 2003, 278, 46869–46877. [Google Scholar] [CrossRef] [Green Version]
- Dixon, D.P.; Davis, B.G.; Edwards, R. Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 30859–30869. [Google Scholar] [CrossRef] [Green Version]
- Ushimaru, T.; Nakagawa, T.; Fujioka, Y.; Daicho, K.; Naito, M.; Yamauchi, Y.; Nonaka, H.; Amako, K.; Yamawaki, K.; Murata, N. Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. J. Plant Physiol. 2006, 163, 1179–1184. [Google Scholar] [CrossRef]
- Shin, S.Y.; Kim, I.S.; Kim, Y.H.; Park, H.M.; Lee, J.Y.; Kang, H.G.; Yoon, H.S. Scavenging reactive oxygen species by rice dehydroascorbate reductase alleviates oxidative stresses in Escherichia coli. Mol. Cells 2008, 26, 616–620. [Google Scholar]
- Jubany-Mari, T.; Munne-Bosch, S.; Alegre, L. Redox regulation of water stress responses in field-grown plants. Role of hydrogen peroxide and ascorbate. Plant Physiol. Biochem. 2010, 48, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Antonio, B.; Namiki, N.; Motoyama, R.; Sugimoto, K.; Takehisa, H.; Minami, H.; Kamatsuki, K.; Kusaba, M.; Hirochika, H.; et al. Field transcriptome revealed critical developmental and physiological transitions involved in the expression of growth potential in japonica rice. BMC Plant Biol. 2011, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Do, H.; Kim, I.S.; Jeon, B.W.; Lee, C.W.; Park, A.K.; Wi, A.R.; Shin, S.C.; Park, H.; Kim, Y.S.; Yoon, H.S.; et al. Structural understanding of the recycling of oxidized ascorbate by dehydroascorbate reductase (OsDHAR) from Oryza sativa L. japonica. Sci. Rep. 2016, 6, 19498. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.I.; Kim, J.J.; Shin, S.Y.; Kim, Y.S.; Yoon, H.S. ASR enhances environmental stress tolerance and improves grain yield by modulating stomatal closure in rice. Front. Plant Sci. 2020, 10, 1752. [Google Scholar] [CrossRef]
- Gillespie, K.M.; Ainsworth, E.A. Measurement of reduced, oxidized and total ascorbate content in plants. Nat. Protoc. 2007, 2, 871–874. [Google Scholar] [CrossRef]
- Eltayeb, A.E.; Kawano, N.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Dalton, D.A.; Langeberg, L.; Robbins, M. Purification and characterization of monodehydroascorbate reductase from soybean root nodules. Arch. Biochem. Biophys. 1992, 292, 281–286. [Google Scholar] [CrossRef]
- Foyer, C.H.; Halliwell, B. The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef]
- Maksimovic, J.J.; Zivanovic, B.D. Quantification of the antioxidant activity in salt-stressed tissues. Methods Mol. Biol. 2012, 913, 237–250. [Google Scholar] [PubMed]
- Nourooz-Zadeh, J. Ferrous ion oxidation in presence of xylenol orange for detection of lipid hydroperoxides in plasma. Methods Enzymol. 1999, 300, 58–62. [Google Scholar] [PubMed]
- Draper, H.H.; Hadley, M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990, 186, 421–431. [Google Scholar] [PubMed]
- Esterbauer, H.; Cheeseman, K.H. Determination of aldehydic lipid peroxidation products: Malonaldehyde and 4-hydroxynonenal. Methods Enzymol. 1990, 186, 407–421. [Google Scholar]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio. Protoc. 2012, 2, e263. [Google Scholar] [CrossRef] [Green Version]
- Hwang, J.E.; Lim, C.J.; Chen, H.; Je, J.; Song, C.; Lim, C.O. Overexpression of Arabidopsis dehydration- responsive element-binding protein 2C confers tolerance to oxidative stress. Mol. Cells 2012, 33, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.R.; Rosenqvist, E. Applications of chlorophyll fluorescence can improve crop production strategies: An examination of future possibilities. J. Exp. Bot. 2004, 55, 1607–1621. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6, 712. [Google Scholar] [CrossRef] [Green Version]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef]
- Roy, S.J.; Negrao, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Abogadallah, G.M. Antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-A potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Noshi, M.; Hatanaka, R.; Tanabe, N.; Terai, Y.; Maruta, T.; Shigeoka, S. Redox regulation of ascorbate and glutathione by a chloroplastic dehydroascorbate reductase is required for high-light stress tolerance in Arabidopsis. Biosci. Biotechnol. Biochem. 2016, 80, 870–877. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.T.; Chiou, C.W.; Chu, Y.L.; Hsiao, Y.; Tseng, Y.F.; Chen, Y.C.; Chen, H.J.; Chang, H.Y.; Lee, T.M. Enhanced ascorbate regeneration via dehydroascorbate reductase confers tolerance to photo-oxidative stress in Chlamydomonas reinhardtii. Plant Cell Physiol. 2016, 57, 2104–2121. [Google Scholar] [CrossRef] [Green Version]
- Gallie, D.R. The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [Green Version]
- Noshi, M.; Yamada, H.; Hatanaka, R.; Tanabe, N.; Tamoi, M.; Shigeoka, S. Arabidopsis dehydroascorbate reductase 1 and 2 modulate redox states of ascorbate-glutathione cycle in the cytosol in response to photooxidative stress. Biosci. Biotechnol. Biochem. 2017, 81, 523–533. [Google Scholar] [CrossRef] [Green Version]
