Identifying Redox-Sensitive Cysteine Residues in Mitochondria
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological and Chemical Materials
2.2. Mammalian Cell Culture
2.3. Isolation of Mitochondria
2.4. Western Blot Analysis of GAPDH, CALR, Histone H3 and ATPIF1
2.5. Treatment of HEK293T Cells with Antimycin A
2.6. Mass Spectrometry Sample Preparation
2.6.1. isoTOP-ABPP Analysis
2.6.2. OxICAT Studies and Cysteine Oxidation Analysis
2.7. Tandem MS Analysis
2.8. LC/LC-MS/MS Data Processing
2.8.1. MS Data Analysis: isoTOP-ABPP for Peroxide Treatment
2.8.2. MS Data Analysis: isoTOP-ABPP for AMA Treatment
2.8.3. MS Data Analysis: OxICAT
2.9. Statistical Analysis of Overrepresented GO Biological Processes Using Panther
3. Results
3.1. Differential Centrifugation to Increase Coverage of Mitochondrial Cysteines
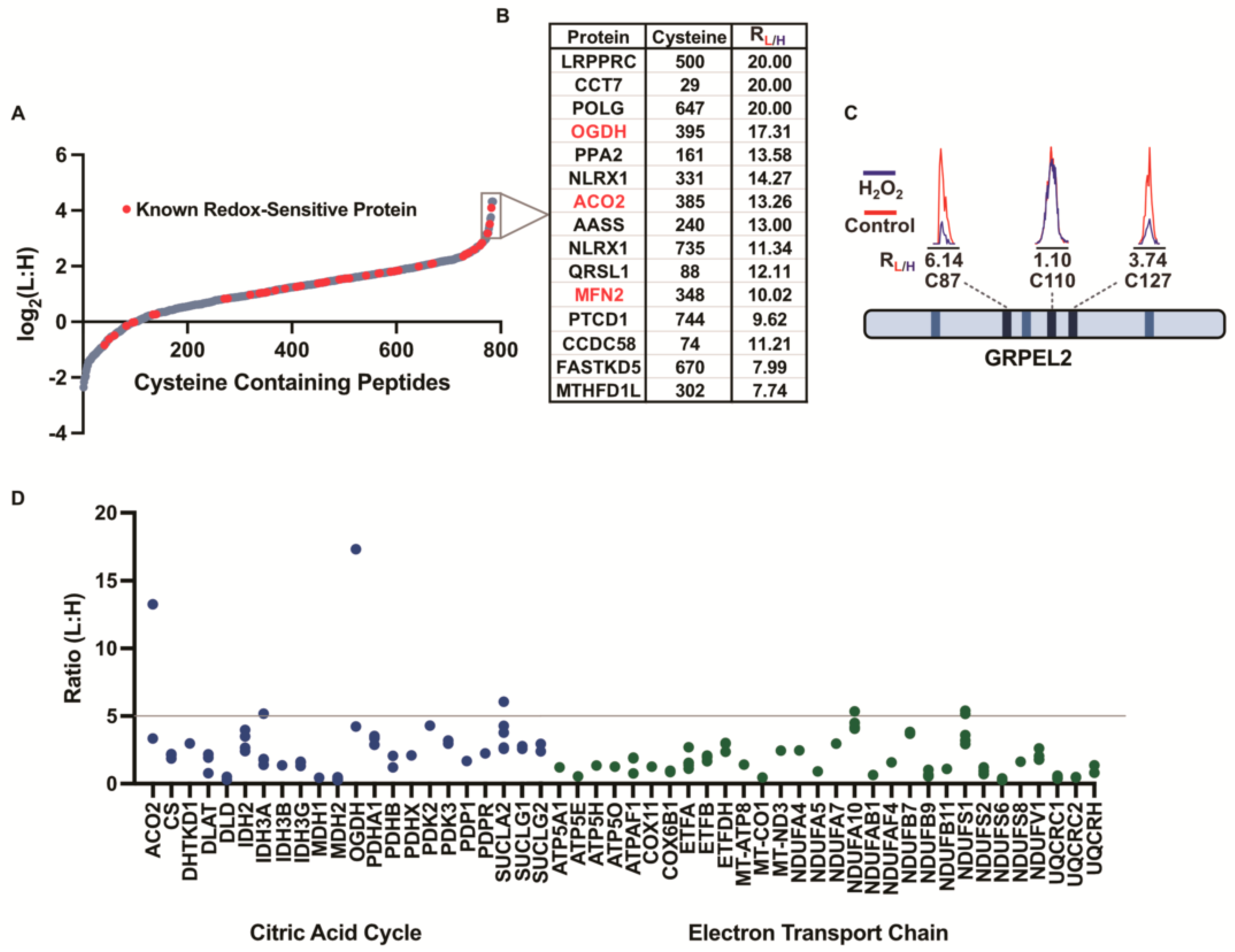
3.2. Monitoring the Redox Sensitivity of Mitochondrial Cysteines
3.3. Cysteine-Reactivity Changes of Known Redox-Sensitive Proteins
3.4. Levels of Cysteine Oxidation within Key Mitochondrial Pathways
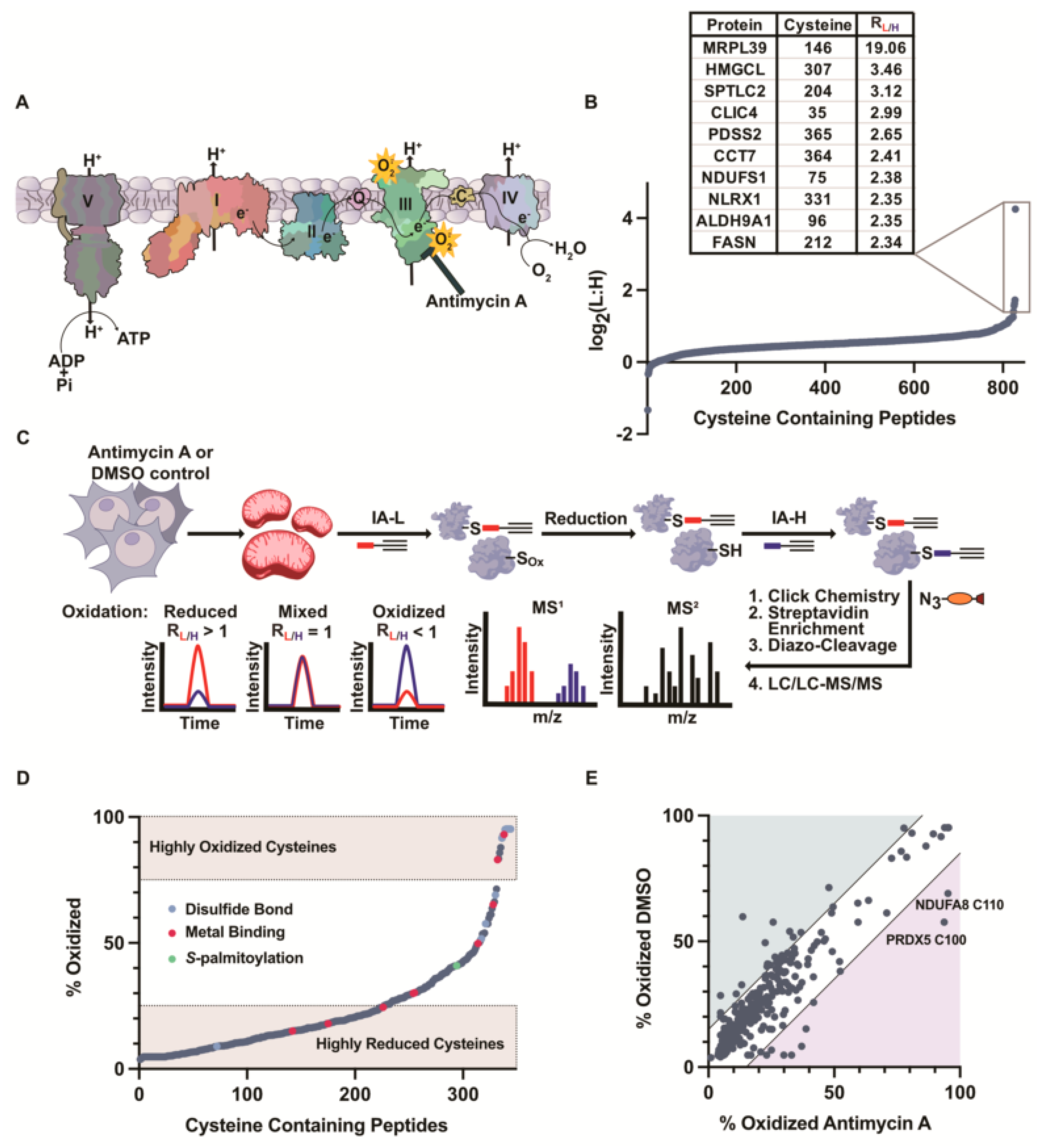
3.5. Inhibition of the Electron Transport Chain Induces ROS Generation and Cysteine Oxidation
3.6. Monitoring Mitochondrial Cysteine Oxidation with OxICAT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FFriedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Riemer, J.; Bulleid, N.; Herrmann, J.M. Disulfide Formation in the ER and Mitochondria: Two Solutions to a Common Process. Science 2009, 324, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Hell, K. The Erv1–Mia40 disulfide relay system in the intermembrane space of mitochondria. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2008, 1783, 601–609. [Google Scholar] [CrossRef]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Wang, C.; Youle, R.J. The Role of Mitochondria in Apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nature 2018, 20, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Lane, N.; Martin, W. The energetics of genome complexity. Nature 2010, 467, 929–934. [Google Scholar] [CrossRef]
