Silibinin Inhibits Cell Ferroptosis and Ferroptosis-Related Tissue Injuries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Culture
2.3. Chemicals
2.4. Cell Viability Assay
2.5. Lipid Peroxidation Measurement
2.6. Labile Iron Pool Measurement
2.7. Prussian Blue Staining
2.8. 2,2-Diphenyl-1-Picrylhydrazyl (DPPH) Assay
2.9. Kidney IRI
2.10. Hematoxylin–Eosin (H&E) Staining
2.11. Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick End Labeling (TUNEL) Staining
2.12. Q-PCR
2.13. Western Blot
2.14. Malondialdehyde (MDA) Assay
2.15. GSH Assay
2.16. Statistics
3. Results
3.1. Silibinin Inhibits Cell Ferroptosis
3.2. Silibinin Inhibits Ferroptosis through Antioxidant Activity
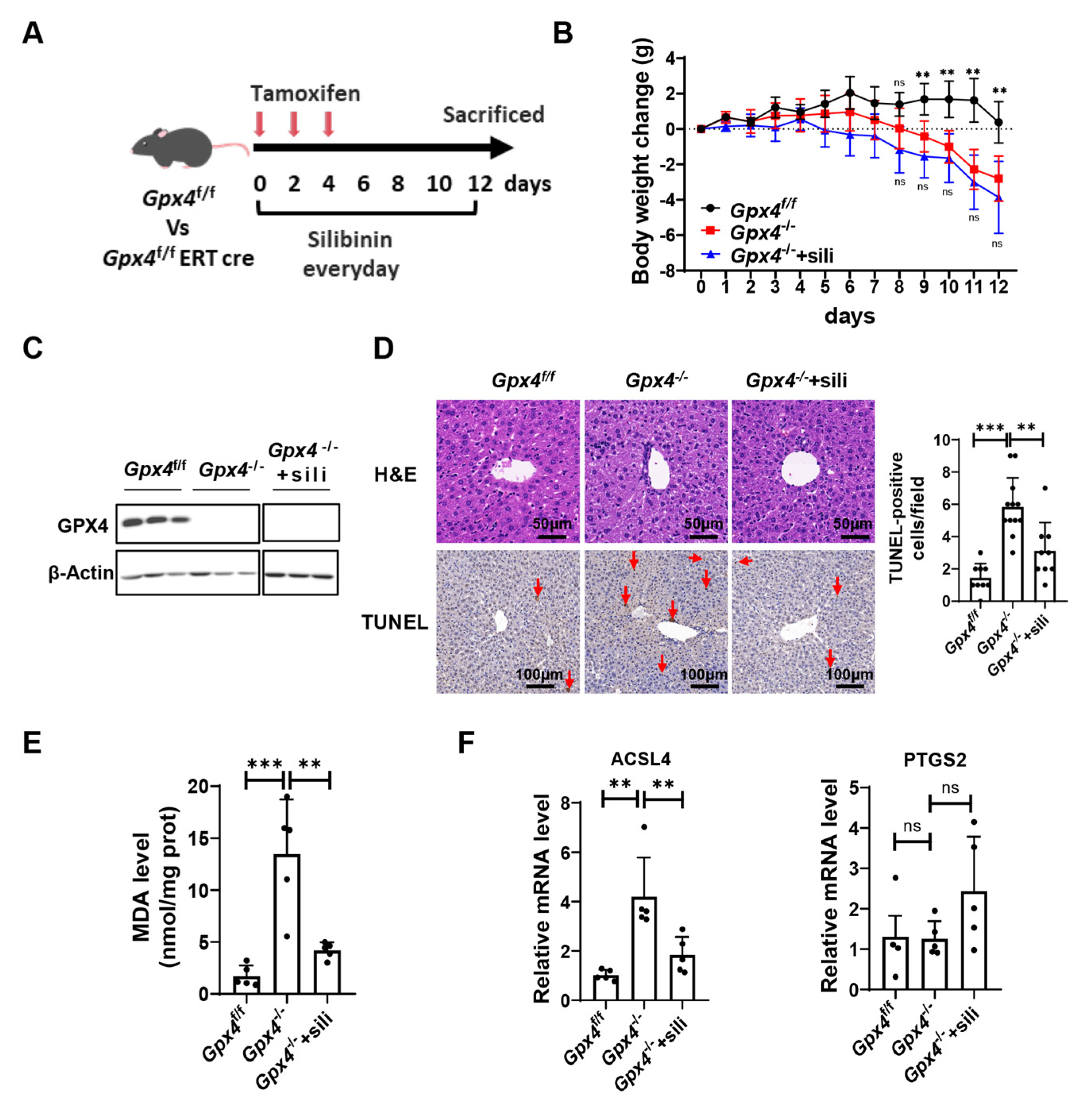
3.3. Silibinin Attenuates Ferroptosis-Induced Liver Injury in GPX4-Deficient Mice
3.4. Silibinin Attenuates Ferroptosis in the Kidney of GPX4-Deficient Mice
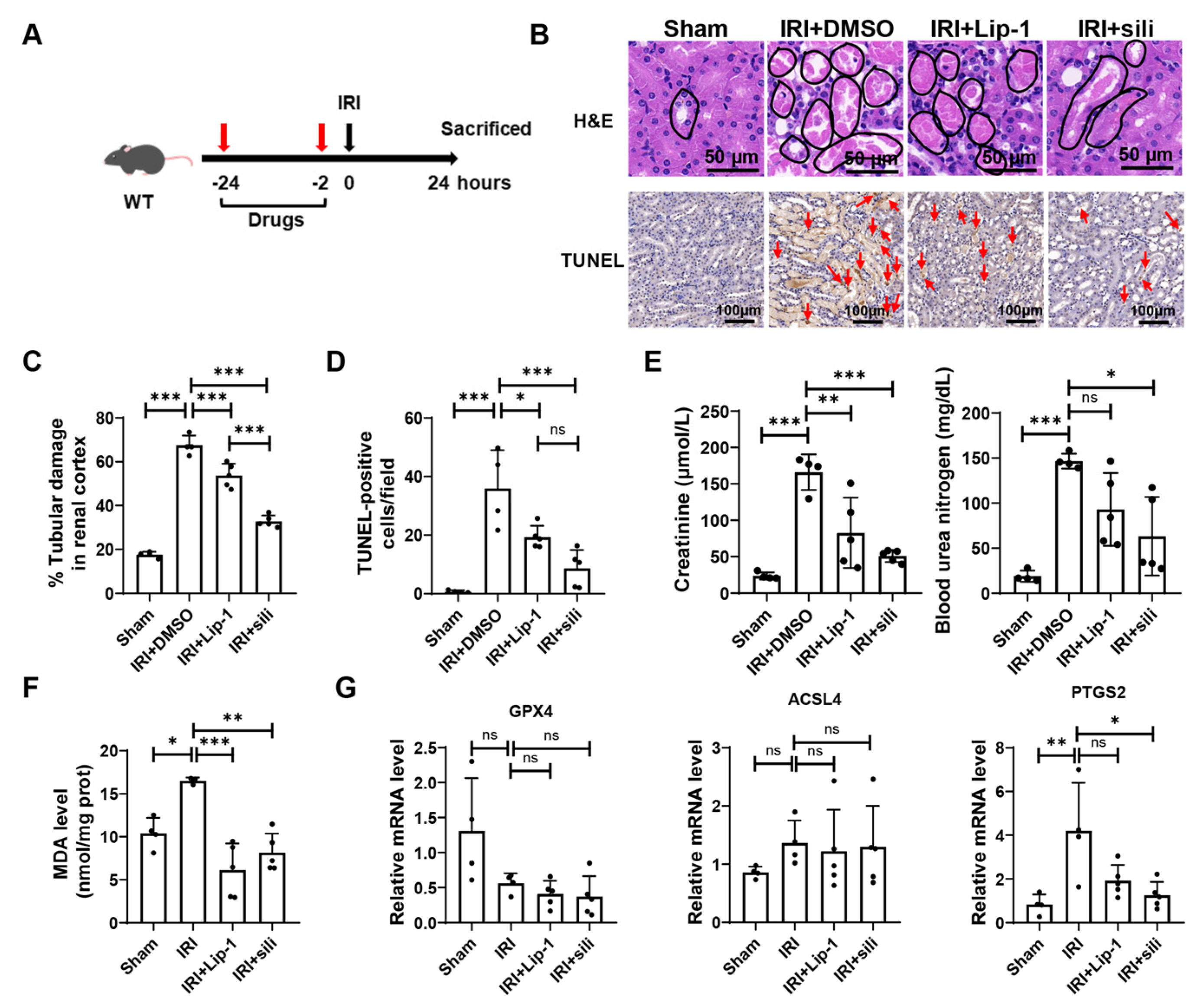
3.5. Silibinin Alleviates Renal Ischemia-Reperfusion Injury
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Jiang, X. The Chemistry and Biology of Ferroptosis. Cell Chem. Biol. 2020, 27, 365–375. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular Mechanisms and Health Implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Friedmann Angeli, J.P.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the Ferroptosis Regulator Gpx4 Triggers Acute Renal Failure in Mice. Nat. Cell Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Yant, L.J.; Ran, Q.; Rao, L.; Van Remmen, H.; Shibatani, T.; Belter, J.G.; Motta, L.; Richardson, A.; Prolla, T.A. The Selenoprotein GPX4 Is Essential for Mouse Development and Protects from Radiation and Oxidative Damage Insults. Free Radic. Biol. Med. 2003, 34, 496–502. [Google Scholar] [CrossRef]
