Characterization of the Metabolic Profile of Olive Tissues (Roots, Stems and Leaves): Relationship with Cultivars’ Resistance/Susceptibility to the Soil Fungus Verticillium dahliae
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Standards
2.2. Plant Material and Samples Pretreatment
2.3. Secondary Metabolites Extraction and LC-MS Analysis
2.4. Data Treatment
3. Results and Discussion
3.1. Qualitative Characterization of Plant Tissue Metabolic Profiles
| Rt/min | Experimental m/z * | Error/mDa | iFIT | Molecular Formula | Name of the Compound | Chemical Family | References | Quantified in: | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | Stem | Root | ||||||||
| 0.8 | 191.0557 | 0.1 | 418.2 | C7H12O6 | quinic acid | organic acids | standard | x | x | x |
| 1.0 | 389.1083 | −0.1 | 266.9 | C16H22O11 | oleoside | secoiridoids and derivatives | [33,34] | x | x | x |
| 1.3 | 315.1078 | −0.2 | 373.2 | C14H20O8 | hydroxytyrosol glucoside | simple phenols and derivatives | [28,30,38,39] | x | x | |
| 1.4 | 153.0551 | −0.1 | 260.2 | C8H10O4 | hydroxytyrosol | simple phenols and derivatives | standard | x | ||
| 1.8 | 465.1035 | 0.2 | 277.6 | C21H22O12 | dihydroquercetin 3-O-glucoside | flavonoids | [28] | x | ||
| 2.3 | 389.1082 | −0.2 | 527.9 | C16H22O11 | secologanoside | secoiridoids and derivatives | [28,33,34] | x | x | x |
| 2.4 | 625.1977 | −0.3 | 376.0 | C25H38O18 | unknown 1 | unknown | - | x | x | x |
| 3.0 | 305.0670 | −0.28 | 313.2 | C15H14O7 | gallocatechin | flavonoids | [30,40,41] | x | ||
| 3.0 | 449.1086 | 0.2 | 122.6 | C21H22O11 | cyanidin O-glucoside | flavonoids | [30,42] | x | ||
| 3.5 | 403.1236 | −0.4 | 479.2 | C17H24O11 | elenolic acid glucoside (isomer 1) | secoiridoids and derivatives | [28,33,39,43] | x | x | x |
| 3.9 | 377.1447 | −0.1 | 337.0 | C16H26O10 | aldehydic form of decarboxymethyl elenolic acid glucoside | secoiridoids and derivatives | [44] | x | x | |
| 3.9 | 537.1974 | 0.2 | 275.9 | C26H34O12 | cycloolivil glucoside (isomer 1) | lignans | [41,45,46] | x | ||
