Fucoxanthin’s Optimization from Undaria pinnatifida Using Conventional Heat Extraction, Bioactivity Assays and In Silico Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Conventional Kinetic Heat Extraction
2.3. Fucoxanthin Detection and Quantification through HPLC
2.4. Statistical Analysis, Mathematical Modeling and Graphical Representation
- Determination of the coefficients: the parametric estimates were obtained by minimizing the sum of the quadratic differences between the experimental values and those predicted by the model, using the non-linear method of least squares (quasi-Newton) provided by the macro Solver in Microsoft Excel 2003 [29], which allows for the rapid analysis of a hypothesis and its consequences [30].
- Significance of the coefficients: the determination of the confidence intervals of the parameters was carried out using ‘SolverAid’ [31]. The model was simplified, discarding the terms that were not statistically significant for the p-value (p > 0.05).
- Model consistency: Fisher’s F test (α = 0.05) was used to determine the adequacy of the models built for the data obtained [32].
- Other statistical evaluation criteria: to re-verify the uniformity of the model, the following criteria were applied: (i) the macro ‘SolverStat’ was used [33] to evaluate the prediction uncertainties of the parameters and models; (ii) the R2 was interpreted as the proportion of versatility of each dependent variable that was described in the model; (iii) the adjusted coefficient of determination (R2adj) corrected R2, taking into account the number of variables used in the model.
2.5. Evaluation of the Biological Properties of the Optimized Extract
2.5.1. Antioxidant Activity
DPPH Radical-Scavenging Activity
ABTS Radical-Scavenging Activity
Crocin Bleaching Assay (CBA)
2.5.2. Antimicrobial Activity
2.5.3. Neuroprotective Activity
2.6. In Silico Studies: Molecular Docking and Pharmacokinetic Study
2.6.1. Molecular Docking
Fucoxanthin (Ligand) Preparation
Protein Preparation
Molecular Docking
Results Analysis and Visualization
2.6.2. Pharmacokinetics Study
3. Results and Discussion
3.1. HPLC Results
3.2. Analysis of the Kinetic Parameters and a Search for Optimal Conditions
3.3. Evaluation of the Antioxidant Response
3.4. Evaluation of the Neuroprotective Activity
3.5. Evaluation of the Antimicrobial Activity
3.6. Validation of the Biological Properties through In Silico Studies
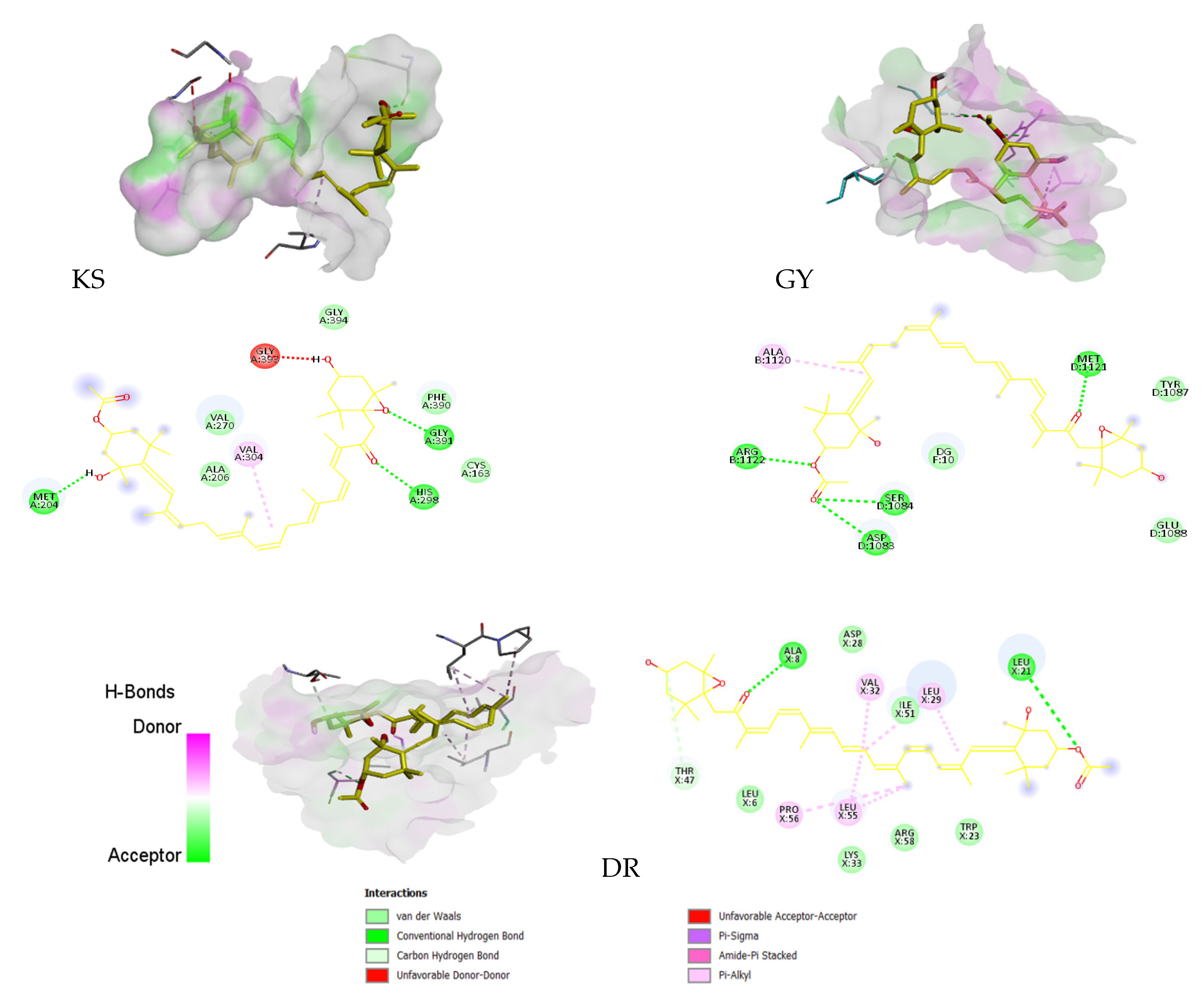
3.6.1. Molecular Docking
3.6.2. Pharmacokinetic Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Generic | |
| ARE | Antioxidant-response element |
| E | Extract |
| EC50 | Half maximal effective concentration |
| Fx | Fucoxanthin |
| CHE | Convectional heat extraction |
| HPLC-DAD | High Performance Liquid Chromatography - Photodiode array detector |
| AS dw | Algae sample dry weight |
| MAE | Mean absolute error |
| PDB | Protein Data Bank |
| RMSE | Root mean squared error |
| ROS | Reactive oxygen species |
| SMILES | Simplified Molecular Input Line Entry |
| S | Solvent concentration |
| t | Time |
| T | Temperature |
| UV | Ultraviolet |
| Compounds | |
| AAPH | 2,2′-azobis-2-amidinopropane: RN = NR |
| ABTS | 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid |
| ALA | Alanine |
| DMSO | Dimethyl sulfoxide |
| DNA | Deoxyribonucleic acid |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| HCl | Hydrochloric acid |
| LEU | Leucine |
| K2S2O8 | Potassium persulfate |
| THR | Threonine |
| TYR | Tyrosine |
| Neuroprotective activity | |
| I/R | Ischemic/reperfusion |
| MCAO | Middle cerebral artery occlusion |
| MHB | Mueller Hinton Broth |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| Nrf2-ARE | Nuclear factor erythroid 2-related factor 2 antioxidant-response element |
| OGD/R | Oxygen-glucose deprivation and reoxygenation |
| TBI | Traumatic brain injury |
| Proteins | |
| AChE | Acetylcholinesterase |
| BuChE | Butyrylcholinesterase |
| DR | Dihydrofolate reductase |
| GY | Gyrase |
| KS | Beta-ketoacyl-(acyl carrier protein) synthase I |
| In silico studies | |
| ADME | Absorption, distribution, metabolism and excretion |
| BBB | Blood–brain barrier permeability |
| CYP | Cytochrome P interaction |
| GI | Gastrointestinal absorption |
| Kp | Permeability coefficient |
| Pgp | Glycoprotein-P interaction |
| RMSD | Root-mean-square deviation |
Appendix A
| Extraction Variables | Extraction Responses for Acetone Solvent in Different Concentrations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y1 | Y2 | ||||||||||||
| T (°C) | t (min) | 50% | 60% | 70% | 80% | 90% | 100% | 50% | 60% | 70% | 80% | 90% | 100% |
| 5 | 30 | 239.2 | 2057.4 | 3193.6 | 2533.7 | 1016.0 | 668.5 | 433.7 | 369.7 | 363.3 | 335.3 | 143.9 | 12.0 |
| 120 | 297.2 | 2840.7 | 3790.7 | 3322.0 | 1405.9 | 1034.7 | 390.5 | 379.7 | 382.6 | 369.9 | 143.9 | 24.4 | |
| 480 | 137.8 | 3228.9 | 4298.1 | 3844.8 | 1530.0 | 1190.8 | 470.3 | 415.0 | 439.7 | 397.9 | 173.3 | 35.3 | |
