The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions
Abstract
:1. Introduction
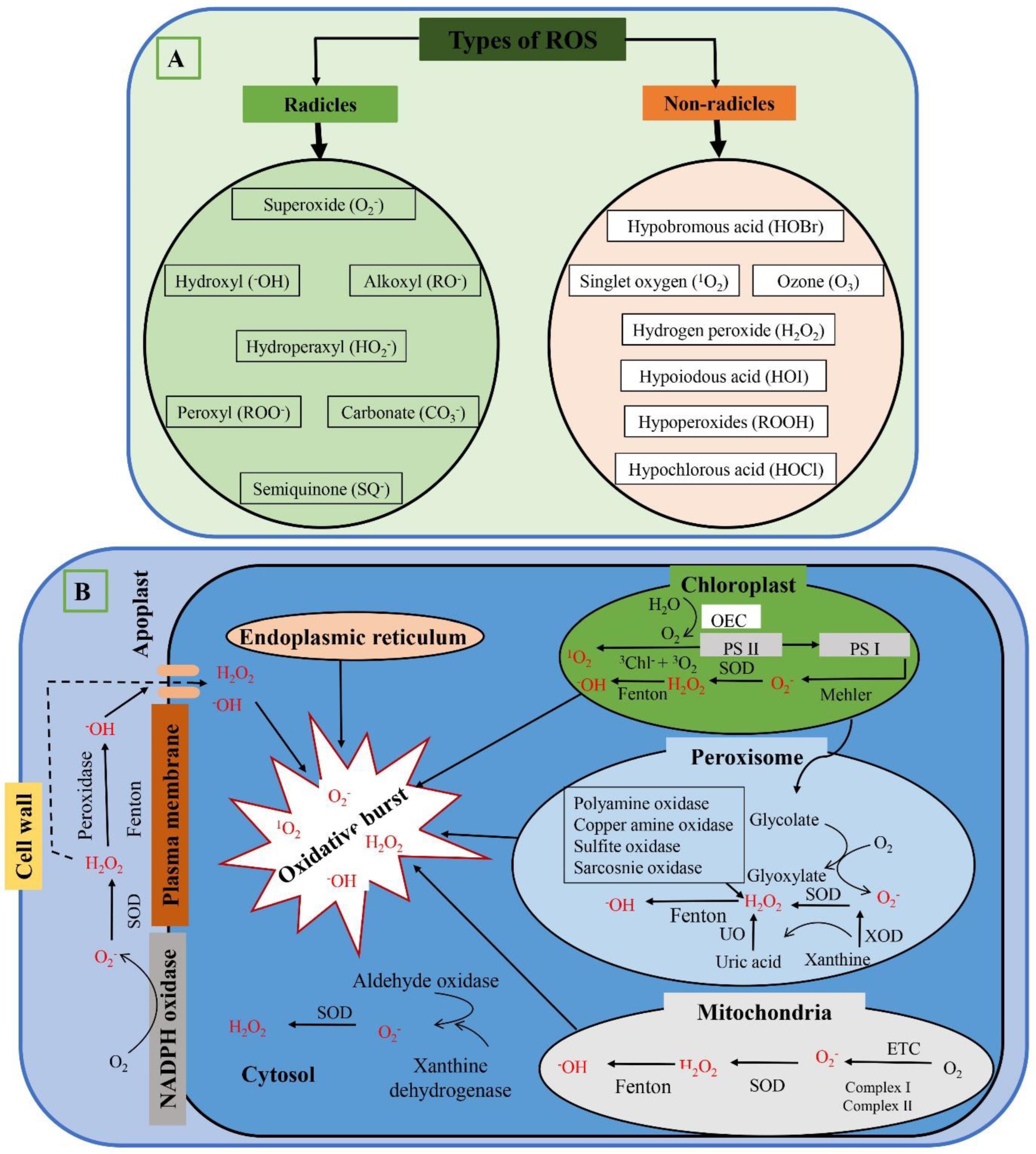
2. Types of ROS
2.1. Superoxide Anions (O2−)
2.2. Singlet Oxygen (1O2−)
2.3. Hydrogen Peroxide (H2O2)
2.4. Hydroxyl Radical (−OH)
2.5. Peroxyl Radical (ROO−)
2.6. Alkoxy Radical (RO−)
3. ROS Production Locations
3.1. Chloroplast
3.2. Mitochondria
3.3. Apoplast
3.4. Plasma Membranes
3.5. Cell wall and Endoplasmic Reticulum
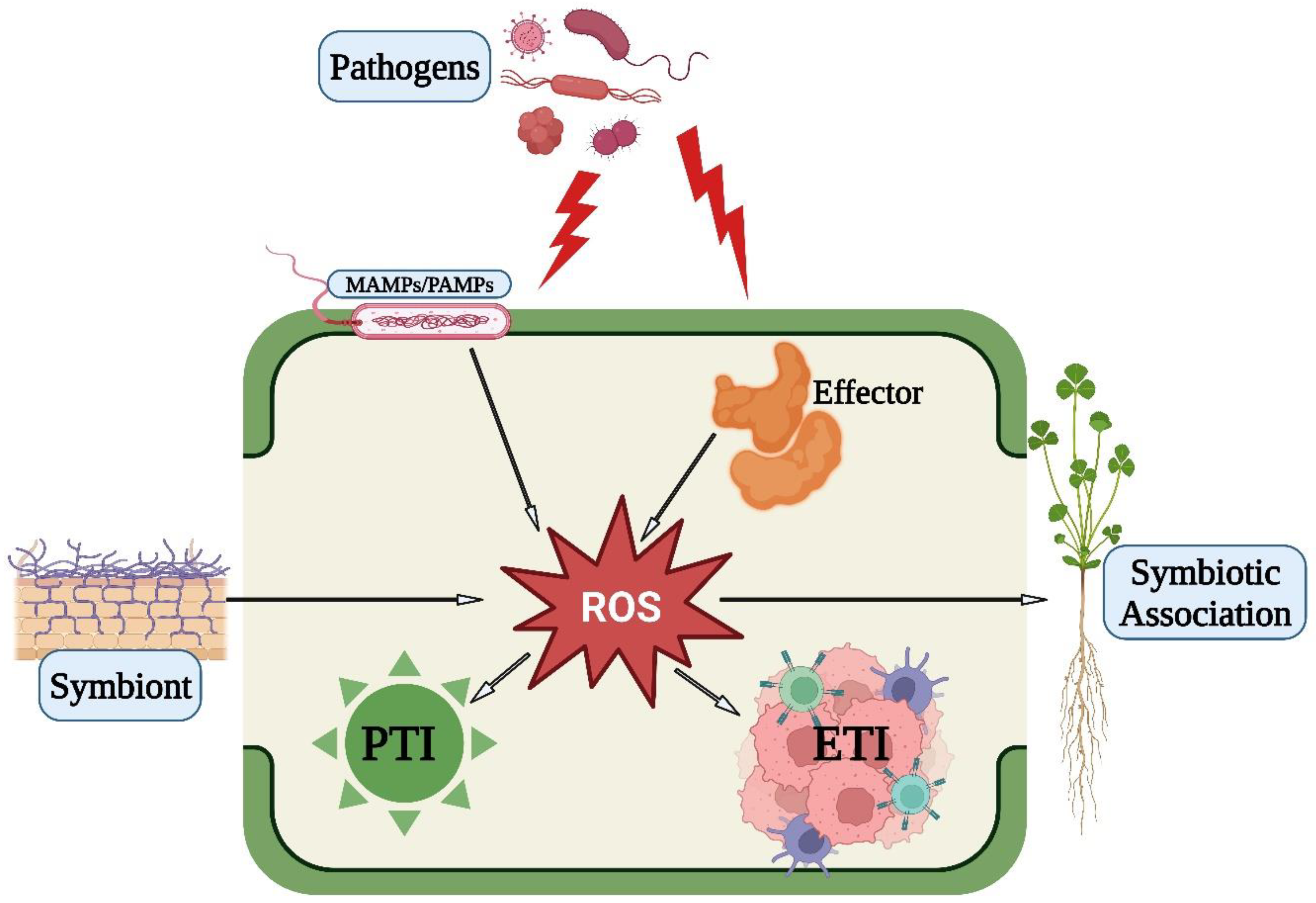
4. ROS Plays a Significant Role in Plant–Microbe Interactions
4.1. Role of ROS in PTI
4.2. Role of ROS in ETI
4.3. Role of ROS in Symbiotic Association
5. Plants Antioxidant Defense System
5.1. Enzymatic Antioxidants
5.1.1. Superoxide Dismutase
5.1.2. Catalase and Peroxidase
5.2. Non-Enzymatic Antioxidants
5.2.1. Ascorbic Acid
5.2.2. Glutathione
5.2.3. Carotenoids
5.2.4. α-Tocopherols
5.2.5. Phenolic Compounds
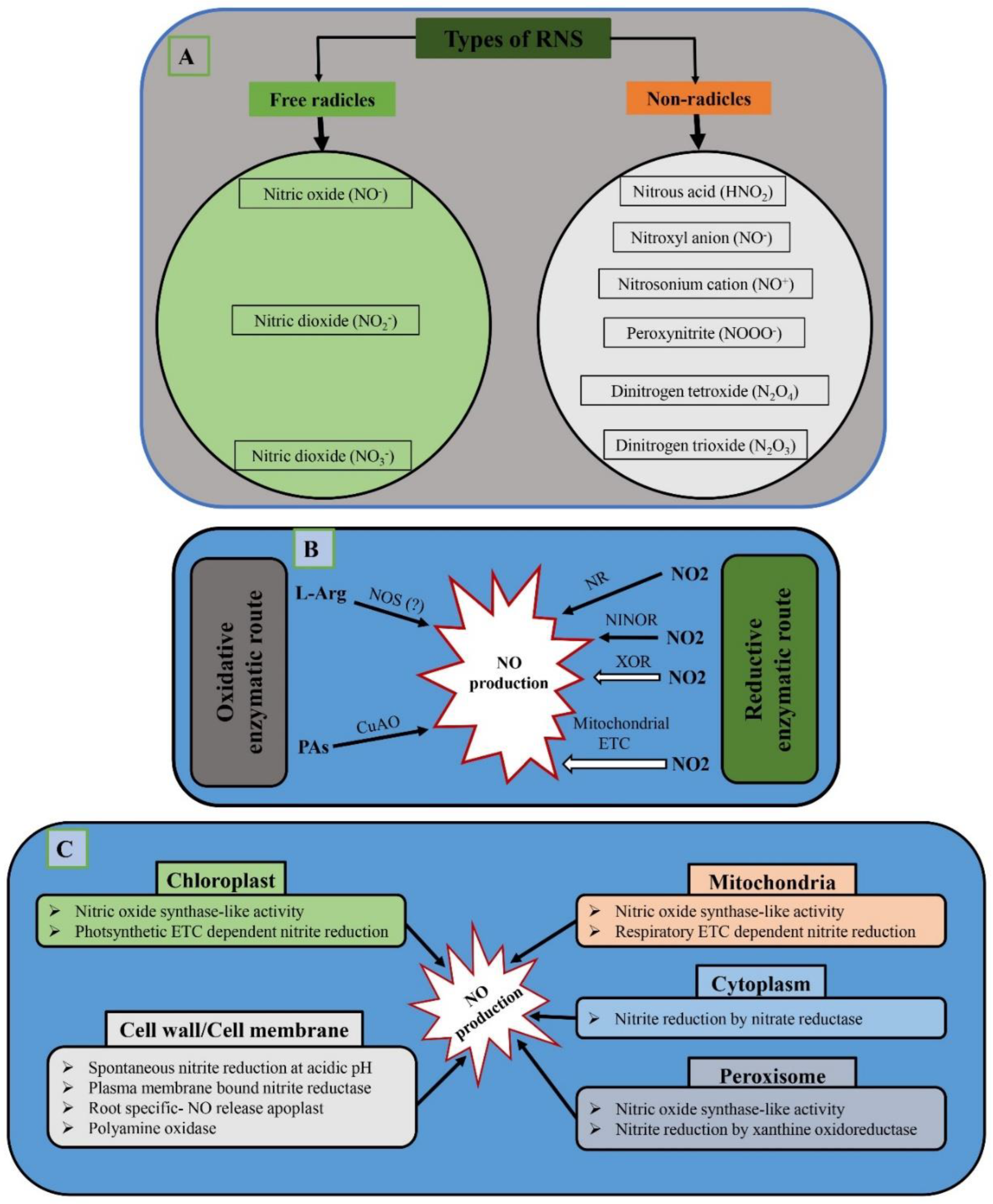
6. RNS Production in Plants
7. Role of RNS in Plant–Microbe Interactions
7.1. Role of NO in PTI, ETI, and Symbiotic Association in Plants
7.2. S-Nitrosylation of NPR1, SAB3, PAL, and CHS Proteins
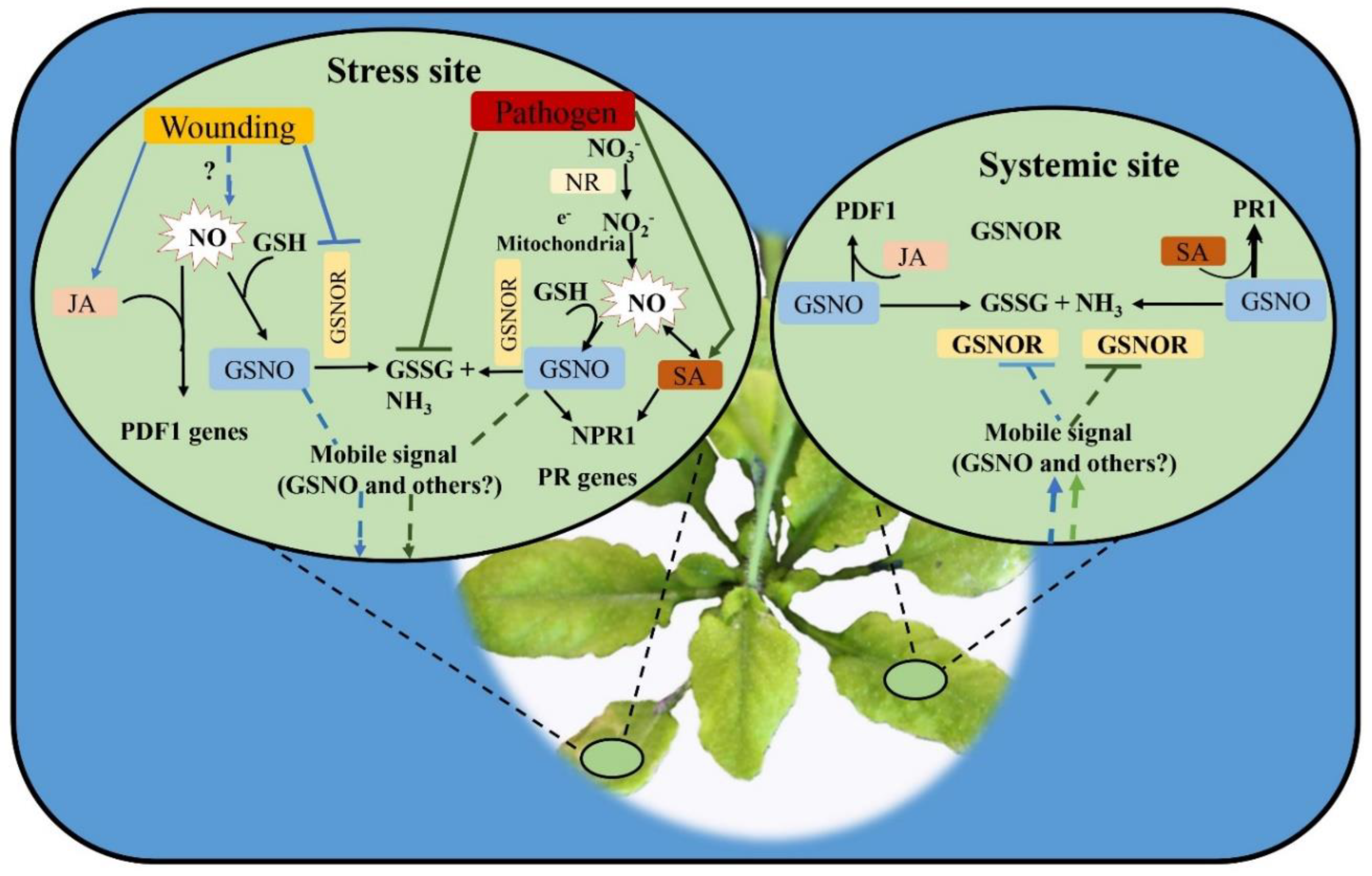
7.3. Role of GSNOR and NR in NO Homeostasis and Plant Immunity
8. Cross-Talk of ROS and RNS
9. Role of ROS and RNS in Protein Modifications
10. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Al Azzawi, T.N.I.; Khan, M.; Hussain, A.; Shahid, M.; Imran, Q.M.; Mun, B.-G.; Lee, S.-U.; Yun, B.-W. Evaluation of Iraqi rice cultivars for their tolerance to drought stress. Agronomy 2020, 10, 1782. [Google Scholar] [CrossRef]
- Khan, M.; Al Azzawi, T.N.I.; Imran, M.; Hussain, A.; Mun, B.-G.; Pande, A.; Yun, B.-W. Effects of lead (Pb)-induced oxidative stress on morphological and physio-biochemical properties of rice. Biocell 2021, 45, 1413. [Google Scholar] [CrossRef]
- Imran, M.; Shazad, R.; Bilal, S.; Imran, Q.M.; Khan, M.; Kang, S.-M.; Khan, A.L.; Yun, B.-W.; Lee, I.-J. Exogenous Melatonin mediates the regulation of endogenous nitric oxide in Glycine max L. to reduce effects of drought stress. Environ. Exp. Bot. 2021, 188, 104511. [Google Scholar] [CrossRef]
- Khan, M.; Ali, S.; Manghwar, H.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. Melatonin function and crosstalk with other phytohormones under normal and stressful conditions. Genes 2022, 13, 1699. [Google Scholar] [CrossRef] [PubMed]
- Pande, A.; Mun, B.-G.; Lee, D.-S.; Khan, M.; Lee, G.-M.; Hussain, A.; Yun, B.-W. No network for plant–microbe communication underground: A review. Front. Plant Sci. 2021, 12, 658679. [Google Scholar] [CrossRef]
- Pande, A.; Mun, B.-G.; Khan, M.; Rahim, W.; Lee, D.-S.; Lee, G.-M.; Al Azawi, T.N.I.; Hussain, A.; Yun, B.-W. Nitric Oxide Signaling and Its Association with Ubiquitin-Mediated Proteasomal Degradation in Plants. Int. J. Mol. Sci. 2022, 23, 1657. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Oelmüller, R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012, 7, 1621–1633. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Nazar, T.; Pande, A.; Mun, B.-G.; Lee, D.; Hussain, A.; Lee, B.-H.; Yun, B.-W. The Role of Nitric Oxide-Induced ATILL6 in Growth and Disease Resistance in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 1314. [Google Scholar] [CrossRef]
- Rahim, W.; Khan, M.; Al Azzawi, T.N.I.; Pande, A.; Methela, N.J.; Ali, S.; Imran, M.; Lee, D.-S.; Lee, G.-M.; Mun, B.-G. Exogenously Applied Sodium Nitroprusside Mitigates Lead Toxicity in Rice by Regulating Antioxidants and Metal Stress-Related Transcripts. Int. J. Mol. Sci. 2022, 23, 9729. [Google Scholar] [CrossRef]
- Ali, S.; Moon, Y.-S.; Hamayun, M.; Khan, M.A.; Bibi, K.; Lee, I.-J. Pragmatic role of microbial plant biostimulants in abiotic stress relief in crop plants. J. Plant Interact. 2022, 17, 705–718. [Google Scholar] [CrossRef]
- Jan, F.G.; Hamayun, M.; Moon, Y.-S.; Jan, G.; Shafique, M.; Ali, S. Endophytic aspergillus oryzae reprograms Abelmoschus esculentus L. to higher growth under salt stress via regulation of physiochemical attributes and antioxidant system. Biologia 2022, 77, 1–14. [Google Scholar] [CrossRef]
- Sarkar, B.; Savita, S.K.; Varalaxmi, Y.; Vanaja, M.; Kumar, N.R.; Sathish, P.; Lakshmi, N.J.; Prabhakar, M.; Shanker, A.K.; Yadav, S.K. Stress Reactions of Maize Genotypes to Drought Stress at Different Phenophases and Recovery. Russ. J. Plant Physiol. 2022, 69, 1–10. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [Green Version]