| 4.7 | 403.1239 | −0.1 | 458.3 | C17H24O11 | elenolic acid glucoside (isomer 2) | secoiridoids and derivatives | [28,33,39,43] | x | x | x |
| 4.8 | 537.1976 | 0.4 | 125.2 | C26H34O12 | cycloolivil glucoside (isomer 2) | lignans | [46] | x | ||
| 5.1 | 415.1607 | 0.3 | 529.8 | C19H28O10 | phenylethyl primeveroside | simple phenols and derivatives | [30] | x | ||
| 5.6 | 525.1604 | −0.4 | 639.1 | C24H30O13 | demethyl oleuropein | secoiridoids and derivatives | [33,41,44] | x | x | |
| 5.7 | 609.1453 | −0.3 | 427.3 | C27H30O16 | rutin | flavonoids | standard | x | x | |
| 5.8 | 359.1341 | −0.1 | 513.8 | C16H24O9 | 7-deoxyloganic acid | iridoid | [40,41,46] | x | ||
| 6.1 | 555.1711 | −0.3 | 302.3 | C25H32O14 | hydroxy oleuropein | secoiridoids and derivatives | [28,30,39,41] | x | x | |
| 6.2 | 303.0506 | 0.1 | 166.6 | C15H12O7 | taxifolin | flavonoids | [30,38,47,48] | x | ||
| 6.2 | 463.0874 | −0.3 | 393.1 | C21H20O12 | quercetin O-glucoside (isomer 1) | flavonoids | [30,41,48] | x | ||
| 6.3 | 375.1444 | 0.0 | 113.0 | C20H24O7 | cycloolivil | lignans | [42,45,46] | x | ||
| 6.4 | 701.2291 | −0.2 | 570.0 | C31H42O18 | neonuzhenide/oleuropein glucoside (isomer 1) | secoiridoids and derivatives | [28,41] | x | x | x |
| 6.4 | 447.0923 | −0.4 | 308.2 | C21H20O11 | luteolin 7-O-glucoside (isomer 1) | flavonoids | standard | x | x | |
| 6.7 | 511.3484 | 0.2 | 161.5 | C25H52O10 | unknown 2 | unknown | - | x | x | x |
| 6.8 | 623.1977 | 0.1 | 450.0 | C29H36O15 | verbascoside | simple phenols and derivatives | standard | x | x | x |
| 7.4 | 577.1561 | 0.4 | 403.9 | C27H30O14 | apigenin O-rutinoside | flavonoids | [41,43] | x | ||
| 7.5 | 623.1975 | −0.1 | 323.9 | C29H36O15 | isoverbascoside | simple phenols and derivatives | [33,34,41] | x | ||
| 7.8 | 447.0925 | −0.2 | 407.7 | C21H20O11 | luteolin O-glucoside (isomer 2) | flavonoids | [39,41] | x | x | |
| 7.9 | 535.1810 | −0.6 | 257.2 | C26H32O12 | hydroxypinoresinol glucoside | lignans | [28,41,45,46] | x | x | |