- Carlson, B.A.; Tobe, R.; Yefremova, E.; Tsuji, P.A.; Hoffmann, V.J.; Schweizer, U.; Gladyshev, V.N.; Hatfield, D.L.; Conrad, M. Glutathione Peroxidase 4 and Vitamin E Cooperatively Prevent Hepatocellular Degeneration. Redox Biol. 2016, 9, 22–31. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, Y.; Lou, H.; Ou, Z.; Liu, J.; Duan, W.; Wang, H.; Ge, Y.; Min, J.; Wang, F.; et al. GPX4 and Vitamin E Cooperatively Protect Hematopoietic Stem and Progenitor Cells from Lipid Peroxidation and Ferroptosis. Cell Death Dis. 2021, 12, 706. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Jiang, X.; Gu, W. Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cell Biol. 2020, 30, 478–490. [Google Scholar] [CrossRef]
- Yan, H.; Zou, T.; Tuo, Q.; Xu, S.; Li, H.; Belaidi, A.A.; Lei, P. Ferroptosis: Mechanisms and Links with Diseases. Signal Transduct. Target. Ther. 2021, 6, 49. [Google Scholar] [CrossRef]
- Angeli, J.P.F.; Shah, R.; Pratt, D.A.; Conrad, M. Ferroptosis Inhibition: Mechanisms and Opportunities. Trends Pharmacol. Sci. 2017, 38, 489–498. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, Biology and Role in Disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Soleimani, V.; Delghandi, P.S.; Moallem, S.A.; Karimi, G. Safety and Toxicity of Silymarin, the Major Constituent of Milk Thistle Extract: An Updated Review. Phytother. Res. 2019, 33, 1627–1638. [Google Scholar] [CrossRef]
- Bijak, M. Silybin, a Major Bioactive Component of Milk Thistle (Silybum marianum L. Gaernt.)—Chemistry, Bioavailability, and Metabolism. Molecules 2017, 22, 1942. [Google Scholar] [CrossRef]
- Trappoliere, M.; Caligiuri, A.; Schmid, M.; Bertolani, C.; Failli, P.; Vizzutti, F.; Novo, E.; di Manzano, C.; Marra, F.; Loguercio, C.; et al. Silybin, a Component of Sylimarin, Exerts Anti-Inflammatory and Anti-Fibrogenic Effects on Human Hepatic Stellate Cells. J. Hepatol. 2009, 50, 1102–1111. [Google Scholar] [CrossRef]
- Gillessen, A.; Schmidt, H.H.J. Silymarin as Supportive Treatment in Liver Diseases: A Narrative Review. Adv. Ther. 2020, 37, 1279–1301. [Google Scholar] [CrossRef]