- Ott, M.; Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria, oxidative stress and cell death. Apoptosis 2007, 12, 913–922. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Bechtel, T.J.; Weerapana, E. From structure to redox: The diverse functional roles of disulfides and implications in disease. Proteomics 2017, 17, 1600391. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.-G. Crosstalk of reactive oxygen species and NF-kappaκB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Canet-Avilés, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef]
- Aiken, C.T.; Kaake, R.M.; Wang, X.; Huang, L. Oxidative Stress-Mediated Regulation of Proteasome Complexes. Mol. Cell. Proteom. 2011, 10, R110.006924. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.A.; Chandel, N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 2012, 48, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Groitl, B.; Jakob, U. Thiol-based redox switches. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2014, 1844, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Schaack, J.; Patel, M. Oxidative Inactivation of Mitochondrial Aconitase Results in Iron and H2O2-Mediated Neurotoxicity in Rat Primary Mesencephalic Cultures. PLoS ONE 2009, 4, e7095. [Google Scholar] [CrossRef]
- Lushchak, O.V.; Piroddi, M.; Galli, F.; Lushchak, V.I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep. 2014, 19, 8–15. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Methner, C.; Nadtochiy, S.M.; Logan, A.; Pell, V.R.; Ding, S.; James, A.M.; Cochemé, H.M.; Reinhold, J.; Lilley, K.S.; et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat. Med. 2013, 19, 753–759. [Google Scholar] [CrossRef]
- Burger, N.; James, A.M.; Mulvey, J.F.; Hoogewijs, K.; Ding, S.; Fearnley, I.M.; Loureiro-López, M.; Norman, A.A.; Arndt, S.; Mottahedin, A.; et al. ND3 Cys39 in complex I is exposed during mitochondrial respiration. Cell Chem. Biol. 2021, 29, 636–649.e14. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Kazak, L.; Jedrychowski, M.P.; Lu, G.Z.; Erickson, B.K.; Szpyt, J.; Pierce, K.A.; Laznik-Bogoslavski, D.; Vetrivelan, R.; Clish, C.B.; et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 2016, 532, 112–116. [Google Scholar] [CrossRef]
- Ježek, P.; Jabůrek, M.; Porter, R.K. Uncoupling mechanism and redox regulation of mitochondrial uncoupling protein 1 (UCP1). Biochim. Biophys. Acta (BBA)—Bioenerg. 2019, 1860, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Chandel, N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Stimpson, S.; Fernandez-Bueno, G.A.; Mathews, C.E. Mitochondrial Reactive Oxygen Species and Type 1 Diabetes. Antioxid. Redox Signal. 2018, 29, 1361–1372. [Google Scholar] [CrossRef] [PubMed]