| 1200 | 157.0 | 2637.1 | 5165.2 | 4196.8 | 2016.7 | 1597.6 | 452.4 | 400.4 | 433.7 | 397.2 | 167.3 | 28.6 | |
| 2640 | 167.0 | 2524.3 | 4487.8 | 4464.3 | 2863.3 | 2195.7 | 461.0 | 437.0 | 413.8 | 425.2 | 189.3 | 36.6 | |
| 4200 | 149.2 | 1871.2 | 4846.9 | 4546.0 | 3075.2 | 3679.3 | 456.4 | 402.4 | 383.3 | 394.6 | 194.6 | 43.3 | |
| 5700 | 114.5 | 2043.8 | 4975.1 | 4550.9 | 3890.6 | 3673.2 | 431.1 | 425.7 | 447.0 | 348.7 | 201.2 | 48.6 | |
| 9680 | 79.8 | 2305.4 | 4957.5 | 4594.8 | 4418.8 | 5318.1 | 471.7 | 424.3 | 412.5 | 429.2 | 203.9 | 51.3 | |
| 25 | 15 | 462.1 | 2511.4 | 2938.1 | 2500.1 | 1114.6 | 1004.1 | 427.5 | 393.4 | 398.0 | 392.6 | 154.4 | 23.9 |
| 30 | 355.6 | 2692.6 | 3244.5 | 2830.2 | 1204.1 | 1148.4 | 358.9 | 388.0 | 414.7 | 368.6 | 151.1 | 23.2 | |
| 120 | 507.7 | 3284.8 | 3881.2 | 3394.3 | 1415.6 | 1396.8 | 423.5 | 421.3 | 419.3 | 389.9 | 146.4 | 26.6 | |
| 480 | 455.7 | 4081.2 | 4482.4 | 4495.1 | 1730.4 | 2306.9 | 421.5 | 414.0 | 425.9 | 406.6 | 148.4 | 41.8 | |
| 1200 | 375.5 | 4593.3 | 4773.3 | 4925.0 | 2095.6 | 2995.2 | 399.6 | 393.4 | 402.0 | 396.6 | 155.1 | 31.9 | |
| 1680 | 645.5 | 3915.4 | 4466.6 | 4275.9 | 2022.4 | 3055.0 | 423.5 | 415.3 | 416.0 | 382.6 | 169.1 | 38.5 | |
| 2640 | 321.8 | 3415.6 | 3746.5 | 3981.9 | 1735.8 | 3625.9 | 432.9 | 398.0 | 406.7 | 397.3 | 177.1 | 41.2 | |
| 45 | 3 | 118.6 | 2118.9 | 1693.3 | 1238.7 | 1329.4 | 1231.9 | 480.8 | 465.2 | 318.4 | 160.3 | 98.9 | 11.3 |
| 5 | 127.0 | 2932.8 | 2812.3 | 1881.1 | 1418.4 | 1408.4 | 458.9 | 459.2 | 460.7 | 272.8 | 112.9 | 13.3 | |
| 15 | 153.4 | 3620.0 | 3266.8 | 2190.6 | 1613.6 | 1706.3 | 466.2 | 469.2 | 443.4 | 345.9 | 110.9 | 20.0 | |
| 60 | 194.1 | 4881.1 | 4800.9 | 2431.3 | 1712.9 | 2375.1 | 481.4 | 469.2 | 468.0 | 363.2 | 116.9 | 18.6 | |
| 210 | 185.0 | 2456.4 | 4218.6 | 3383.3 | 2179.1 | 2725.0 | 449.6 | 451.2 | 454.7 | 458.4 | 120.8 | 30.6 | |
| 1200 | 161.2 | 4761.4 | 5565.2 | 4418.8 | 2018.8 | 4675.5 | 452.2 | 452.6 | 487.9 | 517.6 | 142.8 | 36.6 | |
| 1680 | 223.3 | 4934.7 | 5465.5 | 4571.4 | 1998.9 | 4960.0 | 417.7 | 436.6 | 443.4 | 465.0 | 138.1 | 41.3 | |
| 2640 | 208.2 | 3689.9 | 4621.5 | 4616.4 | 2345.1 | 4739.4 | 444.2 | 442.6 | 341.7 | 466.4 | 151.4 | 51.3 | |
| 65 | 3 | 315.6 | 2062.8 | 1803.4 | 1559.7 | 1507.3 | 1038.4 | 432.9 | 433.4 | 402.9 | 369.0 | 86.5 | 17.3 |
| 5 | 364.7 | 2325.0 | 2219.5 | 1718.7 | 1771.6 | 1180.1 | 415.5 | 448.1 | 438.1 | 398.9 | 107.1 | 23.2 | |
| 15 | 297.4 | 2300.0 | 3095.0 | 1801.6 | 2286.3 | 1647.0 | 427.5 | 428.7 | 440.8 | 400.2 | 141.0 | 29.2 | |
| 30 | 381.1 | 3669.6 | 3753.6 | 2223.0 | 2580.7 | 2050.3 | 435.1 | 448.3 | 457.6 | 398.2 | 146.3 | 33.2 | |
| 60 | 178.7 | 3434.1 | 3417.4 | 1688.0 | 2548.4 | 2479.6 | 430.6 | 425.0 | 390.0 | 378.3 | 136.3 | 25.4 | |
| 120 | 111.1 | 1673.1 | 3882.6 | 2267.8 | 2497.9 | 2740.9 | 450.6 | 441.6 | 437.6 | 407.1 | 154.1 | 17.7 | |
| 210 | 202.2 | 3566.7 | 4101.7 | 2830.6 | 2834.1 | 3390.1 | 405.1 | 410.6 | 51.0 | 423.7 | 154.1 | 37.6 | |
| 480 | 298.5 | 3221.8 | 4671.2 | 3405.5 | 1581.2 | 3815.0 | 420.9 | 441.4 | 416.8 | 426.8 | 153.0 | 40.5 | |
| 1200 | 43.2 | 1995.0 | 808.9 | 4379.2 | 1814.0 | 4336.8 | 430.2 | 388.1 | 419.5 | 440.3 | 174.0 | 43.1 | |
References
- Lourenço-Lopes, C.; Garcia-Oliveira, P.; Carpena, M.; Fraga-Corral, M.; Jimenez-Lopez, C.; Pereira, A.G.; Prieto, M.A.; Simal-Gandara, J. Scientific approaches on extraction, purification and stability for the commercialization of fucoxanthin recovered from brown algae. Foods 2020, 9, 1113. [Google Scholar] [CrossRef] [PubMed]
- Saet, B.L.; Joo, Y.L.; Song, D.G.; Pan, C.H.; Chu, W.N.; Min, C.K.; Eun, H.L.; Sang, H.J.; Kim, H.S.; Yeong, S.K.; et al. Cancer chemopreventive effects of Korean seaweed extracts. Food Sci. Biotechnol. 2008, 17, 613–622. [Google Scholar]
- Lourenço-Lopes, C.; Fraga-Corral, M.; Jimenez-Lopez, C.; Pereira, A.G.; Garcia-Oliveira, P.; Carpena, M.; Prieto, M.A.; Simal-Gandara, J. Metabolites from macroalgae and its applications in the cosmetic industry: A circular economy approach. Resources 2020, 9, 101. [Google Scholar] [CrossRef]
- Willstatter, R.; Page, H. The pigments of the brown algae. Justus Liebigs Ann. Chem. 1914, 404, 237–271. [Google Scholar]
- Billakanti, J.M.; Catchpole, O.J.; Fenton, T.A.; Mitchell, K.A.; Mackenzie, A.D. Enzyme-assisted extraction of fucoxanthin and lipids containing polyunsaturated fatty acids from Undaria pinnatifida using dimethyl ether and ethanol. Process Biochem. 2013, 48, 1999–2008. [Google Scholar] [CrossRef]
- Kajikawa, T.; Okumura, S.; Iwashita, T.; Kosumi, D.; Hashimoto, H.; Katsumura, S. Stereocontrolled total synthesis of fucoxanthin and its polyene chain-modified derivative. Org. Lett. 2012, 14, 808–811. [Google Scholar] [CrossRef]