| 7.9 | 701.2290 | −0.3 | 275.2 | C31H42O18 | neonuzhenide/oleuropein glucoside (isomer 2) | secoiridoids and derivatives | [28,39,41] | x | x | |
| 8.0 | 463.0882 | 0.5 | 380.5 | C21H20O12 | quercetin O-glucoside (isomer 2) | flavonoids | [30,34] | x | ||
| 8.0 | 431.0976 | −0.2 | 395.2 | C21H20O10 | apigenin 7-O-glucoside | flavonoids | standard | x | ||
| 8.1 | 565.1923 | 0.2 | 283.0 | C27H34O13 | methoxypinoresinol glucoside | lignans | [45] | x | x | |
| 8.2 | 607.1666 | 0.3 | 187.2 | C28H32O15 | diosmin | flavonoids | [40,41] | x | ||
| 8.3 | 287.0551 | −0.5 | 351.0 | C15H12O6 | dihydrokaempferol | flavonoids | [30] | x | ||
| 8.3 | 701.2288 | −0.5 | 333.7 | C31H42O18 | neonuzhenide/oleuropein glucoside (isomer 3) | secoiridoids and derivatives | [28] | x | ||
| 8.6 | 461.1080 | −0.4 | 50.3 | C22H22O11 | chrysoeriol O-glucoside | flavonoids | [30,40,43] | x | ||
| 8.7 | 577.1921 | 0.0 | 165.0 | C28H34O13 | acetoxypinoresinol glucoside | lignans | [41,45] | x | x | |
| 8.8 | 447.0924 | −0.3 | 329.7 | C21H20O11 | luteolin O-glucoside (isomer 3) | flavonoids | [28,30,39] | x | x | |
| 9.0 | 463.0881 | 0.4 | 412.2 | C21H20O12 | quercetin O-glucoside (isomer 3) | flavonoids | [30,34] | x | ||
| 9.2 | 491.1769 | 0.4 | 370.4 | C21H32O13 | unknown 3 | unknown | - | x | ||
| 9.5 | 701.2296 | −0.3 | 43.1 | C31H42O18 | neonuzhenide/oleuropein glucoside (isomer 4) | secoiridoids and derivatives | [28] | x | x | x |
| 9.8 | 539.1762 | −0.2 | 646.3 | C25H32O13 | oleuropein | secoiridoids and derivatives | standard | x | x | x |
| 10.3 | 555.2076 | −0.2 | 352.7 | C26H36O13 | 11-hydroxyiridodial glucoside pentaacetate | iridoid | [49] | x | x | |
| 10.8 | 539.1764 | −0.1 | 513.6 | C25H32O13 | oleuroside | secoiridoids and derivatives | [33,41] | x | x | |
| 11.5 | 583.2023 | −0.4 | 211.3 | C27H36O14 | lucidumoside C | secoiridoids and derivatives | [28,30,46] | x | x | x |
| 11.8 | 523.1817 | 0.1 | 60.1 | C25H32O12 | ligstroside | secoiridoids and derivatives | [28,30,41] | x | x | x |