- Kaludercic, N.; Di Lisa, F. Mitochondrial ROS Formation in the Pathogenesis of Diabetic Cardiomyopathy. Front. Cardiovasc. Med. 2020, 7, 12. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Moris, D.; Spartalis, M.; Spartalis, E.; Karachaliou, G.-S.; Karaolanis, G.I.; Tsourouflis, G.; Tsilimigras, D.I.; Tzatzaki, E.; Theocharis, S. The role of reactive oxygen species in the pathophysiology of cardiovascular diseases and the clinical significance of myocardial redox. Ann. Transl. Med. 2017, 5, 326. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Frezza, C.; Cipolat, S.; Scorrano, L. Organelle isolation: Functional mitochondria from mouse liver, muscle and cultured filroblasts. Nat. Protoc. 2007, 2, 287–295. [Google Scholar] [CrossRef]
- Shi, Y.; Fu, L.; Yang, J.; Carroll, K.S. Wittig reagents for chemoselective sulfenic acid ligation enables global site stoichiometry analysis and redox-controlled mitochondrial targeting. Nat. Chem. 2021, 13, 1140–1150. [Google Scholar] [CrossRef]
- Yasueda, Y.; Tamura, T.; Fujisawa, A.; Kuwata, K.; Tsukiji, S.; Kiyonaka, S.; Hamachi, I. A Set of Organelle-Localizable Reactive Molecules for Mitochondrial Chemical Proteomics in Living Cells and Brain Tissues. J. Am. Chem. Soc. 2016, 138, 7592–7602. [Google Scholar] [CrossRef]
- Alcock, L.J.; Oliveira, B.L.; Deery, M.J.; Pukala, T.L.; Perkins, M.V.; Bernardes, G.J.L.; Chalker, J.M. Norbornene Probes for the Detection of Cysteine Sulfenic Acid in Cells. ACS Chem. Biol. 2019, 14, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Fu, L.; Jung, Y.; Conte, M.L.; Lawson, J.R.; Lowther, W.T.; Sun, R.; Liu, K.; Yang, J.; Carroll, K.S. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Fu, L.; Liu, K.; Tian, C.; Wu, Z.; Jung, Y.; Ferreira, R.B.; Carroll, K.S.; Blackwell, T.K.; Yang, J. Global profiling of distinct cysteine redox forms reveals wide-ranging redox regulation in C. elegans. Nat. Commun. 2021, 12, 1415. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Carroll, K.S. Chemical Dissection of an Essential Redox Switch in Yeast. Chem. Biol. 2009, 16, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Hung, V.; Udeshi, N.D.; Lam, S.S.-M.; Loh, K.H.; Cox, K.J.; Pedram, K.; Carr, S.A.; Ting, Y. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016, 11, 456–475. [Google Scholar] [CrossRef]
