Global Profiling of the Antioxidant Constituents in Chebulae Fructus Based on an Integrative Strategy of UHPLC/IM-QTOF-MS, MS/MS Molecular Networking, and Spectrum-Effect Correlation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Plant Material
2.3. Extraction, Isolation, and Identification of the Main Constituents in CF
2.4. UHPLC-QTOF-MS and UPLC/IM-QTOF-MS Analyses
2.5. GNPS-based MS/MS Classic Molecular Networking
2.6. Establishment of In-House Compound Library
2.7. UPLC Fingerprinting and Multivariate Statistical Analysis
2.8. Antioxidant Assays, Spectrum-Effect Correlation, and DPPH• Pretreated UPLC Analysis
3. Results and Discussion
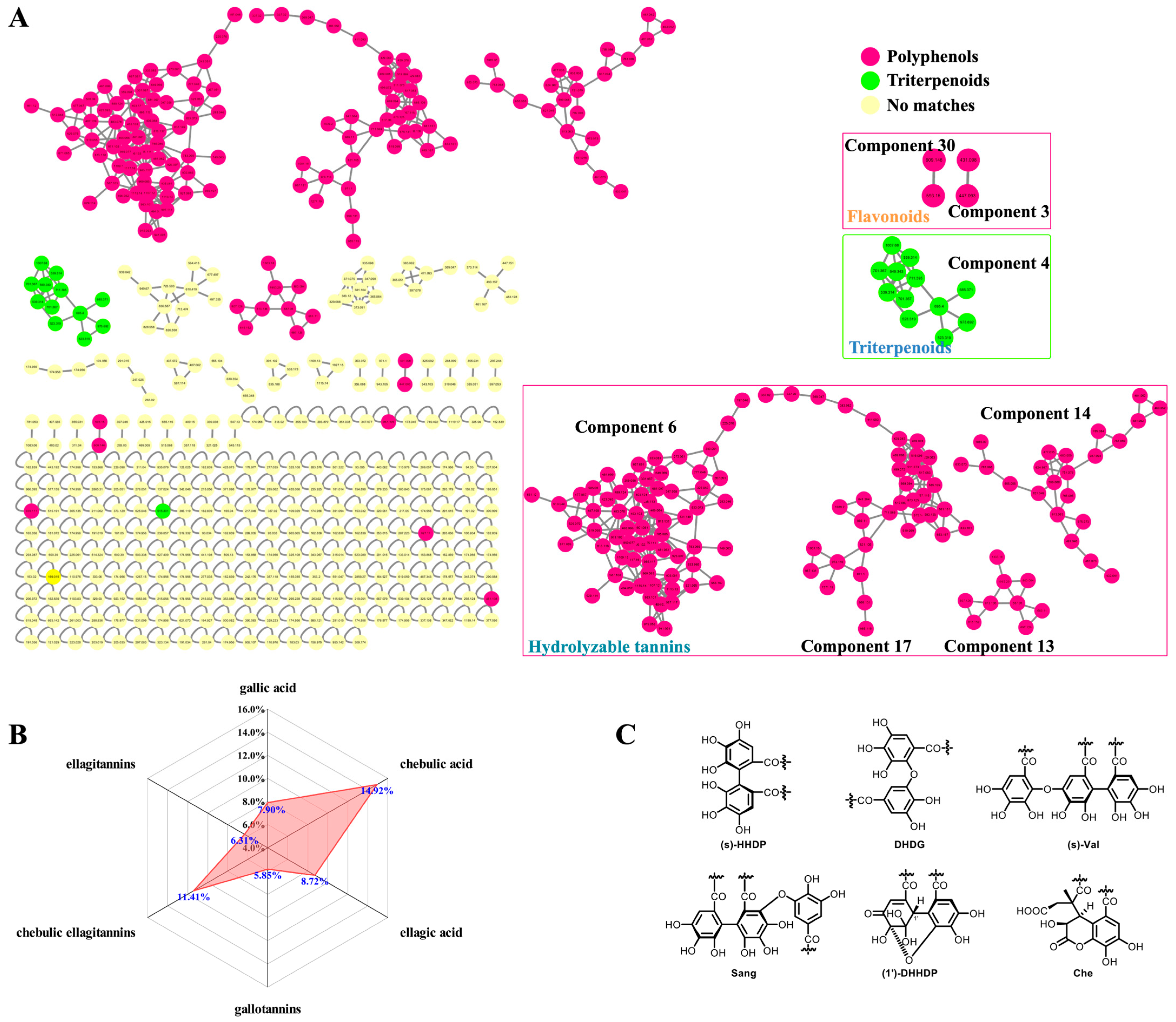
3.1. Integrative Strategy for Global Identification of the Constituents in CF
3.1.1. Identification Based on Molecular Networking and Annotation of the Network Nodes
3.1.2. Identification by in-House Library Search
3.1.3. Identification Based on the Cleavage Law of Standard Constituents
3.1.4. Isomer and Unknown Structure Discrimination Based on Collision Cross-Sections
3.1.5. Global Profiling of the Phenolcarboxylic and Tannic Constituents in CF
3.2. UPLC-PDA Fingerprinting and Multivariate Statistical Analysis
3.3. Antioxidant Evaluation and Screening of the Antioxidant Constituents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nigam, M.; Mishra, A.P.; Adhikari-Devkota, A.; Dirar, A.I.; Hassan, M.M.; Adhikari, A.; Belwal, T.; Devkota, H.P. Fruits of Terminalia chebula Retz.: A review on traditional uses, bioactive chemical constituents and pharmacological activities. Phytother. Res. 2020, 34, 2518–2533. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ding, X.T.; Sharma, A. Exploring scientific validation of Triphala Rasayana in Ayurveda as a source of rejuvenation for contemporary healthcare: An update. J. Ethnopharmacol. 2021, 273, 113829. [Google Scholar] [CrossRef] [PubMed]
- Bidikar, C.M.; Hurkadale, P.J.; Nandanwadkar, S.M.; Hegde, H.V. A validated spectro densitometric regulatory compliant USP-HP-TLC protocol for quantification of polyphenols and antioxidants from polyherbal formulations containing Terminalia species. J. Chromatogr. B 2022, 1207, 123379. [Google Scholar] [CrossRef] [PubMed]
- Pfundstein, B.; El Desouky, S.K.; Hull, W.E.; Haubner, R.; Erben, G.; Owen, R.W. Polyphenolic compounds in the fruits of Egyptian medicinal plants (Terminalia bellerica, Terminalia chebula and Terminalia horrida): Characterization, quantitation and determination of antioxidant capacities. Phytochemistry 2010, 71, 1132–1148. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Ali, Z.; Khan, I.A.; Khan, S.I. Anti-inflammatory activity of constituents isolated from Terminalia chebula. Nat. Prod. Commun. 2014, 9, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.S.; Cheng, B.C.; Zhang, S.F.; Luo, G.; Zhang, C.; Wang, Q.G.; Fu, X.Q.; Wang, Y.W.; Zhang, Y. Tibetan medicine for diabetes mellitus: Overview of pharmacological perspectives. Front. Pharmacol. 2021, 12, 748500. [Google Scholar] [CrossRef]
- Singh, G.; Kumar, P. Extraction, gas chromatography-mass spectrometry analysis and screening of fruits of Terminalia chebula Retz. for its antimicrobial potential. Pharmacogn. Res. 2013, 5, 162–168. [Google Scholar] [CrossRef]
- Patil, V.S.; Harish, D.R.; Vetrivel, U.; Roy, S.; Deshpande, S.H.; Hegde, H.V. Hepatitis C virus NS3/4A inhibition and host immunomodulation by tannins from Terminalia chebula: A structural perspective. Molecules 2022, 27, 1076. [Google Scholar] [CrossRef]
- Melo, L.F.M.; Aquino-Martins, V.G.Q.; Silva, A.P.D.; Oliveira Rocha, H.A.; Scortecci, K.C. Biological and pharmacological aspects of tannins and potential biotechnological applications. Food Chem. 2023, 414, 135645. [Google Scholar] [CrossRef]
- Manosroi, A.; Jantrawut, P.; Akihisa, T.; Manosroi, W.; Manosroi, J. In vitro and in vivo skin anti-aging evaluation of gel containing niosomes loaded with a semi-purified fraction containing gallic acid from Terminalia chebula galls. Pharm. Biol. 2011, 49, 1190–1203. [Google Scholar] [CrossRef]