- Asai, A.; Sugawara, T.; Ono, H.; Nagao, A. Biotransformation of fucoxanthinol into amarouciaxanthin a in mice and HepG2 cells: Formation and cytotoxicity of fucoxanthin metabolites. Drug Metab. Dispos. 2004, 32, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Sangeetha, R.K.; Bhaskar, N.; Divakar, S.; Baskaran, V. Bioavailability and metabolism of fucoxanthin in rats: Structural characterization of metabolites by LC-MS (APCI). Mol. Cell. Biochem. 2010, 333, 299–310. [Google Scholar] [CrossRef]
- Yamamoto, K.; Ishikawa, C.; Katano, H.; Yasumoto, T.; Mori, N. Fucoxanthin and its deacetylated product, fucoxanthinol, induce apoptosis of primary effusion lymphomas. Cancer Lett. 2011, 300, 225–234. [Google Scholar] [CrossRef]
- Sun, P.; Wong, C.C.; Li, Y.; He, Y.; Mao, X.; Wu, T.; Ren, Y.; Chen, F. A novel strategy for isolation and purification of fucoxanthinol and fucoxanthin from the diatom Nitzschia laevis. Food Chem. 2019, 277, 566–572. [Google Scholar] [CrossRef]
- Heo, S.J.; Ko, S.C.; Kang, S.M.; Kang, H.S.; Kim, J.P.; Kim, S.H.; Lee, K.W.; Cho, M.G.; Jeon, Y.J. Cytoprotective effect of fucoxanthin isolated from brown algae Sargassum siliquastrum against H2O2-induced cell damage. Eur. Food Res. Technol. 2008, 228, 145–151. [Google Scholar] [CrossRef]
- D’Orazio, N.; Gemello, E.; Gammone, M.A.; De Girolamo, M.; Ficoneri, C.; Riccioni, G. Fucoxantin: A treasure from the sea. Mar. Drugs 2012, 10, 604–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soo-Jin You-Jin, H.; Seok-Chun, K.; Sung-Myung, K.; Hahk-Soo, K.; Jong-Pyung, K.; Soo-Hyun, K.; Ki-Wan, L.; Man-Gi, C. Jeon Cytoprotective effect of fucoxanthin isolated from brown algae Sargassum siliquastrum against H2O2-induced cell damage. Eur. Food Res. Technol. 2008, 228, 145–151. [Google Scholar]
- Kumar, S.R.; Hosokawa, M.; Miyashita, K. Fucoxanthin: A marine carotenoid exerting anti-cancer effects by affecting multiple mechanisms. Mar. Drugs 2013, 11, 5130–5147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Tang, Y.; Zhang, Y.; Zhang, S.; Qu, J.; Wang, X.; Kong, R.; Han, C.; Liu, Z. Fucoxanthin: A Promising Medicinal and Nutritional Ingredient. Evidence-Based Complement. Altern. Med. 2015, 2015, 723515. [Google Scholar] [CrossRef] [PubMed]
- Van Chuyen, H.; Eun, J.B. Marine carotenoids: Bioactivities and potential benefits to human health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2600–2610. [Google Scholar] [CrossRef]
- Raguraman, V.; Abraham, S.; MubarakAli, D.; Narendrakumar, G.; Thirugnanasambandam, R.; Kirubagaran, R.; Thajuddin, N. Unraveling rapid extraction of fucoxanthin from Padina tetrastromatica: Purification, characterization and biomedical application. Process Biochem. 2018, 73, 211–219. [Google Scholar] [CrossRef]
- Wang, L.; Park, Y.J.; Jeon, Y.J.; Ryu, B.M. Bioactivities of the edible brown seaweed, Undaria pinnatifida: A review. Aquaculture 2018, 495, 873–880. [Google Scholar] [CrossRef]
- Fung, A.; Hamid, N.; Lu, J. Fucoxanthin content and antioxidant properties of Undaria pinnatifida. Food Chem. 2013, 136, 1055–1062. [Google Scholar] [CrossRef]
- Liu, C.L.; Liang, A.L.; Hu, M.L. Protective effects of fucoxanthin against ferric nitrilotriacetate-induced oxidative stress in murine hepatic BNL CL.2 cells. Toxicol. Vitr. 2011, 25, 1314–1319. [Google Scholar] [CrossRef]
- Wang, X.; Cui, Y.J.; Qi, J.; Zhu, M.M.; Zhang, T.L.; Cheng, M.; Liu, S.M.; Wang, G.C. Fucoxanthin Exerts Cytoprotective Effects against Hydrogen Peroxide-induced Oxidative Damage in L02 Cells. Biomed Res. Int. 2018, 2018, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Sachindra, N.M.; Sato, E.; Maeda, H.; Hosokawa, M.; Niwano, Y.; Kohno, M.; Miyashita, K. Radical scavenging and singlet oxygen quenching activity of marine carotenoid fucoxanthin and its metabolites. J. Agric. Food Chem. 2007, 55, 8516–8522. [Google Scholar] [CrossRef] [PubMed]
- Neumann, U.; Derwenskus, F.; Flister, V.F.; Schmid-Staiger, U.; Hirth, T.; Bischoff, S.C. Fucoxanthin, a carotenoid derived from Phaeodactylum tricornutum exerts antiproliferative and antioxidant activities in vitro. Antioxidants 2019, 8, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wang, H.; Fan, Y.; Gao, Y.; Li, X.; Hu, Z.; Ding, K.; Wang, Y.; Wang, X. Fucoxanthin provides neuroprotection in models of traumatic brain injury via the Nrf2-ARE and Nrf2-autophagy pathways. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Rajauria, G.; Foley, B.; Abu-Ghannam, N. Characterization of dietary fucoxanthin from Himanthalia elongata brown seaweed. Food Res. Int. 2017, 99, 995–1001. [Google Scholar] [CrossRef]
- Lourenço-Lopes, C.; Fraga-Corral, M.; Jimenez-Lopez, C.; Carpena, M.; Pereira, A.G.; Garcia-Oliveira, P.; Prieto, M.A.; Simal-Gandara, J. Biological action mechanisms of fucoxanthin extracted from algae for application in food and cosmetic industries. Trends Food Sci. Technol. 2021, 117, 163–181. [Google Scholar] [CrossRef]
- European Parliament and of the Council Directive 2009/32/EC on the approximation of the laws of the Member States on extraction solvents used in the production of foodstuffs and food ingredients. Off. J. Eur. Union 2010, 141, 1–15.