- Hung, V.; Zou, P.; Rhee, H.-W.; Udeshi, N.D.; Cracan, V.; Svinkina, T.; Carr, S.A.; Mootha, V.K.; Ting, A.Y. Proteomic Mapping of the Human Mitochondrial Intermembrane Space in Live Cells via Ratiometric APEX Tagging. Mol. Cell 2014, 55, 332–341. [Google Scholar] [CrossRef]
- Rhee, H.-W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Zeng, K.; Qiang, J.; Cao, Y.; Li, Y.; Fang, Y.; Zhang, Y.; Chen, Y. Selective Mitochondrial Protein Labeling Enabled by Biocompatible Photocatalytic Reactions inside Live Cells. JACS Au 2021, 1, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Bak, D.W.; Pizzagalli, M.D.; Weerapana, E. Identifying Functional Cysteine Residues in the Mitochondria. ACS Chem. Biol. 2017, 12, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Leichert, L.I.; Gehrke, F.; Gudiseva, H.V.; Blackwell, T.; Ilbert, M.; Walker, A.K.; Strahler, J.R.; Andrews, P.C.; Jakob, U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 8197–8202. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, T.J.; Li, C.; Kisty, E.A.; Maurais, A.J.; Weerapana, E. Profiling Cysteine Reactivity and Oxidation in the Endoplasmic Reticulum. ACS Chem. Biol. 2020, 15, 543–553. [Google Scholar] [CrossRef]
- Topf, U.; Suppanz, I.; Samluk, L.; Wrobel, L.; Böser, A.; Sakowska, P.; Knapp, B.; Pietrzyk, M.K.; Chacinska, A.; Warscheid, B. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018, 9, 324. [Google Scholar] [CrossRef]
- Menger, K.E.; James, A.M.; Cochemé, H.M.; Harbour, M.E.; Chouchani, E.T.; Ding, S.; Fearnley, I.M.; Partridge, L.; Murphy, M.P. Fasting, but Not Aging, Dramatically Alters the Redox Status of Cysteine Residues on Proteins in Drosophila melanogaster. Cell Rep. 2015, 11, 1856–1865. [Google Scholar] [CrossRef]
- Abo, M.; Li, C.; Weerapana, E. Isotopically-Labeled Iodoacetamide-Alkyne Probes for Quantitative Cysteine-Reactivity Profiling. Mol. Pharm. 2017, 15, 743–749. [Google Scholar] [CrossRef]
- Weerapana, E.; Wang, C.; Simon, G.M.; Richter, F.; Khare, S.; Dillon, M.B.D.; Bachovchin, D.A.; Mowen, K.; Baker, D.; Cravatt, B.F. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Tabb, D.L.; McDonald, W.H.; Yates, J.R.R. DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics. J. Proteome Res. 2002, 1, 21–26. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, D1251–D1257. [Google Scholar] [CrossRef] [PubMed]