- Kwon, N.; Kim, D.; Swamy, K.M.K.; Yoon, J. Metal-coordinated fluorescent and luminescent probes for reactive oxygen species (ROS) and reactive nitrogen species (RNS). Coord. Chem. Rev. 2021, 427, 213581. [Google Scholar] [CrossRef]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When bad guys become good ones: The key role of reactive oxygen species and nitric oxide in the plant responses to abiotic stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bleau, J.R.; Spoel, S.H. Selective redox signaling shapes plant–pathogen interactions. Plant Physiol. 2021, 186, 53–65. [Google Scholar] [CrossRef]
- Khan, M.A.; Hamayun, M.; Asaf, S.; Khan, M.; Yun, B.-W.; Kang, S.-M.; Lee, I.-J. Rhizospheric bacillus spp. rescues plant growth under salinity stress via regulating gene expression, endogenous hormones, and antioxidant system of Oryza sativa L. Front. Plant Sci. 2021, 12, 1145. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Ma, Z.; Marsolais, F.; Bykova, N.V.; Igamberdiev, A.U. Nitric oxide and reactive oxygen species mediate metabolic changes in barley seed embryo during germination. Front. Plant Sci. 2016, 7, 138. [Google Scholar] [CrossRef] [Green Version]
- Bykova, N.V.; Hoehn, B.; Rampitsch, C.; Hu, J.; Stebbing, J.-A.; Knox, R. Thiol redox-sensitive seed proteome in dormant and non-dormant hybrid genotypes of wheat. Phytochemistry 2011, 72, 1162–1172. [Google Scholar] [CrossRef]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Krieger-Liszkay, A.; Fufezan, C.; Trebst, A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 2008, 98, 551–564. [Google Scholar] [CrossRef]
- Dogra, V.; Kim, C. Singlet oxygen metabolism: From genesis to signaling. Front. Plant Sci. 2020, 10, 1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noctor, G.; Veljovic-Jovanovic, S.; Driscoll, S.; Novitskaya, L.; Foyer, C.H. Drought and oxidative load in the leaves of C3 plants: A predominant role for photorespiration? Ann. Bot. 2002, 89, 841–850. [Google Scholar] [CrossRef]
- Kuźniak, E.; Urbanek, H. The involvement of hydrogen peroxide in plant responses to stresses. Acta Physiol. Plant. 2000, 22, 195–203. [Google Scholar] [CrossRef]
- Bano, A.; Gupta, A.; Rai, S.; Fatima, T. Mechanistic role of reactive oxygen species and its regulation via the antioxidant system under environmental stress. In Plant Stress Physiology—Perspectives in Agriculture; IntechOpen: London, UK, 2021; pp. 1–18. [Google Scholar]
- Richards, S.L.; Wilkins, K.A.; Swarbreck, S.M.; Anderson, A.A.; Habib, N.; Smith, A.G.; McAinsh, M.; Davies, J.M. The hydroxyl radical in plants: From seed to seed. J. Exp. Bot. 2015, 66, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.A.D.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R.O.N. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; De Paepe, R.; Foyer, C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007, 12, 125–134. [Google Scholar] [CrossRef]
- Pastore, D.; Trono, D.; Laus, M.N.; Di Fonzo, N.; Flagella, Z. Possible plant mitochondria involvement in cell adaptation to drought stress: A case study: Durum wheat mitochondria. J. Exp. Bot. 2007, 58, 195–210. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhang, A.; Zhang, J.; Jiang, M. Abscisic acid is a key inducer of hydrogen peroxide production in leaves of maize plants exposed to water stress. Plant Cell Physiol. 2006, 47, 1484–1495. [Google Scholar] [CrossRef] [Green Version]
- Eaton, C.J.; Jourdain, I.; Foster, S.J.; Hyams, J.S.; Scott, B. Functional analysis of a fungal endophyte stress-activated MAP kinase. Curr. Genet. 2008, 53, 163–174. [Google Scholar] [CrossRef]
- Higuchi, T. Look back over the studies of lignin biochemistry. J. Wood Sci. 2006, 52, 2–8. [Google Scholar] [CrossRef]