- López, C.J.; Caleja, C.; Prieto, M.A.; Barreiro, M.F.; Barros, L.; Ferreira, I.C.F.R. Optimization and comparison of heat and ultrasound assisted extraction techniques to obtain anthocyanin compounds from Arbutus unedo L. Fruits. Food Chem. 2018, 264, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Kemmer, G.; Keller, S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protoc. 2010, 5, 267–281. [Google Scholar] [CrossRef]
- Murado, M.A.; Prieto, M.A. Dose-Response Analysis in the Joint Action of Two Effectors. A New Approach to Simulation, Identification and Modelling of Some Basic Interactions. PLoS ONE 2013, 8, e61391. [Google Scholar]
- Prikler, S. Advanced Excel for Scientific Data Analysis, 2nd ed.; de Levie, R., Ed.; Oxford University Press: Oxford, UK, 2009; ISBN 978-0-19-537022-5. [Google Scholar]
- Heleno, S.A.; Diz, P.; Prieto, M.A.; Barros, L.; Rodrigues, A.; Barreiro, M.F.; Ferreira, I.C.F.R. Optimization of ultrasound-assisted extraction to obtain mycosterols from Agaricus bisporus L. by response surface methodology and comparison with conventional Soxhlet extraction. Food Chem. 2016, 197, 1036–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comuzzi, C.; Polese, P.; Melchior, A.; Portanova, R.; Tolazzi, M. SOLVERSTAT: A new utility for multipurpose analysis. An application to the investigation of dioxygenated Co (II) complex formation in dimethylsulfoxide solution. Talanta 2003, 59, 67–80. [Google Scholar] [CrossRef]
- Lopes, C.L.; Pereira, E.; Soković, M.; Carvalho, A.M.; Barata, A.M.; Lopes, V.; Rocha, F.; Calhelha, R.C.; Barros, L.; Ferreira, I.C.F.R. Phenolic Composition and Bioactivity of Lavandula pedunculata (Mill.) Cav. Samples from Different Geographical Origin. Molecules 2018, 23, 1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Viacava, G.E.; Goyeneche, R.; Goñi, M.G.; Roura, S.I.; Agüero, M.V. Natural elicitors as preharvest treatments to improve postharvest quality of Butterhead lettuce. Sci. Hortic. 2018, 228, 145–152. [Google Scholar] [CrossRef]
- Bors, W.; Michel, C.; Saran, M. Inhibition of the bleaching of the carotenoid crocin a rapid test for quantifying antioxidant activity. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 1984, 796, 312–319. [Google Scholar] [CrossRef]
- Sokovic, M.; Glamoclija, J.; Marin, M.D.; Brkic, D.; van Griensven, L.J.L.D. Antibacterial effects of the essential oils of commonly consumed medicinal herbs using an in vitro model. Molecules 2010, 15, 7532–7546. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32, ISBN 1562384856. [Google Scholar]
- Paz, M.; Gúllon, P.; Barroso, M.F.; Carvalho, A.P.; Domingues, V.F.; Gomes, A.M.; Becker, H.; Longhinotti, E.; Delerue-Matos, C. Brazilian fruit pulps as functional foods and additives: Evaluation of bioactive compounds. Food Chem. 2015, 172, 462–468. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.; Silva, S.A.; Lourenço-Lopes, C.; Jimenez-Lopez, C.; Carpena, M.; Gullón, P.; Fraga-Corral, M.; Domingues, V.F.; Fátima Barroso, M.; Simal-Gandara, J.; et al. Antibacterial use of macroalgae compounds against foodborne pathogens. Antibiotics 2020, 9, 712. [Google Scholar] [CrossRef]
- Lynne, S.G. (Ed.) Clinical Microbiology Procedures Handbook, 3rd ed.; American Society of Microbiology: Washington, DC, USA, 2010; ISBN 9781555815271. [Google Scholar]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Ingkaninan, K.; Temkitthawon, P.; Chuenchom, K.; Yuyaem, T.; Thongnoi, W. Screening for acetylcholinesterase inhibitory activity in plants used in Thai traditional rejuvenating and neurotonic remedies. J. Ethnopharmacol. 2003, 89, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Fang, H.; Xie, Q.; Sun, J.; Liu, R.; Hong, Z.; Yi, R.; Wu, H. Comparative evaluation of the radical-scavenging activities of fucoxanthin and its stereoisomers. Molecules 2014, 19, 2100–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, K.; Beppu, F.; Hosokawa, M.; Liu, X.; Wang, S. Bioactive significance of fucoxanthin and its effective extraction. Biocatal. Agric. Biotechnol. 2020, 26, 101639. [Google Scholar] [CrossRef]