| 11.5 | 651.2283 | −0.6 | 374.7 | C31H40O15 | unknown 4 | unknown | - | x | ||
| 12.7 | 415.1392 | −0.1 | 448.8 | C22H24O8 | acetoxipinoresinol | lignans | [33] | x | x | |
| 12.7 | 377.1235 | −0.1 | 350.2 | C19H22O8 | oleuropein aglycone (isomer 1) | secoiridoids and derivatives | [30,41] | x | x | |
| 13.3 | 377.1239 | 0.3 | 54.8 | C19H22O8 | oleuropein aglycone (isomer 2) | secoiridoids and derivatives | [30,41] | x | x | |
| 15.7 | 471.3467 | −0.7 | 607.4 | C30H48O4 | maslinic acid | pentacyclic triterpenes | standard | x | x | x |
| 16.4 | 617.3840 | −0.2 | 101.8 | C39H54O6 | unknown 5 | unknown | - | x | x | |
| 17.5 | 455.3527 | 0.2 | 241.1 | C30H48O3 | betulinic acid | pentacyclic triterpenes | standard | x | x | x |
| 17.8 | 455.3526 | 0.1 | 282.5 | C30H48O3 | oleanolic acid | pentacyclic triterpenes | standard | x | x | x |
3.2. Quantitative Analysis of the Targeted Metabolites
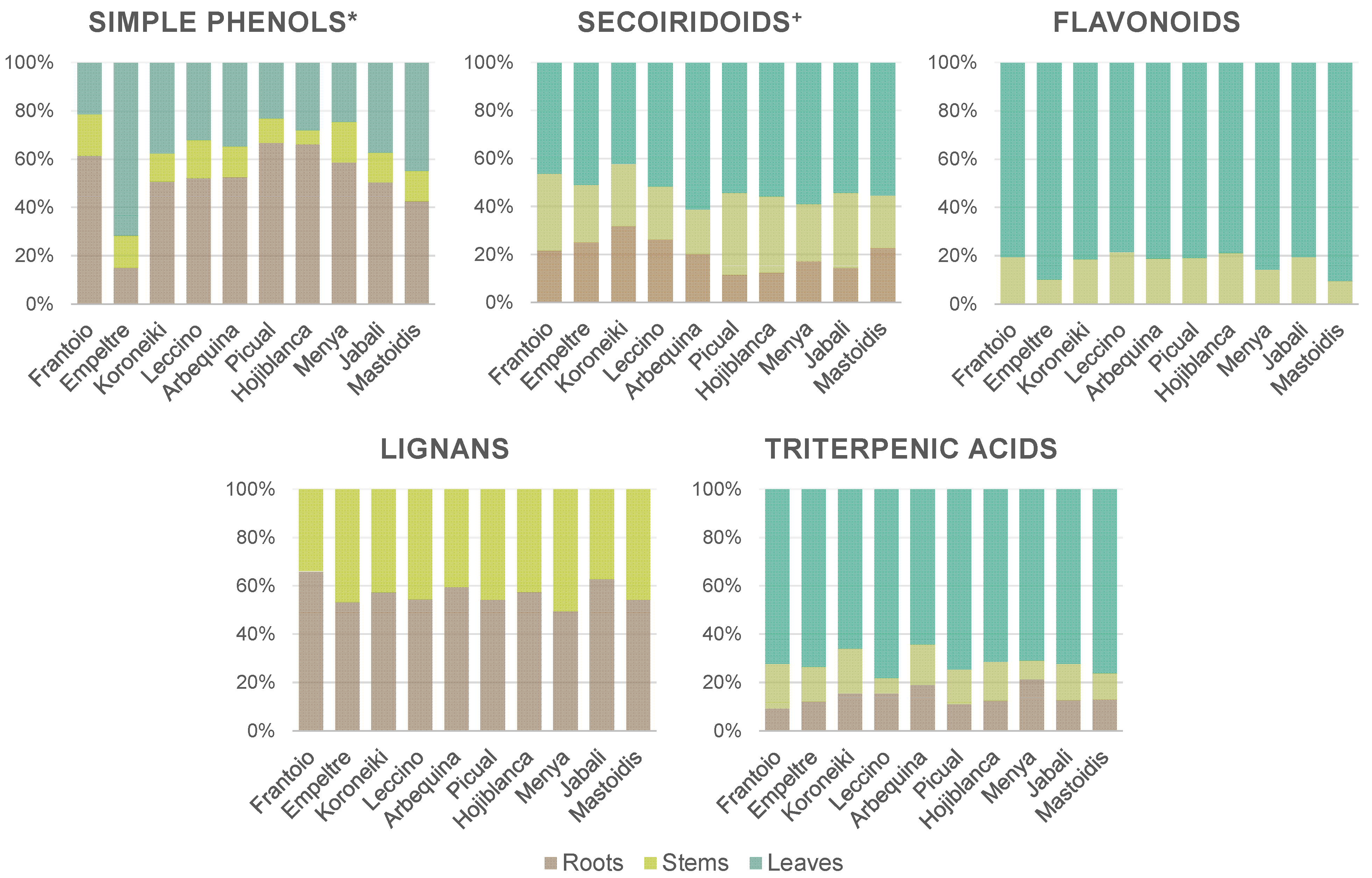
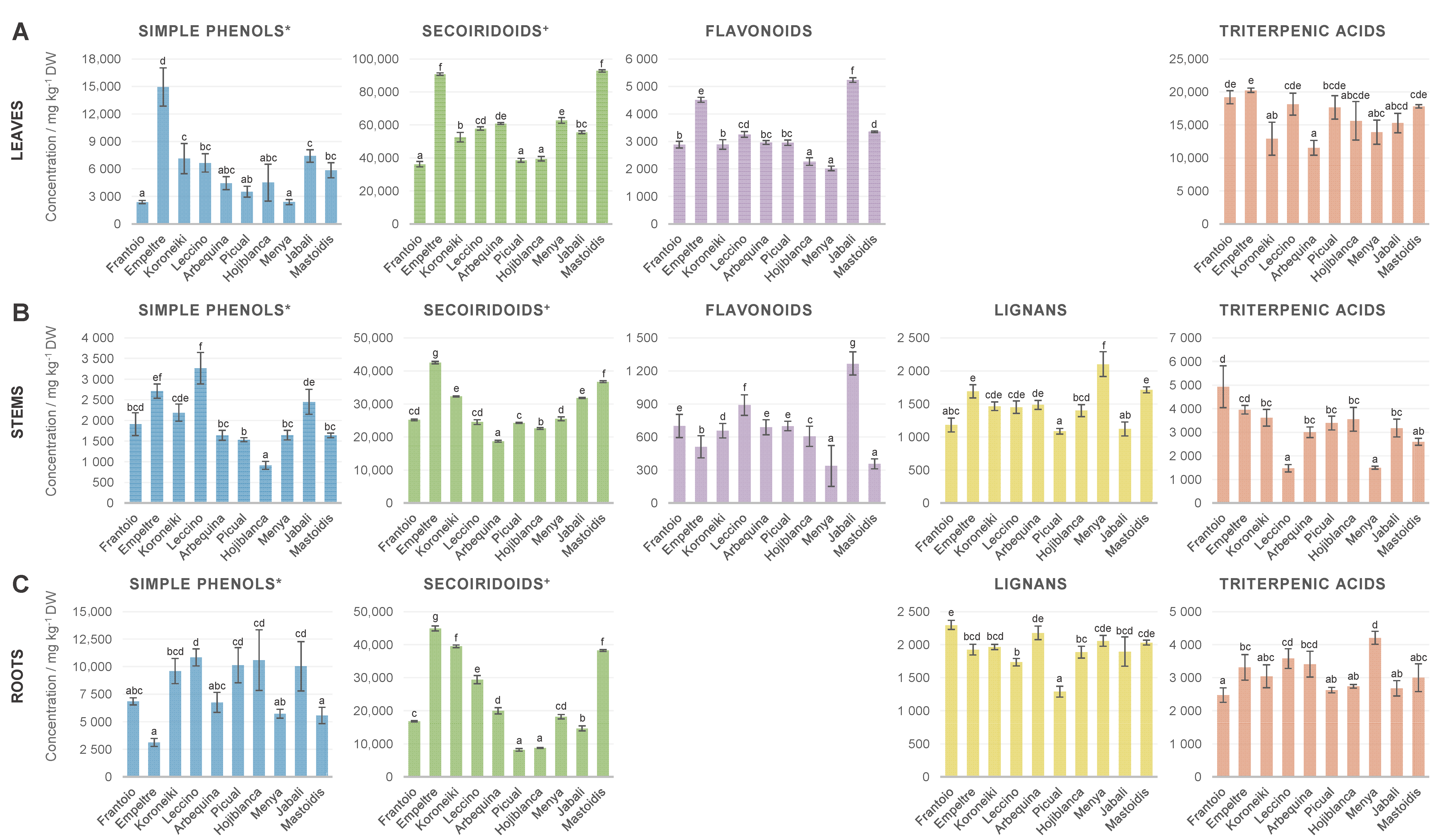
3.2.1. Metabolites Distribution throughout Plant Tissues
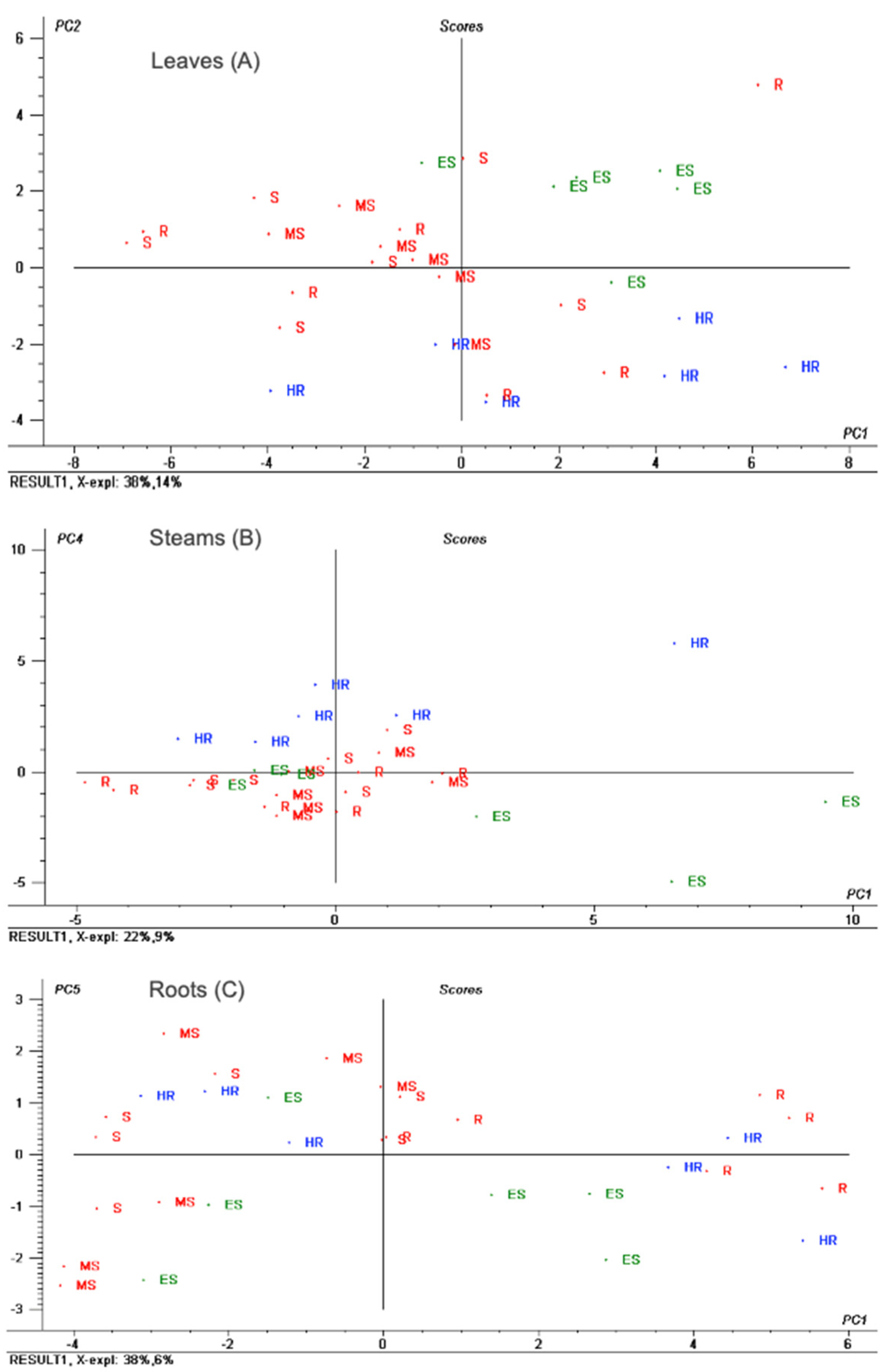
3.2.2. Assessment of Differences in the Metabolic Profiles of the 10 Cultivars under Study
3.3. Relationship between Cultivars Metabolic Profiles and Resistance/Susceptibility to the Soil Fungus Verticillium dahliae
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Martínez-González, M.A.; Salas-Salvadó, J.; Estruch, R.; Corella, D.; Fitó, M.; Ros, E. Benefits of the Mediterranean Diet: Insights From the PREDIMED Study. Prog. Cardiovasc. Dis. 2015, 58, 50–60. [Google Scholar] [CrossRef] [PubMed]
- International Olive Council World Olive Oil Consumption Statistics. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2022/12/IOC-Olive-Oil-Dashboard-2.html#consumption (accessed on 10 September 2023).
- European Commision Market Situation for Olive Oil and Table Olives. Available online: https://agriculture.ec.europa.eu/data-and-analysis/markets/price-data/price-monitoring-sector/olive-oil_en#marketsituation (accessed on 10 September 2023).