- Turkan, I. Emerging roles for ROS and RNS–versatile molecules in plants. J. Exp. Bot. 2017, 68, 4413–4416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabi, R.B.S.; Rolly, N.K.; Tayade, R.; Khan, M.; Shahid, M.; Yun, B.-W. Enhanced Resistance of atbzip62 against Pseudomonas syringae pv. tomato Suggests Negative Regulation of Plant Basal Defense and Systemic Acquired Resistance by AtbZIP62 Transcription Factor. Int. J. Mol. Sci. 2021, 22, 11541. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Choi, J.; Lim, S.-E.; Lee, G.-W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.-S.; Jeon, J. Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A. ROS in biotic interactions. Physiol Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.G.; Shirasu, K.; Menke, F.; Jones, A. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Daudi, A.; Cheng, Z.; O’Brien, J.A.; Mammarella, N.; Khan, S.; Ausubel, F.M.; Bolwell, G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 2012, 24, 275–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.W.; Kim, Y.J.; Hong, L.S.C.; Hong, J.K.; Huang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant aquaporin AtPIP1; 4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 2016, 171, 1635–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jwa, N.-S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, D.; Naumann, M.; Falter, C.; Zwikowics, C.; Jamrow, T.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013, 161, 1433–1444. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, D.; Glöckner, A.; Koch, J.; Naumann, M.; Stürtz, V.; Schütt, K.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Interaction of the Arabidopsis GTPase RabA4c with its effector PMR4 results in complete penetration resistance to powdery mildew. Plant Cell 2014, 26, 3185–3200. [Google Scholar] [CrossRef] [Green Version]
- Deepak, S.; Shailasree, S.; Kini, R.K.; Hause, B.; Shetty, S.H.; Mithöfer, A. Role of hydroxyproline-rich glycoproteins in resistance of pearl millet against downy mildew pathogen Sclerospora graminicola. Planta 2007, 226, 323–333. [Google Scholar] [CrossRef]
- Han, S.W.; Hwang, B.K. Molecular functions of Xanthomonas type III effector AvrBsT and its plant interactors in cell death and defense signaling. Planta 2017, 245, 237–253. [Google Scholar] [CrossRef]
- Singh, R.; Dangol, S.; Chen, Y.; Choi, J.; Cho, Y.-S.; Lee, J.-E.; Choi, M.-O.; Jwa, N.-S. Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cells 2016, 39, 426. [Google Scholar]
- Choi, H.W.; Kim, D.S.; Kim, N.H.; Jung, H.W.; Ham, J.H.; Hwang, B.K. Xanthomonas filamentous hemagglutinin-like protein Fha1 interacts with pepper hypersensitive-induced reaction protein CaHIR1 and functions as a virulence factor in host plants. Mol. Plant-Microbe Interact. 2013, 26, 1441–1454. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, J.T.; Yao, N. The role and regulation of programmed cell death in plant–pathogen interactions. Cell. Microbiol. 2004, 6, 201–211. [Google Scholar] [CrossRef]
- Baltruschat, H.; Fodor, J.; Harrach, B.D.; Niemczyk, E.; Barna, B.; Gullner, G.; Janeczko, A.; Kogel, K.H.; Schafer, P.; Schwarczinger, I.; et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 2008, 180, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Al Mahmud, J.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrowska, E.; Różalska, S.; Kaźmierczak, A.; Nawrocka, J.; Małolepsza, U. Reactive oxygen and nitrogen (ROS and RNS) species generation and cell death in tomato suspension cultures—Botrytis cinerea interaction. Protoplasma 2015, 252, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.; Imran, Q.M.; Shahid, M.; Mun, B.-G.; Lee, S.-U.; Khan, M.A.; Hussain, A.; Lee, I.-J.; Yun, B.-W. Nitric oxide-induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef]
- Hussain, A.; Mun, B.-G.; Imran, Q.M.; Lee, S.-U.; Adamu, T.A.; Shahid, M.; Kim, K.-M.; Yun, B.-W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [Green Version]
- Künstler, A.; Kátay, G.; Gullner, G.; Király, L. Artificial elevation of glutathione contents in salicylic acid-deficient tobacco (Nicotiana tabacum cv. Xanthi NahG) reduces susceptibility to the powdery mildew pathogen Euoidium longipes. Plant Biol. 2020, 22, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Wylie, S.J.; Zhang, C.; Long, V.; Roossinck, M.J.; Koh, S.H.; Jones, M.G.K.; Iqbal, S.; Li, H. Differential responses to virus challenge of laboratory and wild accessions of Australian species of Nicotiana, and comparative analysis of RDR1 gene sequences. PLoS ONE 2015, 10, e0121787. [Google Scholar] [CrossRef]
- Garcia-Mata, C.; Lamattina, L. Abscisic acid (ABA) inhibits light-induced stomatal opening through calcium-and nitric oxide-mediated signaling pathways. Nitric Oxide 2007, 17, 143–151. [Google Scholar] [CrossRef]