- Polachi, N.; Bai, G.; Li, T.; Chu, Y.; Wang, X.; Li, S.; Gu, N.; Wu, J.; Li, W.; Zhang, Y.; et al. Modulatory Effects of Silibinin in Various Cell Signaling Pathways against Liver Disorders and Cancer—A Comprehensive Review. Eur. J. Med. Chem. 2016, 123, 577–595. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Ge, C.; Min, J.; Wang, F. The Multifaceted Role of Ferroptosis in Liver Disease. Cell Death Differ. 2022, 29, 467–480. [Google Scholar] [CrossRef]
- Abenavoli, L.; Capasso, R.; Milic, N.; Capasso, F. Milk Thistle in Liver Diseases: Past, Present, Future. Phytother. Res. 2010, 24, 1423–1432. [Google Scholar] [CrossRef]
- Gharagozloo, M.; Khoshdel, Z.; Amirghofran, Z. The Effect of an Iron (III) Chelator, Silybin, on the Proliferation and Cell Cycle of Jurkat Cells: A Comparison with Desferrioxamine. Eur. J. Pharmacol. 2008, 589, 1–7. [Google Scholar] [CrossRef]
- Tao, Z.-S.; Li, T.-L.; Wei, S. Silymarin Prevents Iron Overload Induced Bone Loss by Inhibiting Oxidative Stress in an Ovariectomized Animal Model. Chem. Biol. Interact. 2022, 366, 110168. [Google Scholar] [CrossRef]
- Hutchinson, C.; Bomford, A.; Geissler, C.A. The Iron-Chelating Potential of Silybin in Patients with Hereditary Haemochromatosis. Eur. J. Clin. Nutr. 2010, 64, 1239–1241. [Google Scholar] [CrossRef]
- Saxena, N.; Dhaked, R.K.; Nagar, D. Silibinin Ameliorates Abrin Induced Hepatotoxicity by Attenuating Oxidative Stress, Inflammation and Inhibiting Fas Pathway. Environ. Toxicol. Pharmacol. 2022, 93, 103868. [Google Scholar] [CrossRef]
- Ajoolabady, A.; Aslkhodapasandhokmabad, H.; Libby, P.; Tuomilehto, J.; Lip, G.Y.H.; Penninger, J.M.; Richardson, D.R.; Tang, D.; Zhou, H.; Wang, S.; et al. Ferritinophagy and Ferroptosis in the Management of Metabolic Diseases. Trends Endocrinol. Metab. 2021, 32, 444–462. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Abenavoli, L.; Izzo, A.A.; Milić, N.; Cicala, C.; Santini, A.; Capasso, R. Milk Thistle (Silybum marianum): A Concise Overview on Its Chemistry, Pharmacological, and Nutraceutical Uses in Liver Diseases. Phytother. Res. 2018, 32, 2202–2213. [Google Scholar] [CrossRef]