- Bhatt, I.D.; Rawat, S.; Badhani, A.; Rawal, R.S. Nutraceutical potential of selected wild edible fruits of the Indian Himalayan region. Food Chem. 2017, 215, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Miao, X.; Wu, M.; Lv, Z.; Bai, Y.; Chang, Y.; Ouyang, H.; He, J. Pharmacokinetics of active compounds of a Terminalia chebula Retz. ethanolic extract after oral administration rats using UPLC-MS/MS. Front. Pharmacol. 2023, 14, 1067089. [Google Scholar] [CrossRef]
- Li, Y.J.; He, F.Q.; Zhao, H.H.; Li, Y.; Chen, J. Screening and identification of acetylcholinesterase inhibitors from Terminalia chebula fruits by immobilized enzyme on cellulose filter paper coupled with ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry and molecular docking. J. Chromatogr. A 2022, 1663, 462784. [Google Scholar] [CrossRef] [PubMed]
- Dhanani, T.; Shah, S.; Kumar, S. A validated high-performance liquid chromatography method for determination of tannin-related marker constituents gallic acid, corilagin, chebulagic acid, ellagic acid and chebulinic acid in four Terminalia species from India. J. Chromatogr. Sci. 2015, 53, 625–632. [Google Scholar] [CrossRef]
- Saha, S.; Imran, I.B. Isolation, detection, and quantification of hydrolyzable tannins of the biosynthetic pathway by liquid chromatography coupled with tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2021, 35, e9005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Bi, J.H.; Xie, M.; Zhang, H.; Shi, Z.Q.; Guo, H.; Yin, H.B.; Zhang, J.N.; Xin, G.Z.; Song, H.P. Classification-based strategies to simplify complex traditional Chinese medicine (TCM) researches through liquid chromatography-mass spectrometry in the last decade (2011–2020): Theory, technical route and difficulty. J. Chromatogr. A 2021, 1651, 462307. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, S.; Zhu, B.; Wang, R.; Cheng, Y. A strategy for identifying effective and risk compounds of botanical drugs with LC-QTOF-MS and network analysis: A case study of Ginkgo biloba preparation. J. Pharm. Biomed. 2021, 193, 113759. [Google Scholar] [CrossRef]
- Tchetan, E.; Ortiz, S.; Olounladé, P.A.; Hughes, K.; Laurent, P.; Azando, E.V.B.; Hounzangbe-Adote, S.M.; Gbaguidi, F.A.; Quetin-Leclercq, J. Fractionation coupled to molecular networking: Towards identification of anthelmintic molecules in Terminalia leiocarpa (DC.) Baill. Molecules 2022, 28, 76. [Google Scholar] [CrossRef]
- Li, C.; Yu, M.; Li, S.; Yang, X.; Qiao, B.; Shi, S.; Zhao, C.; Fu, Y. Valorization of Fig (Ficus carica L.) waste leaves: HPLC-QTOF-MS/MS-DPPH system for online screening and identification of antioxidant compounds. Plants 2021, 10, 2532. [Google Scholar] [CrossRef]
- Dai, Z.; An, L.Y.; Chen, X.Y.; Yang, F.; Zhao, N.; Li, C.C.; Ren, R.; Li, B.Y.; Tao, W.Y.; Li, P.; et al. Target fishing reveals a novel mechanism of 1,2,4-Oxadiazole derivatives targeting Rpn6, a subunit of 26S proteasome. J. Med. Chem. 2022, 65, 5029–5043. [Google Scholar] [CrossRef]
- Chinese Pharmacopoeia Commission. Chinese Pharmacopoeia Part 1, 2020 ed.; China Medical Science Press: Beijing, China, 2020; p. 194. [Google Scholar]
- Sudjaroen, Y.; Hull, W.E.; Erben, G.; Wurtele, G.; Changbumrung, S.; Ulrich, C.M.; Owen, R.W. Isolation and characterization of ellagitannins as the major polyphenolic components of Longan (Dimocarpus longan Lour) seeds. Phytochemistry 2012, 77, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Wang, Q.; Wang, Q.; Yuan, C.; Li, G.; Ji, T. Preparative isolation of antioxidant gallic acid derivatives from Saxifraga tangutica using a class separation method based on medium-pressure liquid chromatography and reversed-phase liquid chromatography. J. Sep. Sci. 2021, 44, 3734–3746. [Google Scholar] [CrossRef] [PubMed]
- Sohretoglu, D.; Sakar, M.K.; Sabuncuoglu, S.A.; Ozgunes, H.; Sterner, O. Polyphenolic Constituents and antioxidant potential of Geranium stepporum Davis. Rec. Nat. Prod. 2011, 5, 22–28. [Google Scholar] [CrossRef]