- Abogadallah, G.M.; Serag, M.M.; Quick, W.P. Fine and coarse regulation of reactive oxygen species in the salt tolerant mutants of barnyard grass and their wild-type parents under salt stress. Physiol. Plant 2010, 138, 60–73. [Google Scholar] [CrossRef]
- Younis, M.E.; Hasaneen, M.N.; Kazamel, A.M. Exogenously applied ascorbic acid ameliorates detrimental effects of NaCl and mannitol stress in Vicia faba seedlings. Protoplasma 2010, 239, 39–48. [Google Scholar] [CrossRef]
- Zhang, J.; Kirkham, M.B. Antioxidant responses to drought in sunflower and sorghum seedlings. New Phytol. 1996, 132, 361–373. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Jubany-Mari, T.; Alegre-Batlle, L.; Jiang, K.; Feldman, L.J. Use of a redox-sensing GFP (c-roGFP1) for real-time monitoring of cytosol redox status in Arabidopsis thaliana water-stressed plants. FEBS Lett. 2010, 584, 889–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallie, D.R. L-ascorbic Acid: A multifunctional molecule supporting plant growth and development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Gallie, D.R. The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 2004, 16, 1143–1162. [Google Scholar] [CrossRef] [Green Version]
- Zha, X.; Luo, X.; Qian, X.; He, G.; Yang, M.; Li, Y.; Yang, J. Over-expression of the rice LRK1 gene improves quantitative yield components. Plant Biotechnol. J. 2009, 7, 611–620. [Google Scholar] [CrossRef]
- Zou, X.; Qin, Z.; Zhang, C.; Liu, B.; Liu, J.; Zhang, C.; Lin, C.; Li, H.; Zhao, T. Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J. Exp. Bot. 2015, 66, 7197–7209. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Lu, Q.; Wen, X.; Lu, C. Enhanced sucrose loading improves rice yield by increasing grain size. Plant Physiol. 2015, 169, 2848–2862. [Google Scholar] [CrossRef] [Green Version]
- Park, K.Y.; Kim, E.Y.; Seo, Y.S.; Kim, W.T. Constitutive expression of CaPLA1 conferred enhanced growth and grain yield in transgenic rice plants. Plant Mol. Biol. 2016, 90, 517–532. [Google Scholar] [CrossRef]
- Li, T.; Jiang, J.; Zhang, S.; Shu, H.; Wang, Y.; Lai, J.; Du, J.; Yang, C. OsAGSW1, an ABC1-like kinase gene, is involved in the regulation of grain size and weight in rice. J. Exp. Bot. 2015, 66, 5691–5701. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. USA 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srikanth, B.; Subhakara Rao, I.; Surekha, K.; Subrahmanyam, D.; Voleti, S.R.; Neeraja, C.N. Enhanced expression of OsSPL14 gene and its association with yield components in rice (Oryza sativa) under low nitrogen conditions. Gene 2016, 576, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Ranathunge, K.; El-Kereamy, A.; Gidda, S.; Bi, Y.M.; Rothstein, S.J. AMT1;1 transgenic rice plants with enhanced NH4(+) permeability show superior growth and higher yield under optimal and suboptimal NH4(+) conditions. J. Exp. Bot. 2014, 65, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.Y.; Chen, H.W.; Ng, C.Y.; Lin, C.Y.; Tseng, T.H.; Li, W.H.; Ku, M.S. Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice 2015, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Potters, G.; Horemans, N.; Caubergs, R.J.; Asard, H. Ascorbate and dehydroascorbate influence cell cycle progression in a tobacco cell suspension. Plant Physiol. 2000, 124, 17–20. [Google Scholar] [CrossRef] [Green Version]
- Horemans, N.; Potters, G.; De Wilde, L.; Caubergs, R.J. Dehydroascorbate uptake activity correlates with cell growth and cell division of tobacco bright yellow-2 cell cultures. Plant Physiol. 2003, 133, 361–367. [Google Scholar] [CrossRef] [Green Version]
- Potters, G.; Horemans, N.; Bellone, S.; Caubergs, R.J.; Trost, P.; Guisez, Y.; Asard, H. Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiol. 2004, 134, 1479–1487. [Google Scholar] [CrossRef] [Green Version]
- Szarka, A.; Lorincz, T. The role of ascorbate in protein folding. Protoplasma 2014, 251, 489–497. [Google Scholar] [CrossRef]
- Liu, L.; Tong, H.; Xiao, Y.; Che, R.; Xu, F.; Hu, B.; Liang, C.; Chu, J.; Li, J.; Chu, C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-S.; Park, S.-I.; Kim, J.-J.; Shin, S.-Y.; Kwak, S.-S.; Lee, C.-H.; Park, H.-M.; Kim, Y.-H.; Kim, I.-S.; Yoon, H.-S. Over-Expression of Dehydroascorbate Reductase Improves Salt Tolerance, Environmental Adaptability and Productivity in Oryza sativa. Antioxidants 2022, 11, 1077. https://doi.org/10.3390/antiox11061077
Kim Y-S, Park S-I, Kim J-J, Shin S-Y, Kwak S-S, Lee C-H, Park H-M, Kim Y-H, Kim I-S, Yoon H-S. Over-Expression of Dehydroascorbate Reductase Improves Salt Tolerance, Environmental Adaptability and Productivity in Oryza sativa. Antioxidants. 2022; 11(6):1077. https://doi.org/10.3390/antiox11061077
Chicago/Turabian StyleKim, Young-Saeng, Seong-Im Park, Jin-Ju Kim, Sun-Young Shin, Sang-Soo Kwak, Choon-Hwan Lee, Hyang-Mi Park, Yul-Ho Kim, Il-Sup Kim, and Ho-Sung Yoon. 2022. "Over-Expression of Dehydroascorbate Reductase Improves Salt Tolerance, Environmental Adaptability and Productivity in Oryza sativa" Antioxidants 11, no. 6: 1077. https://doi.org/10.3390/antiox11061077