- Gomes, M.P.; Smedbol, É.; Carneiro, M.M.L.C.; Garcia, Q.S.; Juneau, P. Reactive oxygen species and plant hormones. In Oxidative Damage to Plants; Elsevier: Amsterdam, The Netherlands, 2014; pp. 65–88. [Google Scholar]
- Zhang, M.; Ahmed Rajput, N.; Shen, D.; Sun, P.; Zeng, W.; Liu, T.; Juma Mafurah, J.; Dou, D. A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Fang, Q.; Zhang, Z.; Wang, Y.; Zheng, X. The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J. Exp. Bot. 2009, 60, 3109–3122. [Google Scholar] [CrossRef] [Green Version]
- Denness, L.; McKenna, J.F.; Segonzac, C.; Wormit, A.; Madhou, P.; Bennett, M.; Mansfield, J.; Zipfel, C.; Hamann, T. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species-and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011, 156, 1364–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Averyanov, A. Oxidative burst and plant disease resistance. Front. Biosci.-Elite 2009, 1, 142–152. [Google Scholar]
- Lee, D.; Lal, N.K.; Lin, Z.-J.D.; Ma, S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houot, V.; Etienne, P.; Petitot, A.S.; Barbier, S.; Blein, J.P.; Suty, L. Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose-dependent manner. J. Exp. Bot. 2001, 52, 1721–1730. [Google Scholar] [PubMed]
- Minina, E.A.; Dauphinee, A.N.; Ballhaus, F.; Gogvadze, V.; Smertenko, A.P.; Bozhkov, P.V. Apoptosis is not conserved in plants as revealed by critical examination of a model for plant apoptosis-like cell death. BMC Biol. 2021, 19, 1–17. [Google Scholar] [CrossRef]
- Lohar, D.P.; Haridas, S.; Gantt, J.S.; VandenBosch, K.A. A transient decrease in reactive oxygen species in roots leads to root hair deformation in the legume–rhizobia symbiosis. New Phytol. 2007, 173, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, L.; Martínez, A.; Sánchez, F.; Quinto, C. Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 2008, 56, 802–813. [Google Scholar] [CrossRef]
- Baptista, P.; Martins, A.; Pais, M.S.; Tavares, R.M.; Lino-Neto, T. Involvement of reactive oxygen species during early stages of ectomycorrhiza establishment between Castanea sativa and Pisolithus tinctorius. Mycorrhiza 2007, 17, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Shahid, M.; Imran, Q.M.; Hussain, A.; Khan, M.; Lee, S.U.; Mun, B.G.; Yun, B.-W. Comprehensive analyses of nitric oxide-induced plant stem cell-related genes in Arabidopsis thaliana. Genes 2019, 10, 190. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Liu, Z.; Shao, Y.; Su, J.; Li, X.; Sun, F.; Zhang, Y.; Li, S.; Zhang, Y.; Cui, J. Nitric oxide enhances rice resistance to rice black-streaked dwarf virus infection. Rice 2020, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Feechan, A.; Kwon, E.; Yun, B.-W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Giudice, J.; Cam, Y.; Damiani, I.; Fung-Chat, F.; Meilhoc, E.; Bruand, C.; Brouquisse, R.; Puppo, A.; Boscari, A. Nitric oxide is required for an optimal establishment of the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 2011, 191, 405–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, M.; Murakami, E.-I.; Shimoda, Y.; Shimoda-Sasakura, F.; Kucho, K.-I.; Suzuki, A.; Abe, M.; Higashi, S.; Uchiumi, T. Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus. Mol. Plant-Microbe Interact. 2008, 21, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Berger, A.; Boscari, A.; Horta Araujo, N.; Maucourt, M.; Hanchi, M.; Bernillon, S.; Rolin, D.; Puppo, A.; Brouquisse, R. Plant nitrate reductases regulate nitric oxide production and nitrogen-fixing metabolism during the Medicago truncatula–Sinorhizobium meliloti symbiosis. Front. Plant Sci. 2020, 11, 1313. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, A.; Sharma, S.K.; Choudhary, D.K.; Sharma, K.P.; Ahmad, E.; Sharma, M.P.; Ramesh, A.; Saxena, A.K. Nitric oxide as a signaling molecule in plant-bacterial interactions. In Plant Microbiome: Stress Response; Springer: Berlin/Heidelberg, Germany, 2018; pp. 183–199. [Google Scholar]