- Keykhasaber, M.; Thomma, B.P.H.J.; Hiemstra, J.A. Verticillium Wilt Caused by Verticillium Dahliae in Woody Plants with Emphasis on Olive and Shade Trees. Eur. J. Plant Pathol. 2018, 150, 21–37. [Google Scholar] [CrossRef]
- Montes-Osuna, N.; Mercado-Blanco, J. Verticillium Wilt of Olive and Its Control: What Did We Learn during the Last Decade? Plants 2020, 9, 735. [Google Scholar] [CrossRef] [PubMed]
- López-Escudero, F.J.; Mercado-Blanco, J. Verticillium Wilt of Olive: A Case Study to Implement an Integrated Strategy to Control a Soil-Borne Pathogen. Plant Soil 2011, 344, 1–50. [Google Scholar] [CrossRef]
- Mulero-Aparicio, A.; Varo, A.; Agustí-Brisach, C.; López-Escudero, F.J.; Trapero, A. Biological Control of Verticillium Wilt of Olive in the Field. Crop. Prot. 2020, 128, 104993. [Google Scholar] [CrossRef]
- Santos-Rufo, A.; Rodríguez-Jurado, D. Unravelling the Relationships among Verticillium Wilt, Irrigation, and Susceptible and Tolerant Olive Cultivars. Plant Pathol. 2021, 70, 2046–2061. [Google Scholar] [CrossRef]
- Belaj, A.; Ninot, A.; Gómez-Gálvez, F.J.; El Riachy, M.; Gurbuz-Veral, M.; Torres, M.; Lazaj, A.; Klepo, T.; Paz, S.; Ugarte, J.; et al. Utility of EST-SNP Markers for Improving Management and Use of Olive Genetic Resources: A Case Study at the Worldwide Olive Germplasm Bank of Córdoba. Plants 2022, 11, 921. [Google Scholar] [CrossRef]
- Serrano, A.; Rodríguez-Jurado, D.; Ramírez-Tejero, J.A.; Luque, F.; López-Escudero, F.J.; Belaj, A.; Román, B.; León, L. Response to Verticillium Dahliae Infection in a Genetically Diverse Set of Olive Cultivars. Sci. Hortic. 2023, 316, 112008. [Google Scholar] [CrossRef]
- Díaz-Rueda, P.; Aguado, A.; Romero-Cuadrado, L.; Capote, N.; Colmenero-Flores, J.M. Wild Olive Genotypes as a Valuable Source of Resistance to Defoliating Verticillium Dahliae. Front. Plant Sci. 2021, 12, 662060. [Google Scholar] [CrossRef]
- Ostos, E.; Garcia-Lopez, M.T.; Porras, R.; Lopez-Escudero, F.J.; Trapero-Casas, A.; Michailides, T.J.; Moral, J. Effect of Cultivar Resistance and Soil Management on Spatial–Temporal Development of Verticillium Wilt of Olive: A Long-Term Study. Front. Plant Sci. 2020, 11, 584496. [Google Scholar] [CrossRef]
- Arias-Calderón, R.; Rodríguez-Jurado, D.; León, L.; Bejarano-Alcázar, J.; De la Rosa, R.; Belaj, A. Pre-Breeding for Resistance to Verticillium Wilt in Olive: Fishing in the Wild Relative Gene Pool. Crop. Prot. 2015, 75, 25–33. [Google Scholar] [CrossRef]
- Ramírez-Tejero, J.A.; Jiménez-Ruiz, J.; Serrano, A.; Belaj, A.; León, L.; de la Rosa, R.; Mercado-Blanco, J.; Luque, F. Verticillium Wilt Resistant and Susceptible Olive Cultivars Express a Very Different Basal Set of Genes in Roots. BMC Genom. 2021, 22, 229. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary Metabolites in Fungus-Plant Interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Gdoura, R.; Triki, M.A. Differential Biochemical and Physiological Responses of Two Olive Cultivars Differing by Their Susceptibility to the Hemibiotrophic Pathogen Verticillium Dahliae. Physiol. Mol. Plant Pathol. 2017, 97, 30–39. [Google Scholar] [CrossRef]
- Cardoni, M.; Mercado-blanco, J.; Villar, R. Functional Traits of Olive Varieties and Their Relationship with the Tolerance Level towards Verticillium Wilt. Plants 2021, 10, 1079. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Pérez, M.D.L.O.; Jiménez-Ruiz, J.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Barroso, J.B.; Luque, F.; Mercado-Blanco, J. Tolerance of Olive (Olea europaea) Cv Frantoio to Verticillium Dahliae Relies on Both Basal and Pathogen-Induced Differential Transcriptomic Responses. New Phytol. 2018, 217, 671–686. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How Do Plants Defend Themselves against Pathogens-Biochemical Mechanisms and Genetic Interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of Secondary Metabolites in Plant Defense against Pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Trabelsi, R.; Sellami, H.; Gharbi, Y.; Cheffi, M.; Chaari, A.; Baucher, M.; El Jaziri, M.; Triki, M.A.; Gdoura, R. Response of Olive Tree (Olea europaea L.Cv. Chemlali) to Infection with Soilborne Fungi. J. Plant Dis. Prot. 2017, 124, 153–162. [Google Scholar] [CrossRef]
- Báidez, A.G.; Gómez, P.; Del Río, J.A.; Ortuño, A. Dysfunctionality of the Xylem in Olea europaea L. Plants Associated with the Infection Process by Verticillium Dahliae Kleb. Role of Phenolic Compounds in Plant Defense Mechanism. J. Agric. Food Chem. 2007, 55, 3373–3377. [Google Scholar] [CrossRef]