- Hirano, Y.; Kondo, R.; Sakai, K. 5α-Reductase inhibitory tannin-related compounds isolated from Shorea laeviforia. J. Wood Sci. 2003, 49, 339–343. [Google Scholar] [CrossRef]
- Zhou, Z.W.; Luo, M.D.; Chen, X.; Yin, Y.D.; Xiong, X.; Wang, R.H.; Zhu, Z.J. Ion mobility collision cross-section atlas for known and unknown metabolite annotation in untargeted metabolomics. Nat. Commun. 2020, 11, 4334. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Ernst, M.; Kang, K.B.; Caraballo-Rodriguez, A.M.; Nothias, L.F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Zhu, J.; Zhong, L.; Kong, S.; Zhang, Y.; Huang, P. Comprehensive evaluation of the antioxidant capacity of Sceptridium ternatum using multiple colorimetric methods and 1,1-diphenyl-2-picrylhydrazyl-high-performance liquid chromatography analysis. J Sep. Sci. 2020, 43, 3615–3624. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Wan, Y.; Xu, J.Y.; Wu, G.H.; Li, L.; Yao, X.H. Ultrasound extraction of polysaccharides from mulberry leaves and their effect on enhancing antioxidant activity. Carbohydr. Polym. 2016, 137, 473–479. [Google Scholar] [CrossRef]
- Piscopo, M.; Tenore, G.C.; Notariale, R.; Maresca, V.; Maisto, M.; de Ruberto, F.; Heydari, M.; Sorbo, S.; Basile, A. Antimicrobial and antioxidant activity of proteins from Feijoa sellowiana Berg. fruit before and after in vitro gastrointestinal digestion. Nat. Prod. Res. 2020, 34, 2607–2611. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.Q.; Zhou, P.; Meng, X.W.; Yang, H.; Li, Y.; Xing, X.D.; Wang, H.Y.; Yan, F.R.; Li, P.; Gao, W. An image-based fingerprint-efficacy screening strategy for uncovering active compounds with interactive effects in Yindan Xinnaotong soft capsule. Phytomedicine 2022, 96, 153911. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.X.; Liu, S.X.; Kwabena Oduro, P.; Ama Mireku-Gyimah, N.; Zhang, L.H.; Wang, Q.L.; Wu, H.H. Vasodilatory constituents of essential oil from Nardostachys jatamansi DC.: Virtual screening, experimental validation and the potential molecular mechanisms. Arab. J. Chem. 2023, 16, 104911. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Castillo, C.M.S.; Caroca, R.; Lazo-Vélez, M.A.; Antonyak, H.; Polishchuk, A.; Lysiuk, R.; Oliinyk, P.; De Masi, L.; et al. Ellagic acid: A review on its natural sources, chemical stability, and therapeuticpotential. Oxid. Med. Cell. Longev. 2022, 2022, 3848084. [Google Scholar] [CrossRef]
- Li, X.H.; Wang, H.D.; Jiang, M.T.; Ding, M.X.; Xu, X.Y.; Xu, B.; Zou, Y.D.; Yu, Y.T.; Yang, W.Z. Collision cross section prediction based on machine learning. Molecules 2023, 28, 4050. [Google Scholar] [CrossRef]
- Song, X.C.; Canellas, E.; Dreolin, N.; Nerin, C.; Goshawk, J. Discovery and Characterization of phenolic compounds in Bearberry (Arctostaphylos uva-ursi) leaves using liquid chromatography-ion mobility-high-resolution mass spectrometry. J. Agric. Food Chem. 2021, 69, 10856–10868. [Google Scholar] [CrossRef]
- de Araújo, F.F.; de Paulo Farias, D.; Neri-Numa, I.A.; Pastore, G.M. Polyphenols and their applications: An approach in food chemistry and innovation potential. Food Chem. 2021, 338, 127535. [Google Scholar] [CrossRef]
- Binette, V.; Côté, S.; Haddad, M.; Nguyen, P.T.; Bélanger, S.; Bourgault, S.; Ramassamy, C.; Gaudreault, R.; Mousseau, N. Corilagin and 1,3,6-tri-O-galloy-β-D-glucose: Potential inhibitors of SARS-CoV-2 variants. Phys. Chem. Chem. Phys. 2021, 23, 14873–14888. [Google Scholar] [CrossRef]
- Kesharwani, A.; Polachira, S.K.; Nair, R.; Agarwal, A.; Mishra, N.N.; Gupta, S.K. Anti-HSV-2 activity of Terminalia chebula Retz extract and its constituents, chebulagic and chebulinic acids. BMC Complement. Altern. Med. 2017, 17, 110. [Google Scholar] [CrossRef]
- Castro, S.M.; Guerrero-Plata, A.; Suarez-Real, G.; Adegboyega, P.A.; Colasurdo, G.N.; Khan, A.M.; Garofalo, R.P.; Casola, A. Antioxidant treatment ameliorates respiratory syncytial virus-induced disease and lung inflammation. Am. J. Respir. Crit. Care Med. 2006, 174, 1361–1369. [Google Scholar] [CrossRef]