- Filadi, R.; Pendin, D.; Pizzo, P. Mitofusin 2: From functions to disease. Cell Death Dis. 2018, 9, 330. [Google Scholar] [CrossRef] [PubMed]
- Thaher, O.; Wolf, C.; Dey, P.N.; Pouya, A.; Wüllner, V.; Tenzer, S.; Methner, A. The thiol switch C684 in Mitofusin-2 mediates redox-induced alterations of mitochondrial shape and respiration. Neurochem. Int. 2018, 117, 167–173. [Google Scholar] [CrossRef]
- Wolf, C.; López Del Amo, V.; Arndt, S.; Bueno, D.; Tenzer, S.; Hanschmann, E.-M.; Berndt, C.; Methner, A. Redox Modifications of Proteins of the Mitochondrial Fusion and Fission Machinery. Cells 2020, 9, 815. [Google Scholar] [CrossRef]
- McLain, A.L.; Szweda, P.A.; Szweda, L.I. α-Ketoglutarate dehydrogenase: A mitochondrial redox sensor. Free. Radic. Res. 2010, 45, 29–36. [Google Scholar] [CrossRef]
- Mailloux, R.J.; Ayre, D.C.; Christian, S.L. Induction of mitochondrial reactive oxygen species production by GSH mediated S-glutathionylation of 2-oxoglutarate dehydrogenase. Redox Biol. 2016, 8, 285–297. [Google Scholar] [CrossRef]
- Murray, C.I.; Kane, L.A.; Uhrigshardt, H.; Wang, S.-B.; Van Eyk, J.E. Site-Mapping of In Vitro S-nitrosation in Cardiac Mitochondria: Implications for Cardioprotection. Mol. Cell. Proteom. 2011, 10, M110.004721. [Google Scholar] [CrossRef]
- Konovalova, S.; Liu, X.; Manjunath, P.; Baral, S.; Neupane, N.; Hilander, T.; Yang, Y.; Balboa, D.; Terzioglu, M.; Euro, L.; et al. Redox regulation of GRPEL2 nucleotide exchange factor for mitochondrial HSP70 chaperone. Redox Biol. 2018, 19, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Jedrychowski, M.P.; Schweppe, D.K.; Huttlin, E.L.; Yu, Q.; Heppner, D.E.; Li, J.; Long, J.; Mills, E.L.; Szpyt, J.; et al. A Quantitative Tissue-Specific Landscape of Protein Redox Regulation during Aging. Cell 2020, 180, 968–983.e24. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, C.L.; Gerencser, A.A.; Treberg, J.R.; Brand, M.D. The Mechanism of Superoxide Production by the Antimycin-inhibited Mitochondrial Q-cycle. J. Biol. Chem. 2011, 286, 31361–31372. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.; Tu, Y.-T.; Amunts, A.; Fontanesi, F.; Barrientos, A. Mitochondrial ribosome assembly in health and disease. Cell Cycle 2015, 14, 2226–2250. [Google Scholar] [CrossRef] [PubMed]