- Guvatova, Z.; Dalina, A.; Marusich, E.; Pudova, E.; Snezhkina, A.; Krasnov, G.; Kudryavtseva, A.; Leonov, S.; Moskalev, A. Protective effects of carotenoid fucoxanthin in fibroblasts cellular senescence. Mech. Ageing Dev. 2020, 189, 111260. [Google Scholar] [CrossRef]
- Ingkaninan, K.; De Best, C.M.; Van Der Heijden, R.; Hofte, A.J.P.; Karabatak, B.; Irth, H.; Tjaden, U.R.; Van Der Greef, J.; Verpoorte, R. High-performance liquid chromatography with on-line coupled UV, mass spectrometric and biochemical detection for identification of acetylcholinesterase inhibitors from natural products. J. Chromatogr. A 2000, 872, 61–73. [Google Scholar] [CrossRef]
- Delerue, T.; Fátima Barroso, M.; Dias-Teixeira, M.; Figueiredo-González, M.; Delerue-Matos, C.; Grosso, C. Interactions between Ginkgo biloba L. and Scutellaria baicalensis Georgi in multicomponent mixtures towards cholinesterase inhibition and ROS scavenging. Food Res. Int. 2021, 140, 109857. [Google Scholar] [CrossRef]
- Hu, L.; Chen, W.; Tian, F.; Yuan, C.; Wang, H.; Yue, H. Neuroprotective role of fucoxanthin against cerebral ischemic/reperfusion injury through activation of Nrf2/HO-1 signaling. Biomed. Pharmacother. 2018, 106, 1484–1489. [Google Scholar] [CrossRef]
- Sivagnanam, S.P.; Yin, S.; Choi, J.H.; Park, Y.B.; Woo, H.C.; Chun, B.S. Biological properties of fucoxanthin in oil recovered from two brown seaweeds using supercritical CO2 extraction. Mar. Drugs 2015, 13, 3422–3442. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Adamczak, A. Fucoxanthin—An antibacterial carotenoid. Antioxidants 2019, 8, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rock, C.O.; Cronan, J.E. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta-Lipids Lipid Metab. 1996, 1302, 1–16. [Google Scholar] [CrossRef]
- Price, A.C.; Choi, K.H.; Heath, R.J.; Li, Z.; White, S.W.; Rock, C.O. Inhibition of β-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin: Structure and mechanism. J. Biol. Chem. 2001, 276, 6551–6559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherer, B.A.; Hull, K.; Green, O.; Basarab, G.; Hauck, S.; Hill, P.; Loch, J.T.; Mullen, G.; Bist, S.; Bryant, J.; et al. Pyrrolamide DNA gyrase inhibitors: Optimization of antibacterial activity and efficacy. Bioorganic Med. Chem. Lett. 2011, 21, 7416–7420. [Google Scholar] [CrossRef]
- Maxwell, A. DNA gyrase as a drug target. Trends Microbiol. 1997, 5, 102–109. [Google Scholar] [CrossRef]
- Li, X.; Hilgers, M.; Cunningham, M.; Chen, Z.; Trzoss, M.; Zhang, J.; Kohnen, L.; Lam, T.; Creighton, C.; Gc, K.; et al. Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines. Bioorganic Med. Chem. Lett. 2011, 21, 5171–5176. [Google Scholar] [CrossRef]
- Andrieu, S.; Coley, N.; Lovestone, S.; Aisen, P.S.; Vellas, B. Prevention of sporadic Alzheimer’s disease: Lessons learned from clinical trials and future directions. Lancet Neurol. 2015, 14, 926–944. [Google Scholar] [CrossRef]
- Anand, P.; Singh, B.; Singh, N. A review on coumarins as acetylcholinesterase inhibitors for Alzheimer’s disease. Bioorganic Med. Chem. 2012, 20, 1175–1180. [Google Scholar] [CrossRef]
- Dos Santos, T.C.; Gomes, T.M.; Pinto, B.A.S.; Camara, A.L.; De Andrade Paes, A.M. Naturally occurring acetylcholinesterase inhibitors and their potential use for Alzheimer’s disease therapy. Front. Pharmacol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Yang, H.; Chen, Y.; Sun, H. Recent progress in the identification of selective butyrylcholinesterase inhibitors for Alzheimer’s disease. Eur. J. Med. Chem. 2017, 132, 294–309. [Google Scholar] [CrossRef]
- Knez, D.; Coquelle, N.; Pišlar, A.; Žakelj, S.; Jukič, M.; Sova, M.; Mravljak, J.; Nachon, F.; Brazzolotto, X.; Kos, J.; et al. Multi-target-directed ligands for treating Alzheimer’s disease: Butyrylcholinesterase inhibitors displaying antioxidant and neuroprotective activities. Eur. J. Med. Chem. 2018, 156, 598–617. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Furnham, N.; Thornton, J.M. The Ramachandran plot and protein structure validation. In Biomolecular Forms and Functions: A Celebration of 50 Years of the Ramachandran Map; World Scientific: Singapore, 2012; pp. 62–75. [Google Scholar]