- Hassan Bulbul, M.R.; Uddin Chowdhury, M.N.; Naima, T.A.; Sami, S.A.; Imtiaj, M.S.; Huda, N.; Uddin, M.G. A comprehensive review on the diverse pharmacological perspectives of Terminalia chebula Retz. Heliyon 2022, 8, e10220. [Google Scholar] [CrossRef]




| No. | Observed tR (min) | Formula | Identification | Measured CCS (Ų) | Predicted CCS (Ų) | Observed Mass (Da) | Mass Error (ppm) | Quasi-Molecular /Adducts Ion | MS/MS Fragmentation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1.04 | C7H11O6 | quinic acid | – | 142.889 | 191.0564 | 4.2 | [M–H]− | 85.0297, 93.0343, 59.0141, 55.0193, 67.0191, 108.0214 |
| 2 | 1.52 | C7H9O5 | shikimic acid | – | 135.297 | 173.0454 | 2.3 | [M–H]− | 93.0344, 65.0399, 55.0194 |
| 17 | 10.11 | C11H11O8 | MN-01 | – | 155.806 | 271.0454 | 0.0 | [M–H]− | 124.0167, 78.0113 |
| phenolcarboxylic acids | |||||||||
| 3 * | 4.15 | C14H11O11 | chebulic acid | – | 174.904 | 355.0329 | 7.9 | [M–H]− | 149.0244, 193.0141, 164.0475, 135.0452, 164.0475, 179.0707, 107.0497 |
| 10 | 6.32 | C17H17O13 | MN-02 | 189.370 | 197.766 | 429.0678 | 2.1 | [M–H]− | 193.0142, 163.0395, 205.0503, 133.0657, 151.0398, 59.0140, 249.0405 |
| 12 * | 8.24 | C7H5O5 | gallic acid | 124.174 | 126.339 | 169.0145 | 4.7 | [M–H]− | 125.0244, 169.0143, 79.0184 |
| 27 | 11.85 | C16H15O11 | MN-03 | 179.820 | 185.931 | 383.0617 | 0.8 | [M–H]− | 193.0138, 205.0508, 163.0399, 135.0450, 233.0062 |
| 30 * | 12.44 | C14H13O9 | 4-O-galloyl-(-)-shikimic acid | 173.481 | 173.304 | 325.0575 | 4.6 | [M–H]− | 125.0168, 93.0344, 169.0144 |
| 33 * | 13.01 | C14H13O9 | 5-O-galloyl-(-)-shikimic acid | 175.504 | 174.817 | 325.0571 | 3.4 | [M–H]− | 125.0244, 107.0137, 93.0344, 169.0138 |
| 34 | 13.38 | C14H13O9 | 3-O-galloyl-(-)-shikimic acid | 174.327 | 175.075 | 325.0572 | 3.7 | [M–H]− | 125.0246, 107.0136, 93.0343, 169.0142 |
| 42 | 15.38 | C12H11O7 | MN-04 | 158.510 | 159.938 | 267.0512 | 2.6 | [M–H]− | 247.9025, 166.8641, 124.0166, 107.0133, 78.0114 |
| 45 | 16.08 | C9H9O5 | ethyl gallate | – | 141.145 | 197.0461 | 5.6 | [M–H]− | 124.0166, 78.0155, 51.0244 |
| 50 | 17.14 | C14H9O9 | digallic acid | – | 166.869 | 321.0254 | 2.2 | [M–H]− | 125.0244, 169.0146, 79.0188 |
| 52 | 17.52 | C21H9O13 | flavogallonic acid | – | 199.719 | 469.0055 | 2.6 | [M–H]− | 299.9913, 379.0090, 407.0039, 300.9979, 351.0145, 335.0195, 425.0136 |
| 58 | 18.78 | C21H17O13 | 3,5-di-O-galloylshikimic acid | 203.970 | 214.557 | 477.0675 | 1.3 | [M–H]− | 125.0244, 93.0343, 123.0083, 263.0552, 201.0547, 169.0143 |
| 64 | 19.95 | C13H7O8 | brevifolin carboxylic acid | 152.493 | 160.170 | 291.0147 | 2.1 | [M–H]− | 145.0293, 190.0268, 247.0238 |
| 67 | 20.40 | C21H17O13 | 4,5-di-O-galloylshikimic acid | 198.260 | 210.711 | 477.0674 | 1.0 | [M–H]− | 169.0143, 125.0244, 201.0547, 93.0343, 51.0245 |
| 74 | 21.41 | C21H17O13 | 3,4-di-O-galloylshikimic acid | 198.420 | 212.094 | 477.0681 | 2.5 | [M–H]− | 169.0143, 125.0244, 263.0552, 201.0547, 93.0343, 51.0245 |
| 82 | 22.70 | C11H13O5 | MN-05 | 153.340 | 151.904 | 225.0762 | –0.4 | [M–H]− | 153.7419, 124.0167, 78.0113 |
| 86 * | 23.03 | C13H7O7 | urolithin M5 | 149.257 | 153.132 | 275.0199 | 2.5 | [M–H]− | 145.0296, 173.0243, 117.0342, 229.0139 |
| 94 | 24.86 | C28H21O17 | 3,4,5-tri-O-galloylshikimic acid | 230.001 | 241.753 | 629.0790 | 1.7 | [M–H]− | 289.0339, 245.0439, 201.0551, 169.0143, 125.0243, 93.0344 |
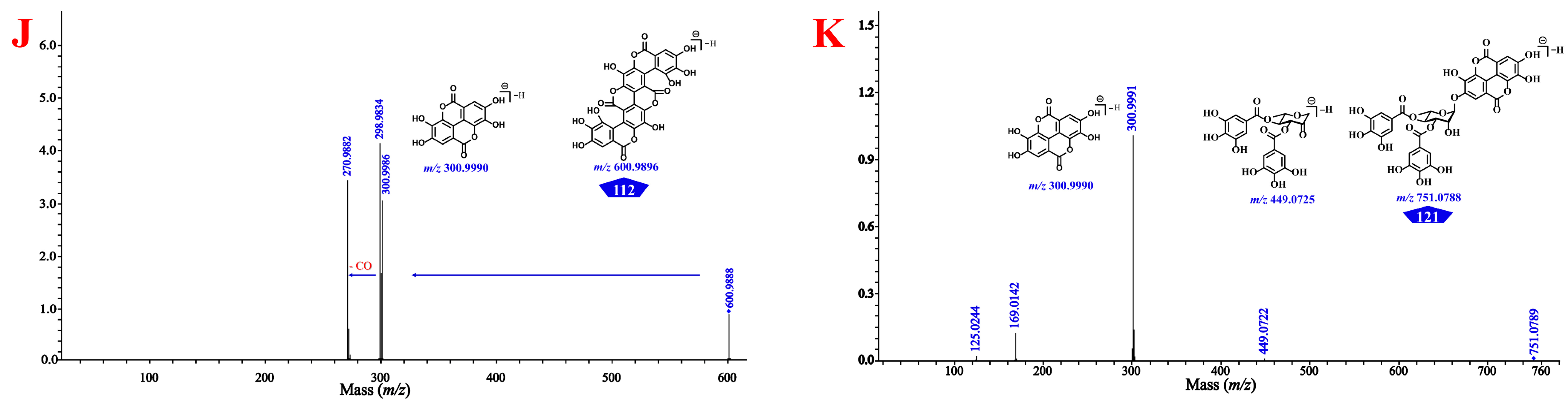
| 110 * | 29.01 | C14H5O8 | ellagic acid | 153.266 | 160.297 | 300.9995 | 3.7 | [M–H]− | 145.0294, 117.0344, 200.0117, 172.0169, 283.9964, 173.0247, 129.0348, 201.018, 299.9915, 300.9988 |
| 112 * | 29.64 | C28H9O16 | terminalin | – | 222.392 | 600.9900 | 1.5 | [M–H]− | 298.9832, 270.9880, 300.9989, 299.9897, 485.0143, 582.9772 |
| gallotannins | |||||||||
| 4 | 4.49 | C13H15O10 | 6-galloyl-β-d-glucose | 172.592 | 171.292 | 331.0667 | 0.6 | [M–H]− | 169.0144, 125.0243, 107.0134, 59.0143, 51.0229 |
| 7 | 5.47 | C13H15O10 | 4-galloyl-β-d-glucose | 173.955 | 174.532 | 331.0669 | 1.2 | [M–H]− | 125.0242, 169.0141, 107.0134, 59.0140 |
| 15 * | 9.27 | C13H15O10 | 1-O-galloyl-β-d-glucose | 172.066 | 170.416 | 331.0669 | 1.2 | [M–H]− | 59.0142, 169.0139, 125.0243, 107.0136, 51.0245 |
