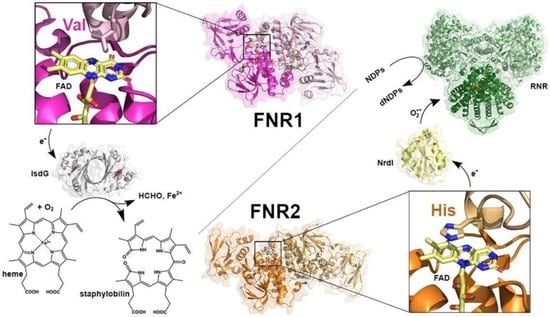
Functional Diversity of Homologous Oxidoreductases—Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Proteins
2.2. Reconstitution of IsdG with Hemin
2.3. Protein Crystallization
2.4. Crystal Data Collection and Processing
2.4.1. FNR2H326V
2.4.2. FNR1V329H
2.4.3. IsdGheme and IsdGapo
2.5. Bioinformatics Analysis—Sequence Similarity Networks
2.6. Investigation of Potential Fur Control in Bc
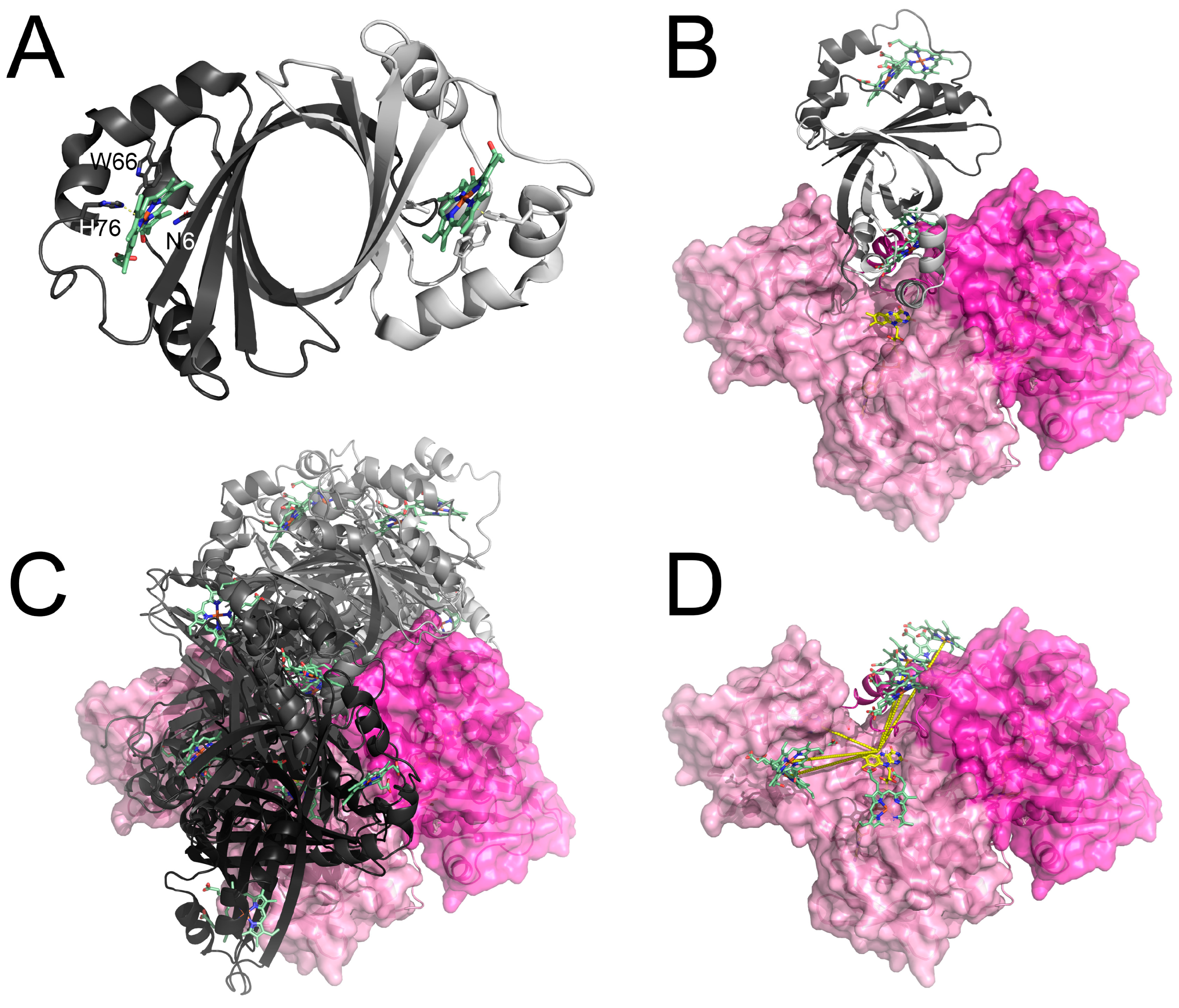
2.7. Protein–Protein Docking of FNR1-IsdG
2.8. Activity Measurements