- Lee, H.; Zandkarimi, F.; Zhang, Y.; Meena, J.K.; Kim, J.; Zhuang, L.; Tyagi, S.; Ma, L.; Westbrook, T.F.; Steinberg, G.R.; et al. Energy-Stress-Mediated AMPK Activation Inhibits Ferroptosis. Nat. Cell Biol. 2020, 22, 225–234. [Google Scholar] [CrossRef]
- Qiu, Y.; Cao, Y.; Cao, W.; Jia, Y.; Lu, N. The Application of Ferroptosis in Diseases. Pharmacol. Res. 2020, 159, 104919. [Google Scholar] [CrossRef]
- Song, X.-Y.; Liu, P.-C.; Liu, W.-W.; Zhou, J.; Hayashi, T.; Mizuno, K.; Hattori, S.; Fujisaki, H.; Ikejima, T. Silibinin Inhibits Ethanol- or Acetaldehyde-Induced Ferroptosis in Liver Cell Lines. Toxicol. In Vitro 2022, 82, 105388. [Google Scholar] [CrossRef]
- Du, Q.; Wu, X.; Ma, K.; Liu, W.; Liu, P.; Hayashi, T.; Mizuno, K.; Hattori, S.; Fujisaki, H.; Ikejima, T. Silibinin Alleviates Ferroptosis of Rat Islet β Cell INS-1 Induced by the Treatment with Palmitic Acid and High Glucose through Enhancing PINK1/Parkin-Mediated Mitophagy. Arch. Biochem. Biophys. 2023, 743, 109644. [Google Scholar] [CrossRef]
- Takke, A.; Shende, P. Nanotherapeutic Silibinin: An Insight of Phytomedicine in Healthcare Reformation. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102057. [Google Scholar] [CrossRef]
- Ripoli, M.; Angelico, R.; Sacco, P.; Ceglie, A.; Mangia, A. Phytoliposome-Based Silibinin Delivery System as a Promising Strategy to Prevent Hepatitis C Virus Infection. J. Biomed. Nanotechnol. 2016, 12, 770–780. [Google Scholar] [CrossRef]



| The PCR Primers | Sequence | |
|---|---|---|
| Mouse-GPX4 | Forward | CCTCTGCTGCAAGAGCCTCCC |
| Mouse-GPX4 | Reverse | CTTATCCAGGCAGACCATGTGC |
| Mouse-ACSL4 | Forward | CCTTTGGCTCATGTGCTGGAAC |