| 18 | 10.12 | C13H15O10 | 3-galloyl-β-d-glucose | 175.413 | 175.534 | 331.0669 | 1.2 | [M–H]− | 125.0243, 107.0134, 169.0140, 59.0143 |
| 19 | 10.12 | C13H17O10 | MN-06 | 175.750 | 174.430 | 333.0830 | 2.4 | [M–H]− | 125.0244, 107.0136, 169.0141, 59.0141 |
| 22 | 10.85 | C13H15O10 | 2-galloyl-β-d-glucose | – | – | 331.0673 | 2.4 | [M–H]− | 169.0137, 125.0243, 107.0134, 59.0143, 211.0248 |
| 28 | 12.05 | C20H19O14 | 2,4-di-O-galloyl-d-glucose | 198.512 | 209.402 | 483.0780 | 1.0 | [M–H]− | 169.0143, 125.0245, 313.0573, 271.0453, 211.0240 |
| 35 | 13.47 | C20H19O14 | 2,3-di-O-galloyl-d-glucose | 197.150 | 206.773 | 483.0780 | 1.0 | [M–H]− | 169.0140, 125.0243, 313.0564, 271.0453, 211.0248 |
| 38 | 14.32 | C20H19O14 | 1,2-di-O-galloyl-d-glucose | 195.054 | 202.905 | 483.0780 | 1.0 | [M–H]− | 169.0144, 125.0245, 271.0452, 211.0245, 59.0142 |
| 41 | 15.23 | C20H19O14 | 1,3-di-O-galloyl-d-glucose | 195.827 | 204.454 | 483.0780 | 1.0 | [M–H]− | 169.0145, 125.0246, 271.0454, 211.0246 |
| 46 | 16.37 | C20H19O14 | 1,6-di-O-galloyl-d-glucose | 194.986 | 200.090 | 483.0780 | 1.0 | [M–H]− | 169.0142, 125.0243, 313.0562, 271.0449, 211.0246 |
| 48 | 16.74 | C20H19O14 | 4,6-di-O-galloyl-d-glucose | 196.386 | 205.002 | 483.0786 | 2.3 | [M–H]− | 169.0142, 125.0241, 313.0544, 271.0455, 211.0240 |
| 49 | 17.10 | C20H19O14 | 3,4-di-O-galloyl-d-glucose | 195.162 | 203.345 | 483.0790 | 3.1 | [M–H]− | 169.0143, 125.0244, 211.0244, 271.0453, 313.0573 |
| 53 | 17.93 | C20H19O14 | 3,6-di-O-galloyl-d-glucose | 196.961 | 205.228 | 483.0779 | 0.8 | [M–H]− | 169.0143, 125.0243, 313.0573, 271.0453, 211.0240, 423.0000, 151.0032 |
| 55 | 18.13 | C27H23O18 | tri-O-galloyl-β-d-glucose | 226.655 | – | 635.0898 | 2.2 | [M–H]− | 169.0144, 465.0668, 423.0566, 313.0567, 211.0243, 271.0453 |
| 63 | 19.58 | C27H23O18 | tri-O-galloyl-β-d-glucose | 225.329 | – | 635.0901 | 2.7 | [M–H]− | 169.0144, 125.0244, 635.0894, 483.0763, 423.0566, 313.0567, 211.0243 |
| 66 | 20.26 | C19H19O11 | MN-07 | 201.510 | 196.492 | 423.0928 | 0.2 | [M–H]− | 169.0136, 109.0289, 125.0244, 203.0373, 299.0122 |
| 68 * | 20.44 | C27H23O18 | 1,3,6-tri-O-galloyl-β-d-glucose | 229.419 | 229.367 | 635.0888 | 0.6 | [M–H]− | 169.0144, 313.0567, 211.0243, 635.0894, 465.0668, 271.0461 |
| 69 | 20.69 | C27H23O18 | tri-O-galloyl-β-d-glucose | 229.174 | – | 635.0889 | 0.8 | [M–H]− | 169.0141, 125.0243, 313.0559, 211.0243, 635.0894, 465.0654, 271.0461, 423.0572 |
| 70 | 20.83 | C27H23O18 | 1,4,6-tri-O-galloyl-β-d-glucose | 228.727 | 228.651 | 635.0895 | 1.7 | [M–H]− | 169.0142, 125.0242, 313.0562, 211.0239, 465.0660, 271.0448, 423.0561 |
| 72 | 21.05 | C27H23O18 | 1,2,6-tri-O-galloyl-β-d-glucose | 227.836 | 227.577 | 635.0894 | 1.6 | [M–H]− | 169.0142, 125.0242, 313.0564, 211.0235, 465.0660 |
| 75 | 21.51 | C34H27O22 | 1,2,4,6-tetra-O-galloyl-β-d-glucose | 252.633 | 255.173 | 787.1000 | 0.8 | [M–H]− | 169.0141, 465.0659, 295.0447, 635.0876, 423.0552, 211.0246, 125.0242 |
| 89 | 23.63 | C34H27O22 | 2,3,4,6-tetra-O-galloyl-β-d-glucose | 266.444 | 259.406 | 787.1004 | 1.3 | [M–H]− | 169.0146, 465.0675, 295.0455, 125.024, 617.0776 |
| 92 | 24.17 | C34H27O22 | 1,2,3,6-tetra-O-galloyl-β-d-glucose | 253.240 | 253.666 | 787.0995 | 0.1 | [M–H]− | 169.0141, 465.0662, 617.0768, 295.0446 |
| 97 | 25.70 | C41H31O26 | 1,2,3,4,6-penta-O-galloyl-β-d-glucose | 296.630 | 282.841 | 939.1114 | 1.1 | [M–H]− | 169.0143, 617.0779, 769.0885, 447.0563, 725.0979, 295.0451 |
| 100 | 26.31 | C34H27O23 | MN-08 | 259.230 | 258.547 | 803.0947 | 0.5 | [M–H]− | 275.0196, 205.0503, 169.0144, 337.0198, 651.0861 |
| 105 | 27.70 | C29H25O15 | di-O-galloyl-2-O-cinnamoyl-β-d-glucose | – | 239.887 | 613.1182 | –1.8 | [M–H]− | 169.0144,125.0243, 71.0140, 21.0239, 401.0860, 443.0983 |
| 106 | 28.04 | C29H25O15 | di-O-galloyl-3-O-cinnamoyl-β-d-glucose | – | 238.022 | 613.1186 | –1.1 | [M–H]− | 169.0140, 125.0240, 451.0789, 211.0221, 71.0136 |
| ellagitannins | |||||||||
| 6 | 5.43 | C20H17O14 | MN-09 | 202.370 | 200.811 | 481.0620 | 0.4 | [M–H]− | 300.9985, 275.0197, 229.0142 |