2.8.1. Steady-State Kinetics of NrdI and Fld1 Reduction by FNR2H326V, and NrdI and Fld2 Reduction by FNR1V329H
2.8.2. Reduction of IsdG and Heme Degradation by FNR1, FNR2, Bdr, FNR1V329H, and FNR2H326V
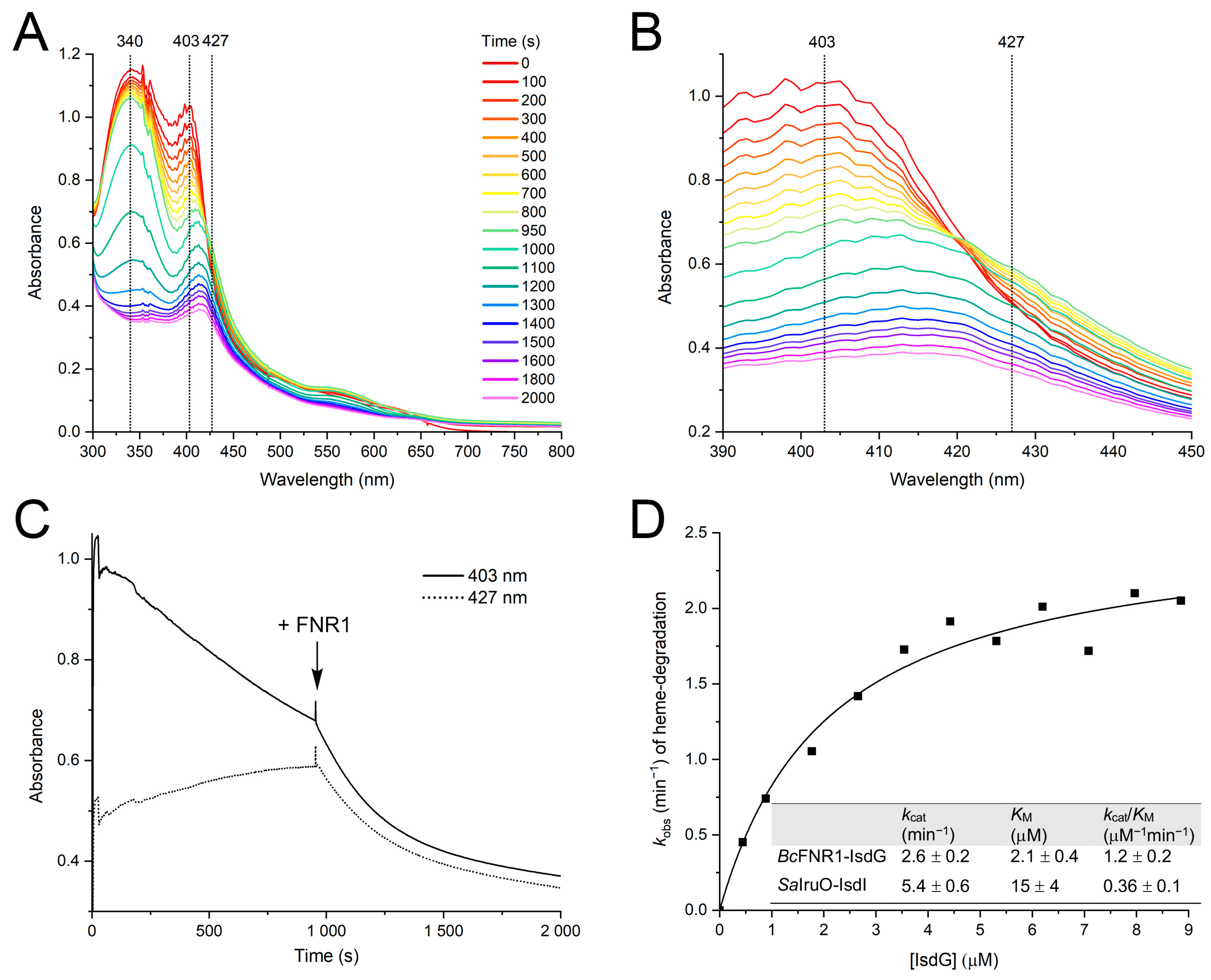
2.8.3. Steady-State Kinetics of IsdG Reduction and Heme Degradation by FNR1