- Cui, B.; Xu, S.; Li, Y.; Umbreen, S.; Frederickson, D.; Yuan, B.; Jiang, J.; Liu, F.; Pan, Q.; Loake, G.J. The Arabidopsis zinc finger proteins SRG2 and SRG3 are positive regulators of plant immunity and are differentially regulated by nitric oxide. New Phytol. 2021, 230, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lin, J.; Chang, Y.; Jiang, C.-Z. Comparative transcriptomic analysis reveals that ethylene/H2O2-mediated hypersensitive response and programmed cell death determine the compatible interaction of sand pear and Alternaria alternata. Front. Plant Sci. 2017, 8, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaurilind, E.; Xu, E.; Brosché, M. A genetic framework for H2O2 induced cell death in Arabidopsis thaliana. BMC Genom. 2015, 16, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, F.; Goodman, R.M. Activity of nitric oxide is dependent on, but is partially required for function of, salicylic acid in the signaling pathway in tobacco systemic acquired resistance. Mol. Plant-Microbe Interact. 2001, 14, 1458–1462. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot 2018, 154, 134–142. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef]
- Pan, S.-M.; Yau, Y.-Y. Characterization of superoxide dismutase in Arabidopsis. Taiwania 1992, 37, 58–66. [Google Scholar]
- Rajput, V.D.; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; Mandzhieva, S. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Sarkar, M.; Khan, A.; Biswas, M.; Masi, A.; Rakwal, R.; Agrawal, G.K.; Srivastava, A.; Sarkar, A. Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS) in Plants–maintenance of structural individuality and functional blend. Adv. Redox Res. 2022, 5, 100039. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Harmens, H.; Zheng, X.; Zhang, C. Melatonin enhances drought resistance by regulating leaf stomatal behaviour, root growth and catalase activity in two contrasting rapeseed (Brassica napus L.) genotypes. Plant Physiol. Biochem. 2020, 149, 86–95. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative damage, lipid peroxidation and antioxidant protection in chloroplasts. Chem. Phys. Lipids 1987, 44, 327–340. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Glušac, J.; Isaschar-Ovdat, S.; Fishman, A.; Kukavica, B. Partial characterization of bean and maize root peroxidases and their ability to crosslink potato protein. Arch. Biol. Sci. 2019, 71, 293–303. [Google Scholar] [CrossRef] [Green Version]
- Soares, C.; Carvalho, M.E.A.; Azevedo, R.A.; Fidalgo, F. Plants facing oxidative challenges—A little help from the antioxidant networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Castro, J.L.S.; Lima-Melo, Y.; Carvalho, F.E.L.; Feitosa, A.G.S.; Lima Neto, M.C.; Caverzan, A.; Margis-Pinheiro, M.; Silveira, J.A.G. Ascorbic acid toxicity is related to oxidative stress and enhanced by high light and knockdown of chloroplast ascorbate peroxidases in rice plants. Theor. Exp. Plant Physiol. 2018, 30, 41–55. [Google Scholar] [CrossRef] [Green Version]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [Green Version]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2014, 79, 597–606. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Munné-Bosch, S.; Alegre, L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002, 21, 31–57. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. Interplay between ascorbic acid and lipophilic antioxidant defences in chloroplasts of water-stressed Arabidopsis plants. FEBS Lett. 2002, 524, 145–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakibara, H.; Honda, Y.; Nakagawa, S.; Ashida, H.; Kanazawa, K. Simultaneous determination of all polyphenols in vegetables, fruits, and teas. J. Agric. Food Chem. 2003, 51, 571–581. [Google Scholar] [CrossRef]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.J. The Role of the Plant Antioxidant System in Drought Tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Halliwell, B.; Gutteridge, J. Antioxidant defences: Endogenous and diet derived. Free. Radic. Biol. Med. 2007, 4, 79–186. [Google Scholar]