| 11 | 8.16 | C20H17O14 | MN-10 | 201.750 | 200.377 | 481.0620 | 0.4 | [M–H]− | 300.9984, 275.0197, 229.0147, 185.0244, 125.0239 |
| 29 | 12.32 | C20H15O13 | MN-11 | 196.640 | 200.247 | 463.0516 | 0.6 | [M–H]− | 300.9990, 275.0204 |
| 31 | 12.65 | C34H21O22 | punicalin A | 247.216 | 240.735 | 781.0539 | 1.9 | [M–H]− | 600.9889, 298.9831, 575.0083, 300.9989, 721.0299, 275.02, 448.9785, 781.0520 |
| 32 | 12.84 | C34H21O22 | punicalin B | 248.928 | 240.735 | 781.0535 | 1.4 | [M–H]− | 600.9896, 298.9831, 575.0039, 721.0300, 781.0514 |
| 36 | 13.86 | C34H23O23 | MN-12 | 242.080 | 246.089 | 783.0672 | –1.1 | [M–H]− | 300.9979, 275.0199, 783.0642, 481.0663 |
| 37 | 14.11 | C27H21O18 | corilagin isomer | – | 223.714 | 633.0738 | 1.6 | [M–H]− | 300.9987, 275.0202, 169.0144, 463.0517 |
| 39 * | 14.77 | C48H27O30 | punicalagin A | 305.520 | 295.162 | 1083.0608 | 1.9 | [M–H]− | 1083.0599, 600.9898, 781.0530, 575.0098, 721.0330, 300.9986 |
| 43 | 15.80 | C34H23O23 | terflavin B | 241.867 | 249.874 | 783.0679 | –0.3 | [M–H]− | 450.9940, 631.0549, 425.0126, 300.997, 783.0695, 169.0143 |
| 44 | 16.03 | C34H25O22 | tercatain or its isomer | 267.387 | 247.710 | 785.0847 | 1.3 | [M–H]− | 300.9987, 275.0198, 249.0406, 633.0736, 169.0143, 785.0817 |
| 47 * | 16.59 | C48H27O30 | punicalagin B | 303.497 | 295.162 | 1083.0608 | 1.9 | [M–H]− | 1083.0599, 600.9898, 781.0523, 721.0313, 300.9987, 549.0295, 448.9775, 249.0400 |
| 56 * | 18.49 | C27H21O18 | corilagin | 224.508 | 224.752 | 633.0739 | 1.7 | [M–H]− | 300.9996, 275.0202, 463.0517, 169.0144 |
| 57 | 18.66 | C34H25O22 | tercatain or its isomer | 268.495 | 249.881 | 785.0837 | 0.0 | [M–H]− | 300.9984, 275.0192, 633.0736, 615.0593, 249.0406, 169.0137 |
| 60 | 19.22 | C48H29O30 | terflavin A or its isomer | 312.025 | 304.291 | 1085.0742 | –0.2 | [M–H]− | 450.9933, 783.0668, 1085.0719, 933.0632, 631.0559, 425.0149, 300.9976 |
| 61 | 19.27 | C34H25O22 | tercatain or its isomer | 268.495 | 249.063 | 785.0851 | 1.8 | [M–H]− | 300.9987, 275.0198, 633.0736, 249.0406, 169.0141 |
| 65 | 19.96 | C34H25O22 | tellimagradin I | 264.090 | 255.565 | 785.0831 | –0.8 | [M–H]− | 300.9987, 275.0198, 633.0723, 169.0141 |
| 71 | 21.02 | C48H29O30 | terflavin A or its isomer | 311.474 | 305.606 | 1085.0747 | 0.3 | [M–H]− | 450.9933, 783.0668, 1085.0719, 933.0632, 631.0559, 425.0149, 300.9976 |
| 83 | 22.73 | C41H29O26 | tellimagrandin II | 295.841 | 277.927 | 937.0940 | –0.7 | [M–H]− | 300.9985, 275.0195, 937.0922, 767.0694, 599.0685, 465.0654, 419.0603, 169.0138 |
| 101 | 26.53 | C20H13O14 | MN-13 | 198.090 | 201.575 | 477.0348 | 9.0 | [M–H]− | 300.9988, 125.024, 302.002, 169.014 |
| 108 | 28.57 | C21H15O14 | MN-14 | 208.100 | 210.883 | 491.0460 | –0.4 | [M–H]− | 299.9907, 270.9857, 169.0138, 125.0242 |
| 111 * | 29.32 | C20H15O12 | eschweilenol C | 200.297 | 197.035 | 447.0571 | 1.6 | [M–H]− | 299.9941, 300.9978, 271.9950 |
| 116 | 31.12 | C21H17O12 | MN-15 | 205.440 | 202.894 | 461.0736 | 3.5 | [M–H]− | 299.9917, 315.0143, 270.9882 |
| 118 | 31.54 | C27H19O16 | 4-O-(4″-O-galloyl-α-l-rhamnosyl)ellagic acid | 213.728 | 228.827 | 599.0686 | 2.2 | [M–H]− | 300.9989, 169.0141, 125.0242 |
| 121 * | 32.49 | C34H23O20 | 4-O-(3″,4″-di-O-galloyl-α-l-rhamnosyl)ellagic acid | 238.780 | 253.167 | 751.0789 | 0.8 | [M–H]− | 300.9986, 169.0138, 449.0716 |
| 122 | 32.99 | C34H23O20 | 4-O-(2″,4″-di-O-galloyl-α-l-rhamnosyl)ellagic acid | 244.308 | 253.300 | 751.0783 | 0.0 | [M–H]− | 300.9986, 169.0138, 449.0716, 599.0658 |
| chebulic ellagitannins | |||||||||
| 5 | 4.71 | C20H23O16 | MN-16 | 205.480 | 206.812 | 519.0996 | 1.9 | [M–H]− | 205.0508, 163.0398, 193.0141, 177.0551, 133.0657, 187.0397, 249.0400, 293.0317, 337.0199 |
| 8 | 5.47 | C20H23O16 | MN-17 | 205.220 | 205.690 | 519.1001 | 1.9 | [M–H]− | 205.0510, 163.0399, 193.0141, 133.0657, 187.0397, 249.0403, 293.0303, 337.0195 |
| 9 | 5.96 | C20H21O16 | MN-18 | – | 207.783 | 517.0844 | 2.7 | [M–H]− | 205.0507, 193.0139, 163.0400, 133.0656, 177.0552, 249.0405, 337.0188, 293.0305, 275.0186 |