- Wróbel, A.; Arciszewska, K.; Maliszewski, D.; Drozdowska, D. Trimethoprim and other nonclassical antifolates an excellent template for searching modifications of dihydrofolate reductase enzyme inhibitors. J. Antibiot. 2020, 73, 5–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, F.R.; Guimarães, A.P.; Cuya, T.; de Freitas, M.P.; da Gonçalves, A.S.; Forgione, P.; Costa França, T.C. Analysis of Coxiela burnetti dihydrofolate reductase via in silico docking with inhibitors and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2017, 35, 2975–2986. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhao, J.; Fang, C.; Cao, Q.; Xing, M.; Li, X.; Hou, J.; Ji, A.; Song, S. Advances in Studies on the Pharmacological Activities of Fucoxanthin. Mar. Drugs 2020, 18, 634. [Google Scholar] [CrossRef]
- Waqar, M.; Batool, S. In silico analysis of binding of neurotoxic venom ligands with acetylcholinesterase for therapeutic use in treatment of Alzheimer’s disease. J. Theor. Biol. 2015, 372, 107–117. [Google Scholar] [CrossRef]
- Shiri, F.; Pirhadi, S.; Ghasemi, J.B. Dynamic structure based pharmacophore modeling of the Acetylcholinesterase reveals several potential inhibitors. J. Biomol. Struct. Dyn. 2019, 37, 1800–1812. [Google Scholar] [CrossRef]
- Xiang, S.; Liu, F.; Lin, J.; Chen, H.; Huang, C.; Chen, L.; Zhou, Y.; Ye, L.; Zhang, K.; Jin, J.; et al. Fucoxanthin Inhibits β-Amyloid Assembly and Attenuates β-Amyloid Oligomer-Induced Cognitive Impairments. J. Agric. Food Chem. 2017, 65, 4092–4102. [Google Scholar] [CrossRef]
- Alghazwi, M.; Smid, S.; Musgrave, I.; Zhang, W. In vitro studies of the neuroprotective activities of astaxanthin and fucoxanthin against amyloid beta (Aβ 1-42) toxicity and aggregation. Neurochem. Int. 2019, 124, 215–224. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Huang, L.; Yu, J.; Xiang, S.; Wang, J.; Zhang, J.; Yan, X.; Cui, W.; He, S.; Wang, Q. Fucoxanthin, a marine carotenoid, reverses scopolamine-induced cognitive impairments in mice and inhibits acetylcholinesterase in vitro. Mar. Drugs 2016, 14, 67. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [Green Version]
- Paudel, P.; Seong, S.H.; Jung, H.A.; Choi, J.S. Characterizing fucoxanthin as a selective dopamine D3/D4 receptor agonist: Relevance to Parkinson’s disease. Chem. Biol. Interact. 2019, 310, 108757. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Ozaki, Y.; Taminato, M.; Das, S.K.; Mizuno, M.; Yoshimura, K.; Maoka, T.; Kanazawa, K. The distribution and accumulation of fucoxanthin and its metabolites after oral administration in mice. Br. J. Nutr. 2009, 102, 242–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
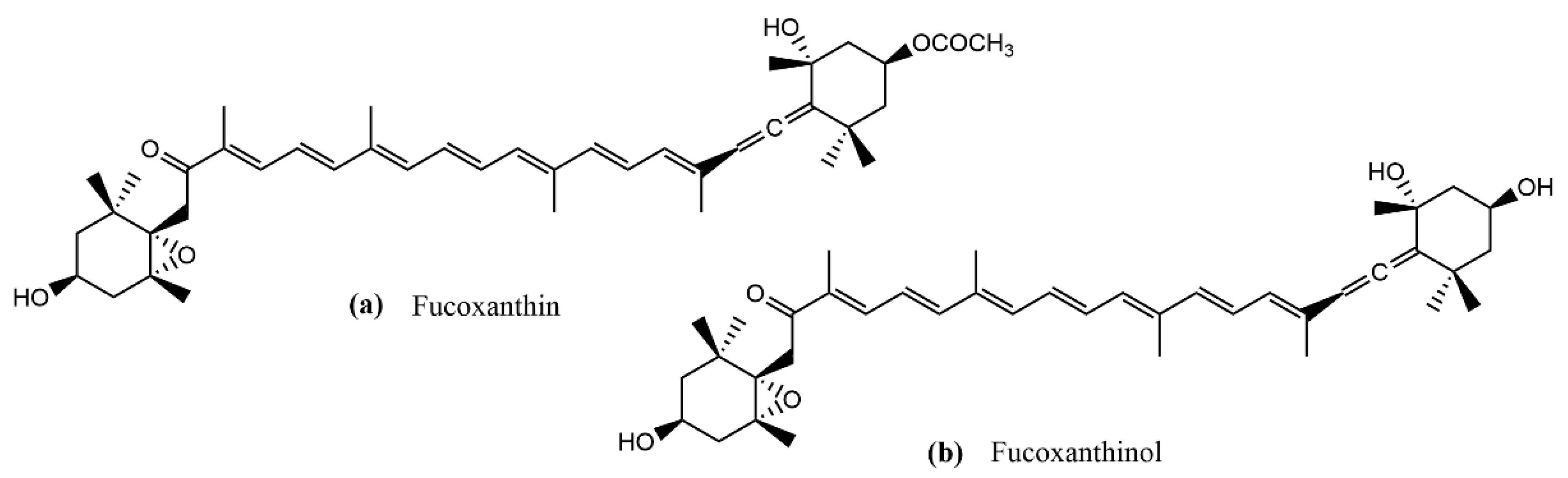

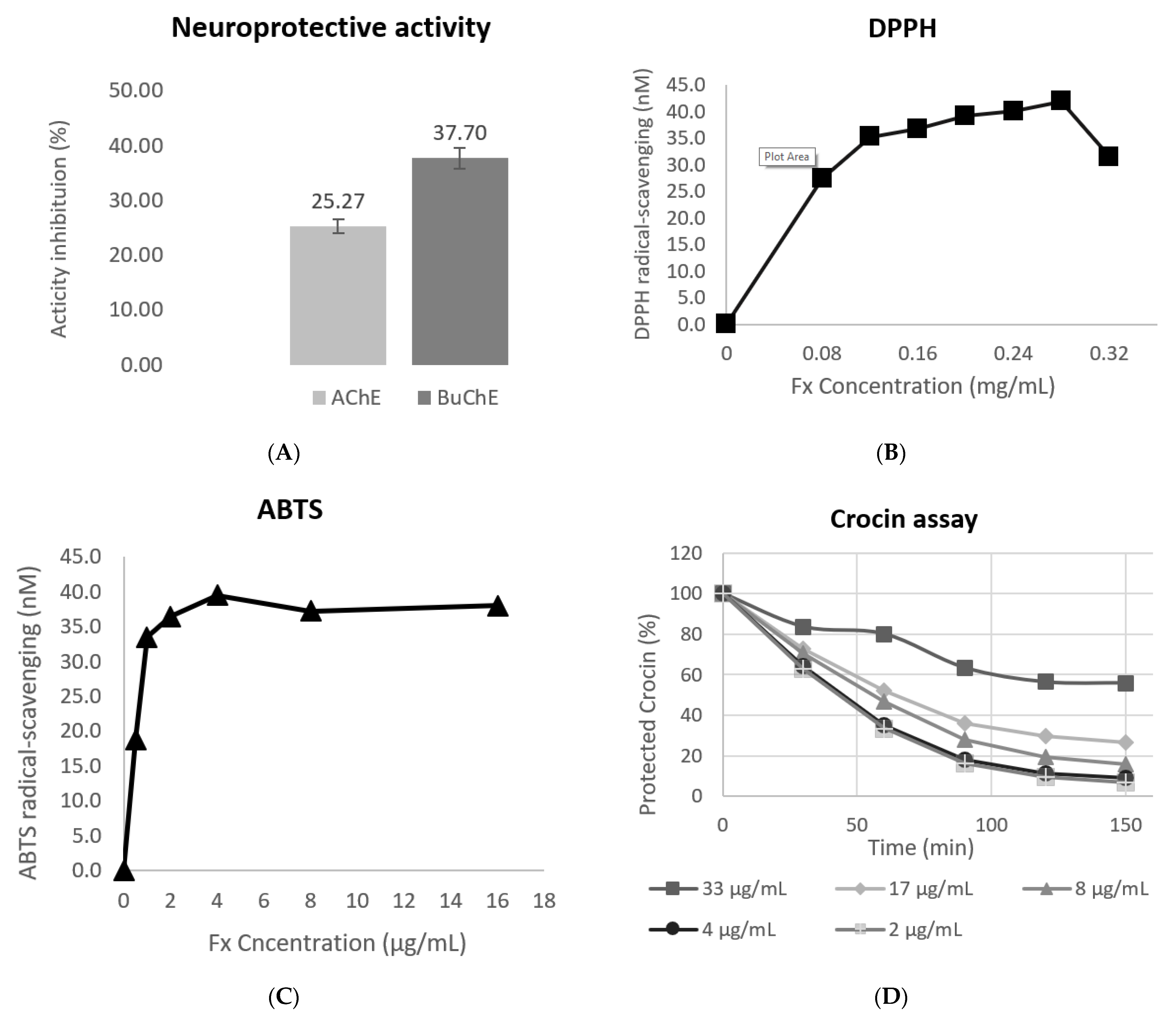

| Variables | Y1 (µg Fx/g AS) | Y2 (mg E/g AS) | Y3 (mg Fx/g E) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | S | k | r | R2 | K | r | R2 | k | r | R2 | ||||||
| (°C) | (%) | (µg Fx/g AS) | (min−1) | (mg E/g AS) | (min−1) | (mg Fx/g E) | (min−1) | |||||||||
| 5 | 50 | 191 | ns | 0.295 | ns | 0.6120 | 444 | ±295.0 | 0.212 | ns | 0.9765 | 0.6 | ns | 0.261 | ns | 0.5024 |
| 60 | 2708 | ±297.6 | 0.048 | ±0.036 | 0.9825 | 407 | ±297.6 | 0.080 | ±0.036 | 0.9825 | 6.4 | ±6.4 | 0.073 | ns | 0.9825 | |
| 70 | 4618 | ±274.5 | 0.037 | ±0.015 | 0.9731 | 487 | ±274.5 | 0.043 | ±0.015 | 0.9731 | 11.2 | ±5.8 | 0.050 | ns | 0.9731 | |
| 80 | 4227 | ±282.3 | 0.025 | ±0.011 | 0.9657 | 462 | ±282.3 | 0.041 | ±0.011 | 0.9657 | 10.8 | ±5.9 | 0.038 | ns | 0.9657 | |
| 90 | 2407 | ±343.6 | 0.008 | ±0.006 | 0.9441 | 172 | Ns | 0.070 | ±0.006 | 0.9441 | 12.7 | ±6.9 | 0.020 | ns | 0.9441 | |
| 100 | 2887 | ±504.0 | 0.001 | ±0.001 | 0.8202 | 38 | Ns | 0.005 | ±0.001 | 0.8202 | 56.0 | ±6.4 | 0.090 | ns | 0.8202 | |