- Gupta, K.J.; Igamberdiev, A.U. Compartmentalization of reactive oxygen species and nitric oxide production in plant cells: An overview. In Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–14. [Google Scholar]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Calo, G.; Salerno, G.; Lamattina, L. Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus tauri Is Light Irradiance and Growth Phase Dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mur, L.A.J.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.M.; Hebelstrup, K.H.; Gupta, K.J. Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 2013, 5, pls052. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Chaki, M.; Airaki, M.; Valderrama, R.; Palma, J.M.; Barroso, J.B.; Corpas, F.J. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal. Behav. 2011, 6, 789–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Park, S.-K.; Kim, W.-C. The pragmatic introduction and expression of microbial transgenes in plants. J. Microbiol. Biotechnol. 2018, 28, 1955–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, B.; Ma, X.; Li, Y.; Zhou, Y.; Ju, X.; Hussain, A.; Umbreen, S.; Yuan, B.; Tabassum, A.; Lubega, J. Perturbations in nitric oxide homeostasis promote Arabidopsis disease susceptibility towards Phytophthora parasitica. Mol. Plant Pathol. 2021, 22, 1134–1148. [Google Scholar] [CrossRef]
- Imran, Q.M.; Falak, N.; Hussain, A.; Mun, B.-G.; Sharma, A.; Lee, S.-U.; Kim, K.-M.; Yun, B.-W. Nitric oxide responsive heavy metal-associated gene AtHMAD1 contributes to development and disease resistance in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1712. [Google Scholar] [CrossRef] [Green Version]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Mou, Z.; Fan, W.H.; Dong, X.N. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindermayr, C.; Sell, S.; Muller, B.; Leister, D.; Durnera, J. Redox Regulation of the NPR1-TGA1 System of Arabidopsis thaliana by Nitric Oxide. Plant Cell 2010, 22, 2894–2907. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Q.; Feechan, A.; Yun, B.W.; Shafiei, R.; Hofmann, A.; Taylor, P.; Xue, P.; Yang, F.Q.; Xie, Z.S.; Pallas, J.A.; et al. S-Nitrosylation of AtSABP3 Antagonizes the Expression of Plant Immunity. J. Biol. Chem. 2009, 284, 2131–2137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Puertas, M.C.; Perazzolli, M.; Zago, E.D.; Delledonne, M. Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol. 2004, 6, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.G.; Magalhaes, J.R.; Salgado, I. Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett. 2005, 579, 3814–3820. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.C.; Saviani, E.E.; Oliveira, J.F.P.; Salgado, I. Nitrate reductase-dependent nitric oxide synthesis in the defense response of Arabidopsis thaliana against Pseudomonas syringae. Trop. Plant Pathol. 2010, 35, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.C.; Justino, G.C.; Sodek, L.; Salgado, I. Amino acid recovery does not prevent susceptibility to Pseudomonas syringae in nitrate reductase double-deficient Arabidopsis thaliana plants. Plant Sci. 2009, 176, 105–111. [Google Scholar] [CrossRef]
- Modolo, L.V.; Augusto, O.; Almeida, I.M.G.; Pinto-Maglio, C.A.F.; Oliveira, H.C.; Seligman, K.; Salgado, I. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 2006, 171, 34–40. [Google Scholar] [CrossRef]
- Rustérucci, C.; Espunya, M.C.; Díaz, M.; Chabannes, M.; Martínez, M.C. S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 2007, 143, 1282–1292. [Google Scholar] [CrossRef] [Green Version]
- Espunya, M.C.; De Michele, R.; Gómez-Cadenas, A.; Martínez, M.C. S-Nitrosoglutathione is a component of wound-and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 3219–3227. [Google Scholar] [CrossRef] [Green Version]
- Yun, B.-W.; Feechan, A.; Yin, M.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-G.; Kwon, E.; Spoel, S.H. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Díaz, M.; Achkor, H.; Titarenko, E.; Martínez, M.C. The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett. 2003, 543, 136–139. [Google Scholar] [CrossRef] [Green Version]
- Salgado, I.; Carmen Martínez, M.; Oliveira, H.C.; Frungillo, L. Nitric oxide signaling and homeostasis in plants: A focus on nitrate reductase and S-nitrosoglutathione reductase in stress-related responses. Braz. J. Bot. 2013, 36, 89–98. [Google Scholar] [CrossRef]
- Wang, Y.; Loake, G.J.; Chu, C. Cross-talk of nitric oxide and reactive oxygen species in plant programed cell death. Front. Plant Sci. 2013, 4, 314. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Wang, Y.; Tang, J.; Xue, P.; Li, C.; Liu, L.; Hu, B.; Yang, F.; Loake, G.J.; Chu, C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012, 158, 451–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lin, A.; Loake, G.J.; Chu, C. H2O2-induced Leaf Cell Death and the Crosstalk of Reactive Nitric/Oxygen Species F. J. Integr. Plant Biol. 2013, 55, 202–208. [Google Scholar] [CrossRef]