| 13 | 8.58 | C20H23O16 | MN-19 | 203.200 | 204.232 | 519.0997 | 2.1 | [M–H]− | 205.0506, 193.0140, 161.0605, 231.0290, 237.0030, 249.04, 59.0144, 293.0301, 337.0190, 401.0722, 275.0188 |
| 14 | 8.88 | C20H21O16 | MN-20 | – | 209.049 | 517.0833 | 0.6 | [M–H]− | 237.0292, 205.0506, 249.0401, 279.0143, 309.0259, 339.0357, 161.0607, 133.0655 |
| 16 | 9.81 | C20H19O15 | MN-21 | 203.370 | 205.632 | 499.0726 | 0.4 | [M–H]− | 163.0401, 193.0131, 151.0382, 231.0302, 205.0506, 177.0544 |
| 20 | 10.59 | C21H19O15 | MN-22 | 207.020 | 211.191 | 511.0739 | 2.9 | [M–H]− | 193.0141, 205.0506, 163.0398, 161.0608, 133.0656, 187.0398, 249.0401, 337.0206, 275.0192 |
| 21 | 10.83 | C20H19O15 | MN-23 | 200.635 | 206.525 | 499.0727 | 0.6 | [M–H]− | 193.0141, 205.0510, 231.0277, 161.0607, 187.0388 |
| 23 | 10.90 | C21H19O15 | MN-24 | – | 212.003 | 511.0736 | 2.3 | [M–H]− | 205.0505, 193.0141, 163.0395, 133.0652, 187.0395, 249.0402, 337.0183, 59.0142, 293.0318, 177.0546 |
| 24 | 11.02 | C21H19O15 | MN-25 | – | 212.647 | 511.0738 | 2.7 | [M–H]− | 205.0505, 193.0140, 163.0398, 133.0655, 187.0393, 249.0398, 337.0192, 393.0455 |
| 25 | 11.32 | C21H19O15 | MN-26 | – | 215.118 | 511.0739 | 2.9 | [M–H]− | 193.0141, 205.0506, 163.0399, 187.0400, 337.0190, 151.0399, 133.0658 |
| 26 | 11.49 | C21H19O15 | MN-27 | – | 216.331 | 511.0739 | 2.9 | [M–H]− | 193.0139, 205.0507, 163.0399, 177.0550, 187.0396, 337.0191, 151.0397, 133.0655 |
| 40 | 14.99 | C27H25O20 | phyllanemblinin E | 233.848 | 233.045 | 669.0955 | 2.4 | [M–H]− | 205.0510, 249.0407, 193.0142, 161.0609, 337.0197, 293.0301, 133.0656 |
| 51 | 17.30 | C28H27O20 | 1′-O-methyl neochebulanin | – | 239.776 | 683.1136 | 5.9 | [M–H]− | 169.0148, 125.0248, 231.0304, 351.0357, 275.0206, 409.0773 |
| 54 | 17.95 | C34H29O24 | MN-28 | 262.480 | 258.372 | 821.1049 | 0.0 | [M–H]− | 337.0191, 483.0759, 293.0314, 249.0394, 205.0511 |
| 59 | 19.04 | C41H31O28 | neochebulagic acid | 284.153 | 281.548 | 971.1003 | 0.1 | [M–H]− | 633.0728, 337.0198, 300.9988, 463.0514, 419.0610, 249.0404, 205.0511 |
| 62 | 19.41 | C28H27O20 | 1′-O-methyl neochebulanin isomer | – | 240.401 | 683.1100 | 0.6 | [M–H]− | 169.0142, 125.0243, 203.0341, 437.0755, 381.0462, 337.0555 |
| 73 | 21.26 | C42H33O28 | methyl neochebulagate | 306.796 | 288.119 | 985.1148 | –1.0 | [M–H]− | 633.0719, 783.0664, 463.0505, 351.0345, 300.9983, 231.0296, 169.0143 |
| 76 | 21.74 | C41H33O28 | neochebulinic acid | 285.501 | 286.651 | 973.1167 | 0.9 | [M–H]− | 635.0887, 337.0194, 465.0652, 249.0396, 293.0292, 205.0495 |
| 77 | 21.75 | C33H35O24 | MN-29 | 275.960 | 258.659 | 815.1520 | 0.2 | [M–H]− | 169.0141, 435.0562, 645.1298, 205.0505, 125.0242, 381.0453, 463.0505 |
| 78 | 22.04 | C33H33O24 | MN-30 | 257.675 | 257.674 | 813.1367 | 0.6 | [M–H]− | 169.0140, 125.0244, 435.05561, 643.1137, 583.0917, 523.0714, 481.0608, 381.0453, 331.0661, 231.0296, |
| 79 | 22.27 | C42H33O28 | 6′-O-methyl neochebulagate | 307.098 | 288.320 | 985.1152 | –0.6 | [M–H]− | 300.9983, 783.0664, 633.0719, 463.0505, 351.0345,231.0296, 169.0143, 953.0876 |
| 80 * | 22.41 | C27H23O19 | chebulanin | 230.233 | 229.479 | 651.0855 | 3.2 | [M–H]− | 169.0144, 125.0245, 275.0196, 203.0344, 337.0560, 409.0770 |
| 81 | 22.58 | C42H35O28 | 1′-methyl neochebulinate | – | 292.165 | 987.1302 | –1.3 | [M–H]− | 635.0885, 465.0654, 351.0345, 169.0142 |
| 84 | 22.75 | C41H33O29 | MN-18 | 299.280 | 288.344 | 989.1107 | –0.1 | [M–H]− | 337.02, 249.04, 205.05, 338.023, 293.03, 193.013, 250.043, 163.039, 161.061, 339.025, 275.018, 481.06, 177.055, 133.065, 319.008, 381.045, 437.071, 483.063, 499.071 |
| 85 | 22.93 | C43H37O28 | dimethyl neochebulinate | 290.933 | 298.271 | 1001.1469 | –0.2 | [M–H]− | 635.0885, 465.0654, 365.0502, 169.0140 |
| 88 | 23.34 | C34H31O23 | MN-31 | 256.171 | 258.627 | 807.1259 | 0.4 | [M–H]− | 169.0144, 275.0193, 481.0611, 435.0552, 231.0290, 637.1017, 125.02433 |
| 90 | 23.75 | C43H35O28 | dimethyl neochebulagate or dimethyl 4’-epi-neochebulagate | 290.933 | 293.830/293.873 | 999.1318 | 0.3 | [M–H]− | 300.9985, 275.0190, 205.0505, 829.1083, 633.0756, 527.1011, 463.0513, 365.0506 |