- Drais, M.I.; Pannucci, E.; Caracciolo, R.; Bernini, R.; Romani, A.; Santi, L.; Varvaro, L. Antifungal Activity of Hydroxytyrosol Enriched Extracts from Olive Mill Waste against Verticillium Dahliae, the Cause of Verticillium Wilt of Olive. Phytopathol. Mediterr. 2021, 60, 139–147. [Google Scholar] [CrossRef]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Hibar, K.; Gdoura, R.; Triki, M.A. Lignification, Phenols Accumulation, Induction of PR Proteins and Antioxidant-Related Enzymes Are Key Factors in the Resistance of Olea europaea to Verticillium Wilt of Olive. Acta Physiol. Plant 2017, 39, 1–15. [Google Scholar] [CrossRef]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Cheffi, M.; Gdoura, R.; Triki, M.A. Differential Fungal Colonization and Physiological Defense Responses of New Olive Cultivars Infected by the Necrotrophic Fungus Verticillium Dahliae. Acta Physiol. Plant 2016, 38, 1–12. [Google Scholar] [CrossRef]
- Markakis, E.A.; Tjamos, S.E.; Antoniou, P.P.; Roussos, P.A.; Paplomatas, E.J.; Tjamos, E.C. Phenolic Responses of Resistant and Susceptible Olive Cultivars Induced by Defoliating and Nondefoliating Verticillium Dahliae Pathotypes. Plant Dis. 2010, 94, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Cardoni, M.; Olmo-García, L.; Serrano-García, I.; Carrasco-Pancorbo, A.; Mercado-Blanco, J. The Roots of Olive Cultivars Differing in Tolerance to Verticillium Dahliae Show Quantitative Differences in Phenolic and Triterpenic Profiles. J. Plant Interact. 2023, 18, 1–14. [Google Scholar] [CrossRef]
- Vergine, M.; Pavan, S.; Negro, C.; Nicolì, F.; Greco, D.; Sabella, E.; Aprile, A.; Ricciardi, L.; De Bellis, L.; Luvisi, A. Phenolic Characterization of Olive Genotypes Potentially Resistant to Xylella. J. Plant Interact. 2022, 17, 462–474. [Google Scholar] [CrossRef]
- Luvisi, A.; Aprile, A.; Sabella, E.; Vergine, M.; Nicolì, F.; Nutricati, E.; Miceli, A.; Negro, C.; De Bellis, L. Xylella Fastidiosa Subsp. Pauca (CoDiRO Strain) Infection in Four Olive (Olea europaea L.) Cultivars: Profile of Phenolic Compounds in Leaves and Progression of Leaf Scorch Symptoms. Phytopathol. Mediterr. 2017, 56, 259–273. [Google Scholar] [CrossRef]
- Olmo-García, L.; Kessler, N.; Neuweger, H.; Wendt, K.; Olmo-Peinado, J.M.; Fernández-Gutiérrez, A.; Baessmann, C.; Carrasco-Pancorbo, A. Unravelling the Distribution of Secondary Metabolites in Olea europaea L.: Exhaustive Characterization of Eight Olive-Tree Derived Matrices by Complementary Platforms (LC-ESI/APCI-MS and GC-APCI-MS). Molecules 2018, 23, 2419. [Google Scholar] [CrossRef]
- Serrano-García, I.; Olmo-García, L.; Polo-Megías, D.; Serrano, A.; León, L.; de la Rosa, R.; Gómez-Caravaca, A.M.; Carrasco-Pancorbo, A. Fruit Phenolic and Triterpenic Composition of Progenies of Olea europaea subsp. Cuspidata, an Interesting Phytochemical Source to Be Included in Olive Breeding Programs. Plants 2022, 11, 1791. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Klen, T.J.; Wondra, A.G.; Vrhovšek, U.; Vodopivec, B.M. Phenolic Profiling of Olives and Olive Oil Process-Derived Matrices Using UPLC-DAD-ESI-QTOF-HRMS Analysis. J. Agric. Food Chem. 2015, 63, 3859–3872. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Aloy, M.; Groff, N.; Masuero, D.; Nisi, M.; Franco, A.; Battelini, F.; Vrhovsek, U.; Mattivi, F. Exploratory Analysis of Commercial Olive-Based Dietary Supplements Using Untargeted and Targeted Metabolomics. Metabolites 2020, 10, 516. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.D.; Wu, D.G.; Cai, X.H.; Kennelly, E.J. New Antioxidant Phenolic Glycosides from Walsura Yunnanensis. Chem. Biodivers. 2006, 3, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vega, J.; Gómez-Alonso, S.; Pérez-Navarro, J.; Alarcón-Enos, J.; Pastene-Navarrete, E. Polyphenolic Compounds Extracted and Purified from Buddleja Globosa Hope (Buddlejaceae) Leaves Using Natural Deep Eutectic Solvents and Centrifugal Partition Chromatography. Molecules 2021, 26, 2192. [Google Scholar] [CrossRef]
- David, J.M.; Barreiros, A.L.B.S.; David, J.P. Antioxidant Phenylpropanoid Esters of Triterpenes from Dioclea Lasiophylla. Pharm. Biol. 2004, 42, 36–38. [Google Scholar] [CrossRef]
- Tóth, G.; Alberti, Á.; Sólyomváry, A.; Barabás, C.; Boldizsár, I.; Noszál, B. Phenolic Profiling of Various Olive Bark-Types and Leaves: HPLC–ESI/MS Study. Ind. Crop. Prod. 2015, 67, 432–438. [Google Scholar] [CrossRef]
- Olmo-García, L.; Bajoub, A.; Benlamaalam, S.; Hurtado-Fernández, E.; Bagur-González, M.G.; Chigr, M.; Mbarki, M.; Fernández-Gutiérrez, A.; Carrasco-Pancorbo, A. Establishing the Phenolic Composition of Olea europaea L. Leaves from Cultivars Grown in Morocco as a Crucial Step Towards Their Subsequent Exploitation. Molecules 2018, 23, 2524. [Google Scholar] [CrossRef]
- Abbattista, R.; Ventura, G.; Calvano, C.D.; Cataldi, T.R.I.; Losito, I. Bioactive Compounds in Waste By-Products from Olive Oil Production: Applications and Structural Characterization by Mass Spectrometry Techniques. Foods 2021, 10, 1236. [Google Scholar] [CrossRef]
- Michel, T.; Khlif, I.; Kanakis, P.; Termentzi, A.; Allouche, N.; Halabalaki, M.; Skaltsounis, A.L. UHPLC-DAD-FLD and UHPLC-HRMS/MS Based Metabolic Profiling and Characterization of Different Olea europaea Organs of Koroneiki and Chetoui Varieties. Phytochem. Lett. 2015, 11, 424–439. [Google Scholar] [CrossRef]