- Delledonne, M.; Zeier, J.; Marocco, A.; Lamb, C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 2001, 98, 13454–13459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaninotto, F.; Camera, S.L.; Polverari, A.; Delledonne, M. Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 2006, 141, 379–383. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.; Adhikari, S.; Tarafdar, A.; Kumar, R.; Saha, S.; Ghosh, P. Reactive Oxygen Species and Antioxidant Defence Systems in Plants: Role and Crosstalk Under Biotic Stress. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer: Cham, Switzerland, 2020; Chapter 12; pp. 265–292. [Google Scholar]
- Lanteri, M.L.; Lamattina, L.; Laxalt, A.M. Mechanisms of xylanase-induced nitric oxide and phosphatidic acid production in tomato cells. Planta 2011, 234, 845–855. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J.r. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, J.T.; Henson, D.; Nyirenda, M.; Desikan, R.; Harrison, J.; Lewis, M.; Hughes, J.; Neill, S.J. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Davis, K.R. The physiology of ozone induced cell death. Planta 2001, 213, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Ahlfors, R.; Brosché, M.; Kollist, H.; Kangasjärvi, J. Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 2009, 58, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Beligni, M.V.; Fath, A.; Bethke, P.C.; Lamattina, L.; Jones, R.L. Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol. 2002, 129, 1642–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, N.M.; Guo, F.-Q. New insights into nitric oxide metabolism and regulatory functions. Trends Plant Sci. 2005, 10, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Grün, S.; Lindermayr, C.; Sell, S.; Durner, J. Nitric oxide and gene regulation in plants. J. Exp. Bot. 2006, 57, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, C.; Loake, G.J.; Chu, C. Nitric oxide: Promoter or suppressor of programmed cell death? Protein Cell 2010, 1, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Sun, S.; Wang, C.; Li, Y.; Liang, Y.; An, F.; Li, C.; Dong, H.; Yang, X.; Zhang, J. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009, 19, 1377–1387. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, K.A.; Bancroft, J.; Bosch, M.; Ings, J.; Smirnoff, N.; Franklin-Tong, V.E. Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of papaver. Plant Physiol. 2011, 156, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, D.; Lachaud, C.; Cotelle, V.; Brière, C.; Grat, S.; Mazars, C.; Thuleau, P. Nitric oxide production is not required for dihydrosphingosine-induced cell death in tobacco BY-2 cells. Plant Signal. Behav. 2011, 6, 736–739. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.U.N.; Li, Z.H.E.; Xing, D.A. Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium-induced Arabidopsis thaliana programmed cell death. Plant Cell Environ. 2013, 36, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.; Desikan, R.; Harrison, J.; Bright, J.; Hooley, R.; Neill, S. Doing the unexpected: Proteins involved in hydrogen peroxide perception. J. Exp. Bot. 2006, 57, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Quintanilha, A.; Moradas-Ferreira, P. Protein oxidation, repair mechanisms and proteolysis in Saccharomyces cerevisiae. IUBMB Life 2007, 59, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Abiotic stress, generation of reactive oxygen species, and their consequences: An overview. In Reactive Oxygen Species in Plants: Boon or Bane-Revisiting the Role of ROS; Wiley: Hoboken, NJ, USA, 2017; pp. 23–50. [Google Scholar]
- Anjum, N.A.; Sofo, A.; Scopa, A.; Roychoudhury, A.; Gill, S.S.; Iqbal, M.; Lukatkin, A.S.; Pereira, E.; Duarte, A.C.; Ahmad, I. Lipids and proteins—Major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. 2015, 22, 4099–4121. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, C.G.; Gómez, F.; Martinez, D.E.; Guiamet, J.J. Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.). J. Exp. Bot. 2004, 55, 1663–1669. [Google Scholar] [CrossRef] [Green Version]
- Lounifi, I.; Arc, E.; Molassiotis, A.; Job, D.; Rajjou, L.; Tanou, G. Interplay between protein carbonylation and nitrosylation in plants. Proteomics 2013, 13, 568–578. [Google Scholar] [CrossRef]