| 25 | 50 | 415 | ±330.6 | 0.065 | ns | 0.8708 | 407 | ±330.6 | 0.332 | ns | 0.9766 | 1.1 | ns | 0.463 | ns | 0.9595 |
| 60 | 3956 | ±381.7 | 0.051 | ±0.020 | 0.9912 | 403 | ±381.7 | 0.319 | ±0.020 | 0.9912 | 9.8 | ±8.2 | 0.055 | ns | 0.9912 | |
| 70 | 4240 | ±292.6 | 0.065 | ±0.024 | 0.9978 | 413 | ±292.6 | 0.246 | ±0.024 | 0.9978 | 10.2 | ±6.3 | 0.067 | ns | 0.9978 | |
| 80 | 4206 | ±296.5 | 0.047 | ±0.016 | 0.9948 | 391 | ±296.5 | 0.402 | ±0.016 | 0.9948 | 10.7 | ±6.4 | 0.050 | ns | 0.9948 | |
| 90 | 1809 | ±331.5 | 0.048 | ±0.039 | 0.9341 | 154 | Ns | 0.309 | ±0.039 | 0.9341 | 11.7 | ±7.1 | 0.049 | ns | 0.9341 | |
| 100 | 2751 | ±393.5 | 0.009 | ±0.005 | 0.7484 | 36 | Ns | 0.033 | ±0.005 | 0.7484 | 70.1 | ±7.1 | 0.049 | ±0.023 | 0.7484 | |
| 45 | 50 | 189 | ns | 0.260 | ns | 0.9120 | 468 | ±401.0 | 2.486 | ±2.107 | 0.9959 | 0.3 | ns | 1.915 | ns | 0.8117 |
| 60 | 4674 | ±428.6 | 0.176 | ±0.057 | 0.9787 | 463 | ±428.6 | 2.129 | ±0.057 | 0.9787 | 7.0 | ns | 1.069 | ns | 0.9787 | |
| 70 | 5029 | ±326.5 | 0.113 | ±0.032 | 0.9594 | 463 | ±326.5 | 0.460 | ±0.032 | 0.9594 | 10.5 | ±6.9 | 0.156 | ns | 0.9594 | |
| 80 | 3667 | ±330.7 | 0.093 | ±0.037 | 0.9862 | 444 | ±330.7 | 0.152 | ±0.037 | 0.9862 | 8.1 | ±6.4 | 0.368 | ns | 0.9862 | |
| 90 | 1968 | ±337.2 | 0.304 | ±0.233 | 0.8638 | 124 | Ns | 0.476 | ±0.233 | 0.8638 | 14.6 | ±6.4 | 1.543 | ns | 0.8638 | |
| 100 | 4063 | ±386.8 | 0.024 | ±0.010 | 0.7838 | 34 | Ns | 0.036 | ±0.010 | 0.7838 | 109.2 | ±5.8 | 4.979 | ns | 0.7838 | |
| 65 | 50 | 347 | ns | 0.872 | ns | 0.9584 | 428 | ±419.4 | 3.008 | ns | 0.9985 | 0.8 | ns | 2.673 | ns | 0.9482 |
| 60 | 3274 | ±338.5 | 0.297 | ±0.173 | 0.9909 | 437 | ±338.5 | 1.990 | ±0.173 | 0.9909 | 6.3 | ns | 3.434 | ns | 0.9909 | |
| 70 | 3914 | ±290.9 | 0.162 | ±0.058 | 0.9906 | 380 | ±290.9 | 2.373 | ±0.058 | 0.9906 | 36.2 | ±12.8 | 0.012 | ±0.011 | 0.9906 | |
| 80 | 2381 | ±274.3 | 0.289 | ±0.174 | 0.9708 | 406 | ±274.3 | 0.807 | ±0.174 | 0.9708 | 5.7 | ns | 0.446 | ns | 0.9708 | |
| 90 | 2597 | ±405.1 | 0.249 | ±0.147 | 0.8919 | 141 | Ns | 0.300 | ±0.147 | 0.8919 | 17.5 | ±7.5 | 1.005 | ns | 0.8919 | |
| 100 | 3174 | ±363.6 | 0.045 | ±0.020 | 0.7444 | 31 | Ns | 0.234 | ±0.020 | 0.7444 | 105.4 | ±7.3 | 0.065 | ±0.019 | 0.7444 | |
| Variables | Y1 (µg Fx/g AS) | Y2 (mg E/g AS) | Y3 (mg Fx/g E) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S | T | MAE | RMSE | RMSE-MAE | MAE | RMSE | RMSE-MAE | MAE | RMSE | RMSE-MAE |
| (%) | (°C) | |||||||||
| 50 | 5 | 43.95 | 53.29 | 9.34 | 17.86 | 24.10 | 6.25 | 0.20 | 0.23 | 0.03 |
| 25 | 44.55 | 58.49 | 13.94 | 18.07 | 23.48 | 5.42 | 0.06 | 0.08 | 0.02 | |
| 45 | 16.16 | 20.74 | 4.58 | 9.13 | 11.20 | 2.07 | 0.05 | 0.06 | 0.01 | |
| 65 | 22.15 | 28.43 | 6.28 | 4.81 | 6.61 | 1.80 | 0.07 | 0.07 | 0.01 | |
| 60 | 5 | 189.55 | 265.20 | 75.65 | 10.97 | 15.88 | 4.91 | 0.74 | 0.97 | 0.23 |
| 25 | 371.33 | 444.06 | 72.73 | 10.02 | 11.63 | 1.62 | 0.97 | 1.24 | 0.27 | |
| 45 | 264.95 | 343.90 | 78.95 | 5.17 | 6.32 | 1.14 | 1.40 | 1.80 | 0.40 | |
| 65 | 319.24 | 440.00 | 120.76 | 7.46 | 8.74 | 1.28 | 1.21 | 1.37 | 0.16 | |
| 70 | 5 | 263.78 | 388.14 | 124.36 | 48.32 | 146.04 | 97.73 | 0.66 | 0.94 | 0.28 |
| 25 | 256.06 | 354.93 | 98.87 | 4.90 | 7.04 | 2.13 | 0.68 | 0.93 | 0.25 | |
| 45 | 364.91 | 483.49 | 118.58 | 16.31 | 23.13 | 6.82 | 0.79 | 1.24 | 0.45 | |
| 65 | 164.07 | 363.35 | 199.29 | 32.29 | 118.69 | 86.40 | 4.17 | 19.21 | 15.04 | |
| 80 | 5 | 244.76 | 356.14 | 111.38 | 41.53 | 123.47 | 81.94 | 0.53 | 1.12 | 0.59 |
| 25 | 324.49 | 440.27 | 115.77 | 6.99 | 10.52 | 3.53 | 0.74 | 1.03 | 0.30 | |
| 45 | 460.77 | 676.96 | 216.19 | 32.58 | 45.80 | 13.22 | 0.84 | 1.32 | 0.49 | |
| 65 | 198.90 | 484.48 | 285.58 | 4.84 | 13.54 | 8.70 | 0.43 | 0.75 | 0.32 | |
| 90 | 5 | 415.83 | 499.51 | 83.68 | 11.53 | 15.45 | 3.92 | 1.87 | 2.26 | 0.39 |
| 25 | 189.80 | 223.55 | 33.75 | 4.93 | 6.94 | 2.01 | 0.90 | 1.18 | 0.28 | |
| 45 | 206.90 | 239.23 | 32.32 | 6.73 | 9.32 | 2.59 | 1.01 | 1.57 | 0.56 | |