| Mouse-ACSL4 | Reverse | GCCATAAGTGTGGGTTTCAGTAC |
| Mouse-PTGS2 | Forward | GCGACATACTCAAGCAGGAGCA |
| Mouse-PTGS2 | Reverse | AGTGGTAACCGCTCAGGTGTTG |
| Mouse-β-Actin | Forward | GGCTGTATTCCCCTCCATCG |
| Mouse-β-Actin | Reverse | CCAGTTGGTAACAATGCCATGT |
| Human-GPX4 | Forward | GAGGCAAGACCGAAGTAAACTAC |
| Human-GPX4 | Reverse | CCGAACTGGTTACACGGGAA |
| Human-CAT | Forward | TGGAGCTGGTAACCCAGTAGG |
| Human-CAT | Reverse | CCTTTGCCTTGGAGTATTTGGTA |
| Human-FSP1 | Forward | GATGAGCAACTTGGACAGCAA |
| Human-FSP1 | Reverse | CTGGGCTGCTTATCTGGGAAG |
| Human-GCH1 | Forward | GCCATGCAGTTCTTCACCAAGG |
| Human-GCH1 | Reverse | ATGGAACCAAGTGATGCTCACAC |
| Human-SLC7A11 | Forward | TCCTGCTTTGGCTCCATGAACG |
| Human-SLC7A11 | Reverse | AGAGGAGTGTGCTTGCGGACAT |
| Human-DHODH | Forward | GAGGACATTGCCAGTGTGGTCA |
| Human-DHODH | Reverse | TTCCCACTCAGCCCTCCTGTTT |
| Human-SOD1 | Forward | GGTGGGCCAAAGGATGAAGAG |
| Human-SOD1 | Reverse | CCACAAGCCAAACGACTTCC |
| Human-β-Actin | Forward | CATGTACGTTGCTATCCAGGC |
| Human-β-Actin | Reverse | CTCCTTAATGTCACGCACGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, W.; Ou, Z.; Huang, Y.; Zhang, Y.; Zhang, L.; Zhao, Y.; He, R.; Zhang, Y.; Ge, Y.; Lou, H.; et al. Silibinin Inhibits Cell Ferroptosis and Ferroptosis-Related Tissue Injuries. Antioxidants 2023, 12, 2119. https://doi.org/10.3390/antiox12122119
Duan W, Ou Z, Huang Y, Zhang Y, Zhang L, Zhao Y, He R, Zhang Y, Ge Y, Lou H, et al. Silibinin Inhibits Cell Ferroptosis and Ferroptosis-Related Tissue Injuries. Antioxidants. 2023; 12(12):2119. https://doi.org/10.3390/antiox12122119
Chicago/Turabian StyleDuan, Wentao, Zexian Ou, Yuxing Huang, Yifan Zhang, Lan Zhang, Yanan Zhao, Ruikun He, Yihan Zhang, Yuanlong Ge, Huiling Lou, and et al. 2023. "Silibinin Inhibits Cell Ferroptosis and Ferroptosis-Related Tissue Injuries" Antioxidants 12, no. 12: 2119. https://doi.org/10.3390/antiox12122119