- Ghanbari, R.; Anwar, F.; Alkharfy, K.M.; Gilani, A.H.; Saari, N. Valuable Nutrients and Functional Bioactives in Different Parts of Olive (Olea europaea L.)—A Review. Int. J. Mol. Sci. 2012, 13, 3291–3340. [Google Scholar] [CrossRef]
- Talhaoui, N.; Gómez-Caravaca, A.M.; León, L.; De la Rosa, R.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Determination of Phenolic Compounds of “Sikitita” Olive Leaves by HPLC-DAD-TOF-MS. Comparison with Its Parents “Arbequina” and “Picual” Olive Leaves. LWT Food Sci. Technol. 2014, 58, 28–34. [Google Scholar] [CrossRef]
- Contreras, M.D.M.; Gomez-Cruz, I.; Romero, I.; Castro, E. Olive Pomace-Derived Biomasses Fractionation through a Two-Step Extraction Based on the Use of Ultrasound: Chemical Characteristics. Foods 2021, 10, 111. [Google Scholar] [CrossRef]
- Al-Warhi, T.; Elmaidomy, A.H.; Maher, S.A.; Abu-Baih, D.H.; Selim, S.; Albqmi, M.; Al-Sanea, M.M.; Alnusaire, T.S.; Ghoneim, M.M.; Mostafa, E.M.; et al. The Wound-Healing Potential of Olea europaea L. Cv. Arbequina Leaves Extract: An Integrated In Vitro, In Silico, and In Vivo Investigation. Metabolites 2022, 12, 791. [Google Scholar] [CrossRef] [PubMed]
- Ammar, S.; Contreras, M.d.M.; Gargouri, B.; Segura-Carretero, A.; Bouaziz, M. RP-HPLC-DAD-ESI-QTOF-MS Based Metabolic Profiling of the Potential Olea europaea by-Product “Wood” and Its Comparison with Leaf Counterpart. Phytochem. Anal. 2017, 28, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, M.A.; Khan, A.; Hanif, M.; Farooq, U.; Perveen, S. Traditional Uses, Phytochemistry, and Pharmacology of Olea europaea (Olive). Evidence-Based Complement. Altern. Med. 2015, 2015, 541591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xin, X.; Zhang, J.; Zhu, S.; Niu, E.; Zhou, Z.; Liu, D. Comparative Evaluation of the Phytochemical Profiles and Antioxidant Potentials of Olive Leaves from 32 Cultivars Grown in China. Molecules 2022, 27, 1292. [Google Scholar] [CrossRef]
- Sánchez-Lucas, R. The Effect of Increasing Temperature on Olive Trees (Olea europaea L. subsp. Europaea) Biology: An Integrated Morphological, Phenological and Biomolecular Study; University of Cordoba: Córdoba, Spain, 2019. [Google Scholar]
- Vidović, N.; Pasković, I.; Lukić, I.; Žurga, P.; Germek, V.M.; Grozić, K.; Cukrov, M.; Marcelić, Š.; Ban, D.; Talhaoui, N.; et al. Biophenolic Profile Modulations in Olive Tissues as Affected by Manganese Nutrition. Plants 2021, 10, 1724. [Google Scholar] [CrossRef]
- Ben Brahim, S.; Priego-Capote, F.; Bouaziz, M. Use of High-Performance Liquid Chromatography/Electrospray Ionization Mass Spectrometry for Structural Characterization of Bioactive Compounds in the Olive Root Bark and Wood of Chemlali Cultivar. ACS Omega 2022, 7, 33873–33883. [Google Scholar] [CrossRef]
- Mechri, B.; Tekaya, M.; Attia, F.; Hammami, M.; Chehab, H. Drought Stress Improved the Capacity of Rhizophagus Irregularis for Inducing the Accumulation of Oleuropein and Mannitol in Olive (Olea europaea) Roots. Plant Physiol. Biochem. 2020, 156, 178–191. [Google Scholar] [CrossRef]
- Skodra, C.; Michailidis, M.; Dasenaki, M.; Ganopoulos, I.; Thomaidis, N.S.; Tanou, G.; Molassiotis, A. Unraveling Salt-Responsive Tissue-Specific Metabolic Pathways in Olive Tree. Physiol. Plant. 2021, 173, 1643–1656. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Zhang, L.; Ma, C.; Abd El-Aty, A.M. Edible Pentacyclic Triterpenes: A Review of Their Sources, Bioactivities, Bioavailability, Self-Assembly Behavior, and Emerging Applications as Functional Delivery Vehicles. Crit. Rev. Food Sci. Nutr. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Machała, P.; Liudvytska, O.; Kicel, A.; Dziedzic, A.; Olszewska, M.A.; Żbikowska, H.M. Valorization of the Photo-Protective Potential of the Phytochemically Standardized Olive (Olea europaea L.) Leaf Extract in UVA-Irradiated Human Skin Fibroblasts. Molecules 2022, 27, 5144. [Google Scholar] [CrossRef] [PubMed]
- Cittan, M.; Çelik, A. Development and Validation of an Analytical Methodology Based on Liquid Chromatography–Electrospray Tandem Mass Spectrometry for the Simultaneous Determination of Phenolic Compounds in Olive Leaf Extract. J. Chromatogr. Sci. 2018, 56, 336–343. [Google Scholar] [CrossRef] [PubMed]
| Leaves | Stems | Roots | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Real/Predicted | Class 1 | Class 2 | Class 3 | Class 1 | Class 2 | Class 3 | Class 1 | Class 2 | Class 3 |
| Class 1 | 6 | 0 | 0 | 6 | 0 | 0 | 5 | 0 | 1 |
| Class 2 | 2 | 15 | 1 | 2 | 16 | 0 | 5 | 9 | 4 |
| Class 3 | 0 | 0 | 6 | 0 | 0 | 6 | 0 | 0 | 6 |
| Components | 3 | 4 | 3 | ||||||
| Error rate | 0.06 | 0.04 | 0.28 | ||||||
| Accuracy | 0.90 | 0.93 | 0.70 | ||||||