- Gupta, K.J.; Kolbert, Z.; Durner, J.; Lindermayr, C.; Corpas, F.J.; Brouquisse, R.; Barroso, J.B.; Umbreen, S.; Palma, J.M.; Hancock, J.T. Regulating the regulator: Nitric oxide control of post-translational modifications. New Phytol. 2020, 227, 1319–1325. [Google Scholar] [CrossRef]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Skelly, M.J.; Malik, S.I.; Le Bihan, T.; Bo, Y.; Jiang, J.; Spoel, S.H.; Loake, G.J. A role for S-nitrosylation of the SUMO-conjugating enzyme SCE1 in plant immunity. Proc. Natl. Acad. Sci. USA 2019, 116, 17090–17095. [Google Scholar] [CrossRef] [Green Version]
- Del Castello, F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The era of nitric oxide in plant biology: Twenty years tying up loose ends. Nitric Oxide 2019, 85, 17–27. [Google Scholar] [CrossRef]
- Besson-Bard, A.; Courtois, C.; Gauthier, A.; Dahan, J.; Dobrowolska, G.; Jeandroz, S.; Pugin, A.; Wendehenne, D. Nitric oxide in plants: Production and cross-talk with Ca2+ signaling. Mol. Plant 2008, 1, 218–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawer, I.; Bucholc, M.; Astier, J.; Anielska-Mazur, A.; Dahan, J.; Kulik, A.; Wysłouch-Cieszynska, A.; Zaręba-Kozioł, M.; Krzywinska, E.; Dadlez, M. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 2010, 429, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, T.; Vandelle, E.; Bellin, D.; Kleinfelder-Fontanesi, K.; Huang, J.J.; Chen, J.; Digby, A.M.; Delledonne, M. Nitric oxide produced during the hypersensitive response modulates the plant signaling network and inhibits the pathogen’s virulence machinery. Nitric Oxide 2012, 27, S9. [Google Scholar] [CrossRef]
- Nakamura, T.; Lipton, S.A. Nitric oxide-dependent protein post-translational modifications impair mitochondrial function and metabolism to contribute to neurodegenerative diseases. Antioxid. Redox Signal. 2020, 32, 817–833. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Mengel, A.; Ageeva, A.; Georgii, E.; Bernhardt, J.; Wu, K.; Durner, J.; Lindermayr, C. Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 2017, 173, 1434–1452. [Google Scholar] [CrossRef] [Green Version]
- Ou, X.; Zhuang, T.; Yin, W.; Miao, Y.; Wang, B.; Zhang, Y.; Lin, X.; Xu, C.; von Wettstein, D.; Rustgi, S. DNA methylation changes induced in rice by exposure to high concentrations of the nitric oxide modulator, sodium nitroprusside. Plant Mol. Biol. Report. 2015, 33, 1428–1440. [Google Scholar] [CrossRef]
| Plant | ROS/RNS | Effects | References |
|---|---|---|---|
| Sand pear | H2O2 | HR-mediated cell death | [80] |
| Arabidopsis | H2O2 | Programmed cell death (PCD) | [81] |
| Nicotiana benthamiana | SOA and H2O2 | Replication of two unrelated RNA viruses | [63] |
| Arabidopsis | ROS- and JA-dependent process | Lignin biosynthesis | [64] |
| Arabidopsis | Oxidative burst | PTI | [65] |
| Arabidopsis | RBOHD | Plant immunity | [66] |
| Tobacco | H2O2 | PCD and HR | [67,68] |
| Medicago truncatula | ROS | Symbiosis | [69] |
| Phaseolus vulgaris | ROS | Symbiosis | [70] |
| Castanea sativa | ROS | Symbiosis | [71] |
| Arabidopsis | NO | Plant immunity | [8,37,72] |
| Arabidopsis | NO | Plant immunity, growth, and many more | [5,6,57] |
| Arabidopsis | NO | Plant immunity | [37] |
| Rice | NO | Resistance to viral infection | [73] |
| Tobaco | NO | Resistance against tobacco mosaic virus | [82] |
| Wheat | SNO | Resistance against powdery mildew invasion | [74] |
| Medicago truncatula | NO | Symbiosis | [75] |
| Lotus japonicus | NO | Symbiosis | [76] |
| Medicago truncatula | NO | Symbiosis | [77] |
| Legumes | NO | Symbiosis | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions. Antioxidants 2023, 12, 268. https://doi.org/10.3390/antiox12020268
Khan M, Ali S, Al Azzawi TNI, Saqib S, Ullah F, Ayaz A, Zaman W. The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions. Antioxidants. 2023; 12(2):268. https://doi.org/10.3390/antiox12020268
Chicago/Turabian StyleKhan, Murtaza, Sajid Ali, Tiba Nazar Ibrahim Al Azzawi, Saddam Saqib, Fazal Ullah, Asma Ayaz, and Wajid Zaman. 2023. "The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions" Antioxidants 12, no. 2: 268. https://doi.org/10.3390/antiox12020268
APA StyleKhan, M., Ali, S., Al Azzawi, T. N. I., Saqib, S., Ullah, F., Ayaz, A., & Zaman, W. (2023). The Key Roles of ROS and RNS as a Signaling Molecule in Plant–Microbe Interactions. Antioxidants, 12(2), 268. https://doi.org/10.3390/antiox12020268