| 91 | 23.92 | C41H33O29 | MN-32 | 297.870 | 286.938 | 989.1112 | 0.4 | [M–H]− | 337.0197, 249.0401, 205.0507, 293.0312 |
| 95 * | 24.97 | C41H29O27 | chebulagic acid | 301.139 | 274.834 | 953.0911 | 1.6 | [M–H]− | 300.9990, 275.0198, 205.0504, 337.0199, 169.0144, 463.0513, 633.0724 |
| 96 | 25.12 | C34H27O23 | MN-33 | 259.196 | 253.905 | 803.0949 | –2.1 | [M–H]− | 169.0143, 275.0201, 481.0621, 633.0727, 293.0283, 337.0189 |
| 98 | 26.21 | C42H35O28 | 6′-methyl neochebulinate | 303.472 | 292.172 | 987.1317 | 0.2 | [M–H]− | 351.0351, 205.0506, 465.0663, 169.0143, 231.0289, 337.0199, 275.0197, 307.0459, 447.0560, 617.0763, 785.0822 |
| 107 * | 28.53 | C41H31O27 | chebulinic acid | 303.706 | 278.017 | 955.1061 | 0.8 | [M–H]− | 275.0199, 205.0506, 337.0200, 319.0094, 169.0143, 465.0668, 293.0402, 617.0781, 249.0402,785.0834, 447.0567 |
| flavonoids | |||||||||
| 87 | 23.29 | C21H19O11 | orientin | – | 200.463 | 447.0926 | –0.2 | [M–H]− | 297.0402, 327.0505, 285.0396, 133.0289, 311.0560, 339.0502, 357.0608 |
| 93 | 24.27 | C21H19O10 | vitexin | – | 198.043 | 431.0991 | 3.0 | [M–H]− | 283.0608, 311.0568, 239.0709, 161.0243, 117.0342 |
| 99 | 26.22 | C27H29O16 | rutin | 235.254 | 230.474 | 609.1460 | 0.7 | [M–H]− | 300.0276, 271.0236, 151.0033 |
| 104 | 27.57 | C27H29O15 | kaempferol-3-O-rutinoside | – | 228.280 | 593.1503 | –0.5 | [M–H]− | 285.0391, 255.0289, 229.0513, 187.0395 |
| triterpenoids | |||||||||
| 102 | 26.92 | C37H59O13 | MN-34 | 271.880 | 259.737 | 711.3989 | 4.6 | [M+HCOOH–H]– | 503.3389, 485.3279, 441.3377, 169.0132 |
| 103 | 27.33 | C37H59O13 | MN-35 | 265.590 | 257.723 | 711.3979 | 3.2 | [M+HCOOH–H]– | 503.3397, 453.2992, 169.0147 |
| 109 | 28.76 | C37H59O13 | MN-36 | 259.720 | 255.801 | 711.3954 | –0.3 | [M+HCOOH–H]– | 503.3372, 169.0143, 113.0226, 59.0133 |
| 113 | 30.32 | C30H47O6 | arjungenin or terminolic acid | 233.955 | 225.260 | 503.3383 | 2.0 | [M–H]− | 409.3106, 453.3002, 421.3100, 379.2992, 300.9970, 485.3248, 503.3364 |
| 114 | 30.49 | C30H47O6 | arjungenin or its isomer | 235.768 | 225.454 | 503.3382 | 1.8 | [M–H]− | 503.3374, 457.3321, 409.3100, 391.2991, 73.0298 |
| 115 | 30.71 | C30H47O6 | arjungenin or its isomer | 232.995 | 224.792 | 503.3383 | 2.0 | [M–H]− | 503.3370, 485.3272,453.3001, 409.3108, 391.3014 |
| 117 | 31.39 | C30H47O6 | arjungenin or its isomer | 238.133 | 227.331 | 503.3388 | 3.0 | [M–H]− | 503.3378, 457.3336, 407.2948, 337.2841 |
| 119 | 32.10 | C30H47O5 | MN-37 | 234.349 | 222.987 | 487.3433 | 2.1 | [M–H]− | 487.3423, 299.9897, 410.3133 |
| 120 | 32.20 | C30H47O5 | MN-38 | 238.780 | 222.913 | 487.3436 | 2.7 | [M–H]− | 487.3417, 393.3158, 423.3254, 300.996, 467.3138, 441.3362 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Xu, J.; Zhang, L.-H.; Yang, W.; Yu, H.; Zhang, M.; Wang, Y.; Wu, H.-H. Global Profiling of the Antioxidant Constituents in Chebulae Fructus Based on an Integrative Strategy of UHPLC/IM-QTOF-MS, MS/MS Molecular Networking, and Spectrum-Effect Correlation. Antioxidants 2023, 12, 2093. https://doi.org/10.3390/antiox12122093
Wang X, Xu J, Zhang L-H, Yang W, Yu H, Zhang M, Wang Y, Wu H-H. Global Profiling of the Antioxidant Constituents in Chebulae Fructus Based on an Integrative Strategy of UHPLC/IM-QTOF-MS, MS/MS Molecular Networking, and Spectrum-Effect Correlation. Antioxidants. 2023; 12(12):2093. https://doi.org/10.3390/antiox12122093
Chicago/Turabian StyleWang, Xiangdong, Jian Xu, Li-Hua Zhang, Wenzhi Yang, Huijuan Yu, Min Zhang, Yuefei Wang, and Hong-Hua Wu. 2023. "Global Profiling of the Antioxidant Constituents in Chebulae Fructus Based on an Integrative Strategy of UHPLC/IM-QTOF-MS, MS/MS Molecular Networking, and Spectrum-Effect Correlation" Antioxidants 12, no. 12: 2093. https://doi.org/10.3390/antiox12122093
APA StyleWang, X., Xu, J., Zhang, L.-H., Yang, W., Yu, H., Zhang, M., Wang, Y., & Wu, H.-H. (2023). Global Profiling of the Antioxidant Constituents in Chebulae Fructus Based on an Integrative Strategy of UHPLC/IM-QTOF-MS, MS/MS Molecular Networking, and Spectrum-Effect Correlation. Antioxidants, 12(12), 2093. https://doi.org/10.3390/antiox12122093