| 65 | 119.74 | 154.98 | 35.24 | 2.71 | 3.27 | 0.56 | 0.71 | 0.86 | 0.15 | |
| 100 | 5 | 464.49 | 546.32 | 81.83 | 4.50 | 5.65 | 1.15 | 10.37 | 14.89 | 4.51 |
| 25 | 353.15 | 403.64 | 50.49 | 4.57 | 5.76 | 1.18 | 10.78 | 13.25 | 2.47 | |
| 45 | 628.71 | 825.88 | 197.17 | 4.71 | 6.46 | 1.75 | 10.56 | 14.92 | 4.36 | |
| 65 | 274.31 | 429.70 | 155.39 | 2.79 | 6.19 | 3.40 | 17.38 | 25.67 | 8.30 | |
| Y1 (µg Fx/g AS) | |||
| Same solvent | k | constant | does not depend on T |
| r | increases with T | depends on T | |
| Same temperature | k | increases and decreases with S (curve) | depends on S |
| r | decreases with S | depends on S | |
| Y2 (mg E/g AS) | |||
| Same solvent | k | constant | does not depend on T |
| r | increases with T | depends on T | |
| Same temperature | k | decreases with S | depends on S |
| r | decreases with S | depends on S | |
| Y3 (mg Fx/g E) | |||
| Same solvent | k | constant | does not depend on T |
| r | not statistically significant * | - | |
| Same temperature | k | increases with S | depends on S |
| r | not statistically significant * | - | |
| Protein | PDB ID | Organism | R | Rp | Ligand Complex |
|---|---|---|---|---|---|
| AChE | 4EY7 | Homo sapiens | 2.35 Å | 90.3% | Donepezil |
| BuChE | 1P0P | Homo sapiens | 2.30 Å | 88.2% | N-acetylglucosamine |
| KS | 1FJ4 | Escherichia coli | 2.35 Å | 88.4% | Thiolactomycin |
| GY | 2XCS | Staphylococcus aureus | 2.10 Å | 92.2% | GSK-299423 |
| DR | 3SRW | Staphylococcus aureus | 1.70 Å | 89.7% | Q27 |
| Protein/Inhibitor | H-B | Be (kcal/mol) | ki (μM) | AA with H-B Interactions |
|---|---|---|---|---|
| Antimicrobial | ||||
| KS | 3 | −8.1 | 1.155 | MET 204, HIS 298, GLY 391 |
| GY | 4 | −7.5 | 3.180 | GLN 91, SER 128, ASP 81 |
| DR | 2 | −9.7 | 0.078 | ALA 8, LEU 21 |
| Neuroprotective | ||||
| AChE | 2 | −11.6 | 0.003 | TYR 72, THR 75 |
| BuChE | 1 | −6 | 39.991 | THR 284 |
| AChE Inhibitors | ||||
| Donepezil | 1 | −11.8 | 0.0022 | SER125 |
| Fx | 2 | −11.6 | 0.0031 | TYR 72, THR 75 |
| Galanyamine | 0 | −8.7 | 0.4196 | - |
| Memantine | 0 | −8.1 | 1.1552 | - |
| Rivastigmine | 1 | −8 | 1.3676 | PHE 295 |
| Formula | C42H58O6 | Pgp substrate | Yes |
| Molecular weight | 658.91 g/mol | CYP1A2 inhibitor | No |
| Heavy atoms | 48 | CYP2C19 inhibitor | No |
| H-B acceptors | 6 | CYP2C9 inhibitor | No |
| H-B donors | 2 | CYP2D6 inhibitor | No |
| GI | Low | CYP3A4 inhibitor | No |
| BBB | No | log Kp (cm/s) | −460 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lourenço-Lopes, C.; Fraga-Corral, M.; Soria-Lopez, A.; Nuñes-Estevez, B.; Barral-Martinez, M.; Silva, A.; Li, N.; Liu, C.; Simal-Gandara, J.; Prieto, M.A. Fucoxanthin’s Optimization from Undaria pinnatifida Using Conventional Heat Extraction, Bioactivity Assays and In Silico Studies. Antioxidants 2022, 11, 1296. https://doi.org/10.3390/antiox11071296
Lourenço-Lopes C, Fraga-Corral M, Soria-Lopez A, Nuñes-Estevez B, Barral-Martinez M, Silva A, Li N, Liu C, Simal-Gandara J, Prieto MA. Fucoxanthin’s Optimization from Undaria pinnatifida Using Conventional Heat Extraction, Bioactivity Assays and In Silico Studies. Antioxidants. 2022; 11(7):1296. https://doi.org/10.3390/antiox11071296
Chicago/Turabian StyleLourenço-Lopes, Catarina, Maria Fraga-Corral, Anton Soria-Lopez, Bernabe Nuñes-Estevez, Marta Barral-Martinez, Aurora Silva, Ningyang Li, Chao Liu, Jesus Simal-Gandara, and Miguel A. Prieto. 2022. "Fucoxanthin’s Optimization from Undaria pinnatifida Using Conventional Heat Extraction, Bioactivity Assays and In Silico Studies" Antioxidants 11, no. 7: 1296. https://doi.org/10.3390/antiox11071296
APA StyleLourenço-Lopes, C., Fraga-Corral, M., Soria-Lopez, A., Nuñes-Estevez, B., Barral-Martinez, M., Silva, A., Li, N., Liu, C., Simal-Gandara, J., & Prieto, M. A. (2022). Fucoxanthin’s Optimization from Undaria pinnatifida Using Conventional Heat Extraction, Bioactivity Assays and In Silico Studies. Antioxidants, 11(7), 1296. https://doi.org/10.3390/antiox11071296