3. Results and Discussion
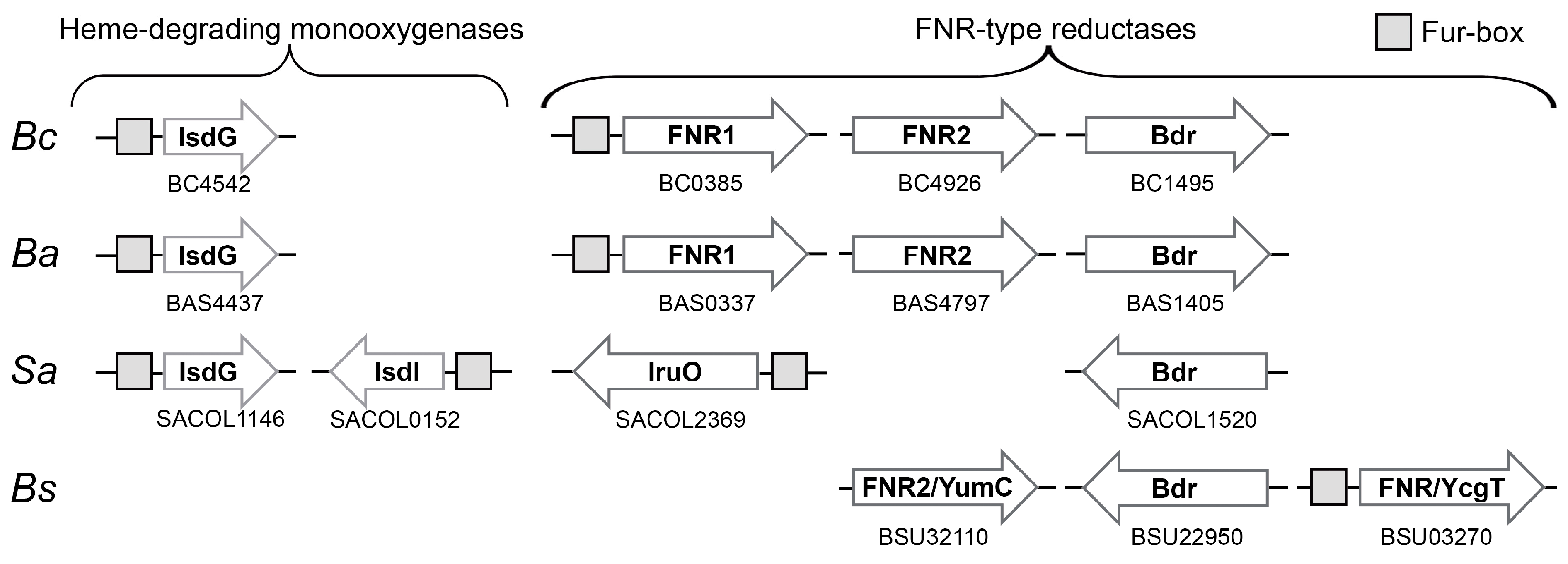
3.1. Comparison of TrxR-Like FNRs in Bc and Related Species
3.2. Bc FNR1 Reduces IsdG
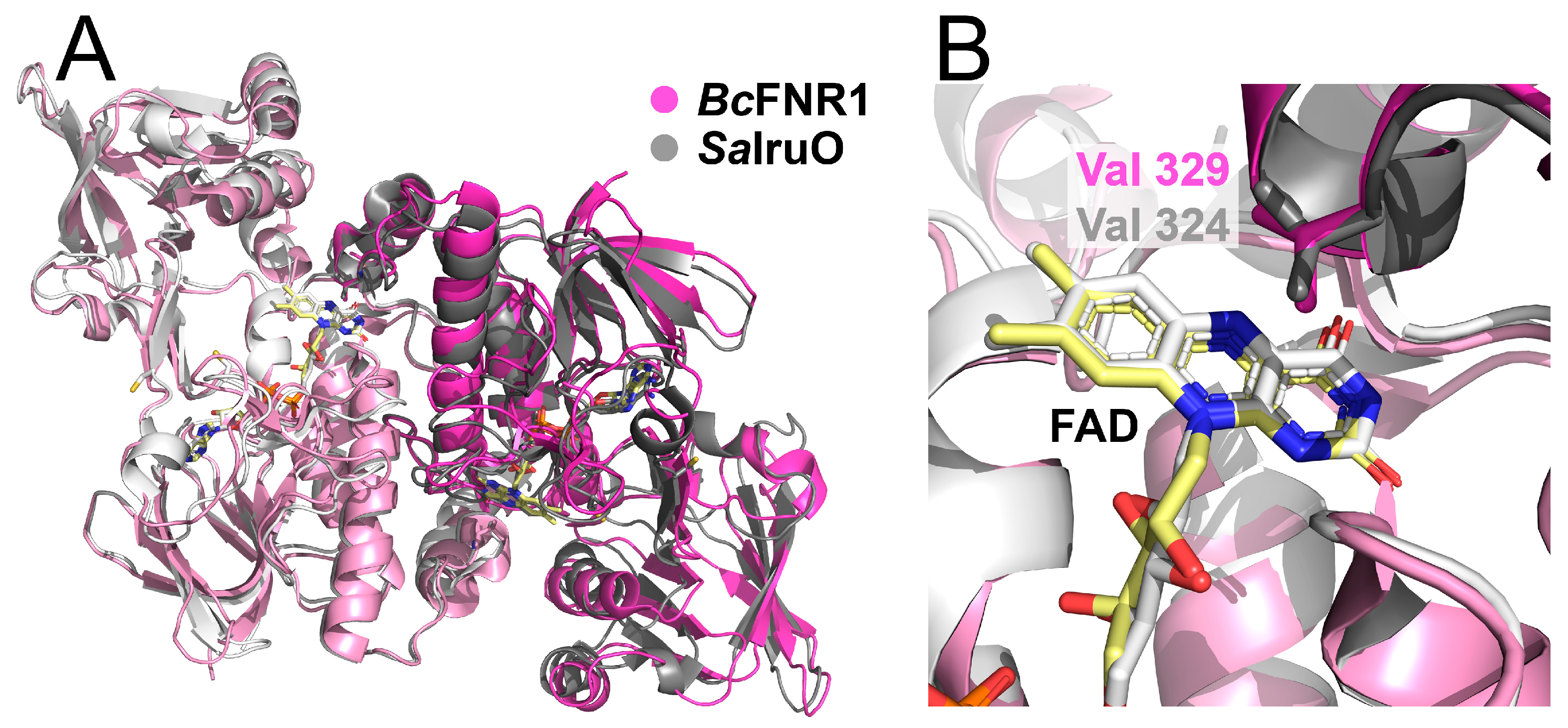
3.3. Structure of IsdG and the Putative IsdG-FNR Complex