| Highly Resistant Cultivars | Medium Susceptibility Cultivars * | Extremely Susceptible Cultivars | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Regression Coefficient | VIP Value | Metabolite | Regression Coefficient | VIP Value | Metabolite | Regression Coefficient | VIP Value | ||
| Leaves | ↑ | Maslinic acid | 0.057 | 1.60 | Gallocatechin | 0.121 | 3.78 | Lucidumoside C | 0.064 | 1.43 |
| Oleuropein aglycone (is 2) | 0.055 | 1.92 | Oleuroside | 0.060 | 1.44 | |||||
| Neonuzhenide/oleuropein glucoside (is 3) | 0.060 | 2.05 | ||||||||
| Hydroxytyrosol glucoside | 0.057 | 1.87 | ||||||||
| Elenolic acid glucoside (is 2) | 0.038 | 1.71 | ||||||||
| Maslinic acid + | 0.035 | 1.82 | ||||||||
| Elenolic acid glucoside (is 1) | 0.019 | 1.11 | ||||||||
| ↓ | Lucidumoside C | −0.002 | 1.35 | Luteolin 7-O-glucoside (is 1) | −0.066 | 1.65 | Oleuropein aglycone (is 2) | −0.001 | 1.49 | |
| Oleuroside | −0.007 | 1.61 | Chrysoeriol O-glucoside | −0.070 | 2.03 | Aldehydic form of DEA glucoside | −0.004 | 1.37 | ||
| Neonuzhenide/oleuropein glucoside (is 3) | −0.030 | 2.83 | Oleuropein aglycone (is 1) | −0.077 | 2.05 | |||||
| Hydroxytyrosol glucoside | −0.032 | 2.62 | Demethyl oleuropein | −0.084 | 2.19 | |||||
| Elenolic acid glucoside (is1) | −0.040 | 1.53 | Maslinic acid + | −0.092 | 2.96 | |||||
| Elenolic acid glucoside (is 2) + | −0.045 | 2.54 | ||||||||
| Aldehydic form of DEA glucoside + | −0.053 | 1.49 | ||||||||
| Stems | ↑ | Unknown 4 | 0.058 | 3.39 | 11-Hydroxyiridodial glucoside pentaacetate + | 0.096 | 2.12 | Oleuroside | 0.094 | 4.04 |
| Quinic acid | 0.051 | 2.49 | Metoxypinoresinol glucoside | 0.067 | 2.14 | |||||
| Demethyl oleuropein | 0.042 | 1.94 | Oleuropein | 0.060 | 3.34 | |||||
| Oleanolic acid | 0.036 | 2.70 | Neonuzhenide/oleuropein glucoside (is 2) | 0.057 | 1.65 | |||||
| Acetoxypinoresinol | 0.029 | 1.64 | Betulinic acid | 0.045 | 1.68 | |||||
| Unknown 5 | 0.022 | 2.06 | Maslinic acid + | 0.023 | 1.63 | |||||
| ↓ | Neonuzhenide/oleuropein glucoside (is 2) | −0.029 | 2.55 | Betulinic acid | −0.059 | 2.21 | 11-Hydroxyiridodial glucoside pentaacetate + | −0.085 | 1.90 | |
| Aldehydic form of DEA glucoside + | −0.038 | 1.54 | Maslinic acid + | −0.063 | 1.92 | |||||
| Elenolic acid glucoside (is 2) + | −0.042 | 2.40 | Metoxypinoresinol glucoside | −0.086 | 2.71 | |||||
| Unknown 2 | −0.045 | 2.96 | Oleuropein | −0.112 | 4.08 | |||||
| Oleuroside | −0.130 | 5.18 | ||||||||
| Roots | ↑ | Cycloolivil glucoside (is 2) | 0.082 | 5.69 | 11-Hydroxyiridodial glucoside pentaacetate + | 0.110 | 2.56 | Acetoxypinoresinol glucoside | 0.109 | 3.04 |
| Betulinic acid | 0.047 | 1.94 | Hydroxypinoresinol glucoside | 0.092 | 1.92 | Unknown 1 | 0.083 | 1.74 | ||
| Elenolic acid glucoside (is 1) | 0.034 | 1.42 | Betulinic acid | 0.052 | 1.70 | |||||
| ↓ | Verbascoside | −0.047 | 1.75 | Acetoxypinoresinol glucoside | −0.110 | 2.20 | Cycloolivil glucoside (is 2) | −0.015 | 1.67 | |
| Cycloolivil glucoside (is 2) | −0.067 | 2.56 | Oleanolic acid | −0.076 | 2.17 | |||||
| Hydroxypinoresinol glucoside | −0.088 | 2.78 | ||||||||
| Betulinic acid | −0.099 | 3.06 | ||||||||
| 11-Hydroxyiridodial glucoside pentaacetate + | −0.113 | 3.85 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano-García, I.; Olmo-García, L.; Monago-Maraña, O.; de Alba, I.M.C.; León, L.; de la Rosa, R.; Serrano, A.; Gómez-Caravaca, A.M.; Carrasco-Pancorbo, A. Characterization of the Metabolic Profile of Olive Tissues (Roots, Stems and Leaves): Relationship with Cultivars’ Resistance/Susceptibility to the Soil Fungus Verticillium dahliae. Antioxidants 2023, 12, 2120. https://doi.org/10.3390/antiox12122120
Serrano-García I, Olmo-García L, Monago-Maraña O, de Alba IMC, León L, de la Rosa R, Serrano A, Gómez-Caravaca AM, Carrasco-Pancorbo A. Characterization of the Metabolic Profile of Olive Tissues (Roots, Stems and Leaves): Relationship with Cultivars’ Resistance/Susceptibility to the Soil Fungus Verticillium dahliae. Antioxidants. 2023; 12(12):2120. https://doi.org/10.3390/antiox12122120
Chicago/Turabian StyleSerrano-García, Irene, Lucía Olmo-García, Olga Monago-Maraña, Iván Muñoz Cabello de Alba, Lorenzo León, Raúl de la Rosa, Alicia Serrano, Ana María Gómez-Caravaca, and Alegría Carrasco-Pancorbo. 2023. "Characterization of the Metabolic Profile of Olive Tissues (Roots, Stems and Leaves): Relationship with Cultivars’ Resistance/Susceptibility to the Soil Fungus Verticillium dahliae" Antioxidants 12, no. 12: 2120. https://doi.org/10.3390/antiox12122120
APA StyleSerrano-García, I., Olmo-García, L., Monago-Maraña, O., de Alba, I. M. C., León, L., de la Rosa, R., Serrano, A., Gómez-Caravaca, A. M., & Carrasco-Pancorbo, A. (2023). Characterization of the Metabolic Profile of Olive Tissues (Roots, Stems and Leaves): Relationship with Cultivars’ Resistance/Susceptibility to the Soil Fungus Verticillium dahliae. Antioxidants, 12(12), 2120. https://doi.org/10.3390/antiox12122120