- Bonn, F.; Tatsuta, T.; Petrungaro, C.; Riemer, J.; Langer, T. Presequence-dependent folding ensures MrpL32 processing by them-AAA protease in mitochondria. EMBO J. 2011, 30, 2545–2556. [Google Scholar] [CrossRef]
- Nagai-Singer, M.A.; Morrison, H.A.; Allen, I.C. NLRX1 Is a Multifaceted and Enigmatic Regulator of Immune System Function. Front. Immunol. 2019, 10, 2419. [Google Scholar] [CrossRef]
- De Simoni, S.; Linard, D.; Hermans, E.; Knoops, B.; Goemaere, J. Mitochondrial peroxiredoxin-5 as potential modulator of mitochondria-ER crosstalk in MPP+-induced cell death. J. Neurochem. 2013, 125, 473–485. [Google Scholar] [CrossRef]
- Seo, M.S.; Kang, S.W.; Kim, K.; Baines, I.C.; Lee, T.H.; Rhee, S.G. Identification of a New Type of Mammalian Peroxiredoxin That Forms an Intramolecular Disulfide as a Reaction Intermediate. J. Biol. Chem. 2000, 275, 20346–20354. [Google Scholar] [CrossRef]
- Smeets, A.; Marchand, C.; Linard, D.; Knoops, B.; Declercq, J.-P. The crystal structures of oxidized forms of human peroxiredoxin 5 with an intramolecular disulfide bond confirm the proposed enzymatic mechanism for atypical 2-Cys peroxiredoxins. Arch. Biochem. Biophys. 2008, 477, 98–104. [Google Scholar] [CrossRef]
- Szklarczyk, R.; Wanschers, B.F.; Nabuurs, S.B.; Nouws, J.; Nijtmans, L.G.; Huynen, M.A. NDUFB7 and NDUFA8 are located at the intermembrane surface of complex I. FEBS Lett. 2011, 585, 737–743. [Google Scholar] [CrossRef]
- Fischer, M.; Horn, S.; Belkacemi, A.; Kojer, K.; Petrungaro, C.; Habich, M.; Ali, M.; Küttner, V.; Bien, M.; Kauff, F.; et al. Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol. Biol. Cell 2013, 24, 2160–2170. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kisty, E.A.; Saart, E.C.; Weerapana, E. Identifying Redox-Sensitive Cysteine Residues in Mitochondria. Antioxidants 2023, 12, 992. https://doi.org/10.3390/antiox12050992
Kisty EA, Saart EC, Weerapana E. Identifying Redox-Sensitive Cysteine Residues in Mitochondria. Antioxidants. 2023; 12(5):992. https://doi.org/10.3390/antiox12050992
Chicago/Turabian StyleKisty, Eleni A., Emma C. Saart, and Eranthie Weerapana. 2023. "Identifying Redox-Sensitive Cysteine Residues in Mitochondria" Antioxidants 12, no. 5: 992. https://doi.org/10.3390/antiox12050992
APA StyleKisty, E. A., Saart, E. C., & Weerapana, E. (2023). Identifying Redox-Sensitive Cysteine Residues in Mitochondria. Antioxidants, 12(5), 992. https://doi.org/10.3390/antiox12050992