3.4. Difference in Regulation of FNR1 and FNR2-Fur Control of Reductases Involved in Iron Acquisition in Firmicutes
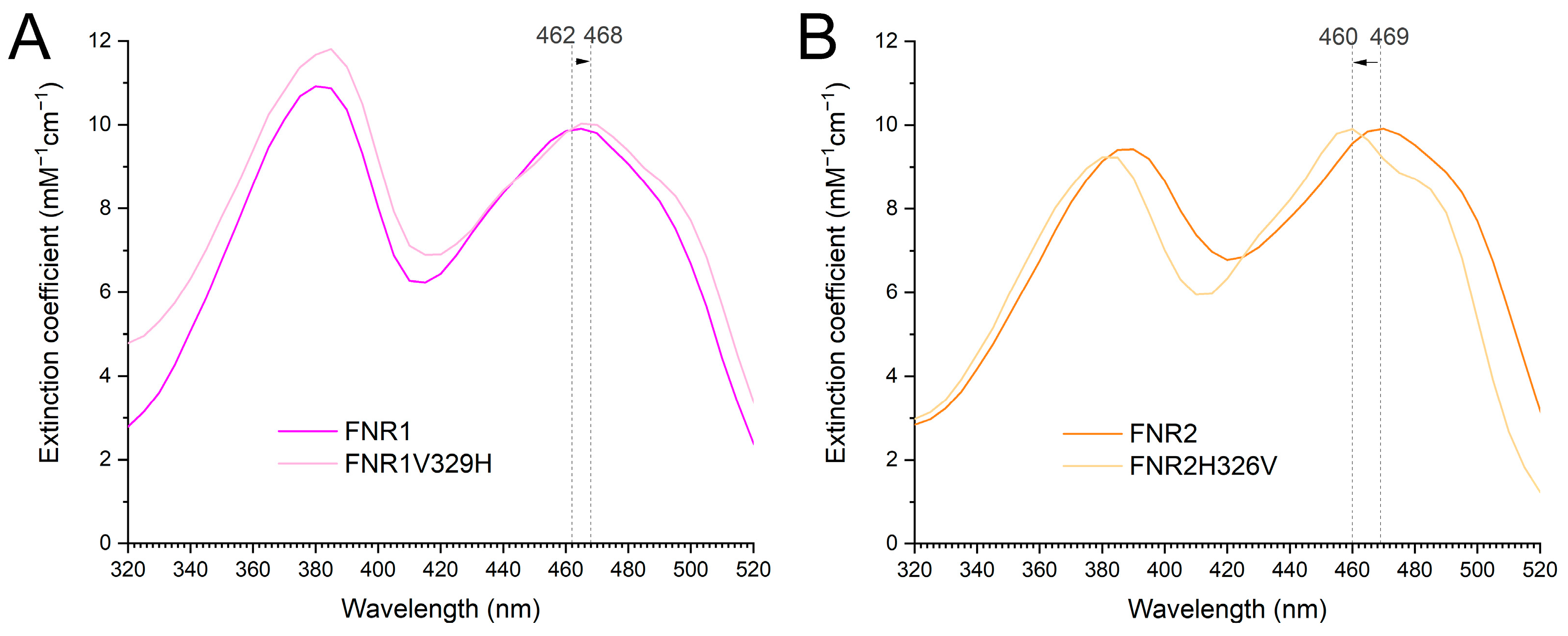
3.5. The Effect of the Conserved Val and His in FNR1 and FNR2 on Reduction Rates
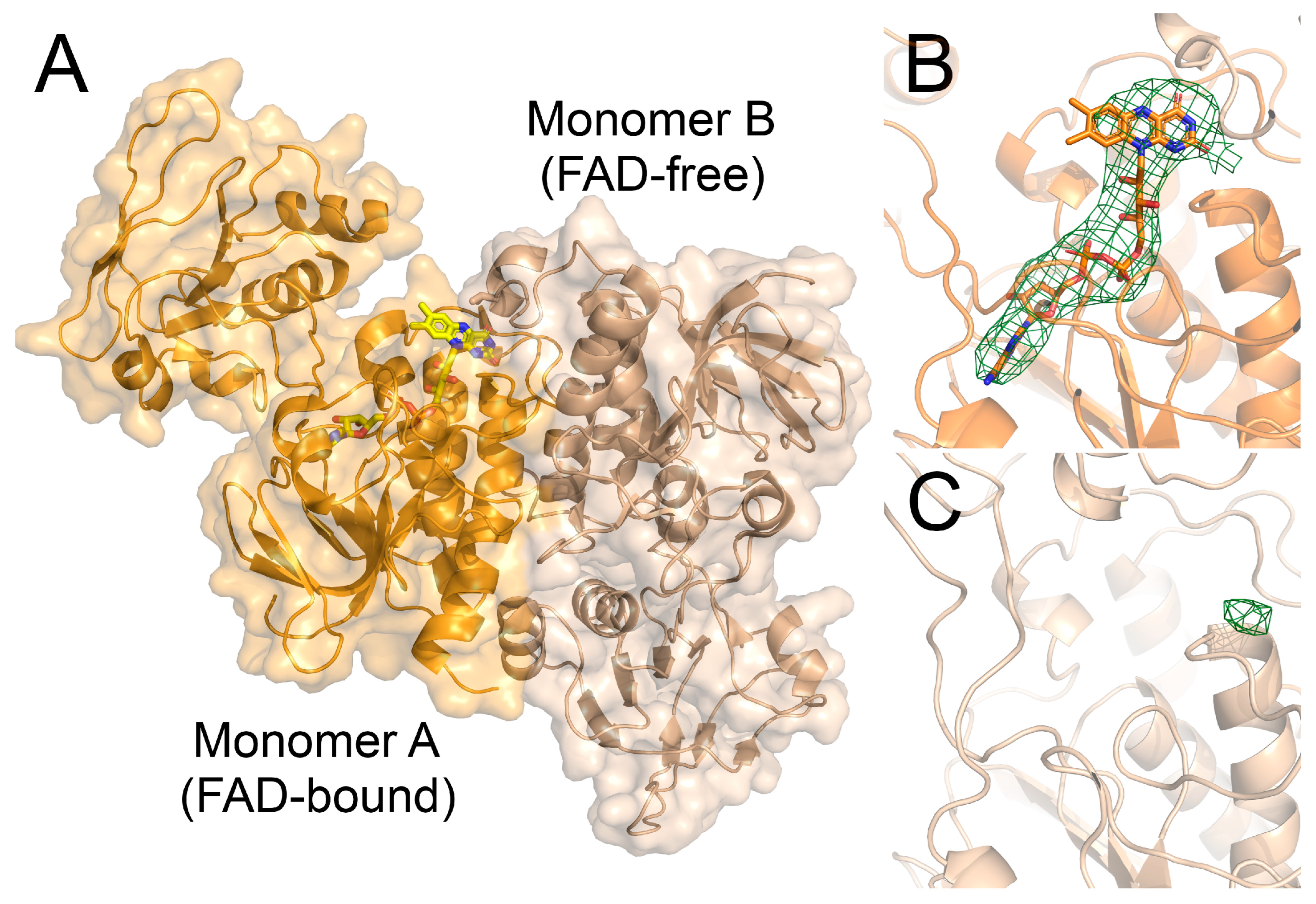
3.6. The Crystal Structure of the FNR2H326V Mutant Can Explain the Lowered Reduction Rate towards Flds
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Macheroux, P.; Kappes, B.; Ealick, S.E. Flavogenomics—A genomic and structural view of flavin-dependent proteins. FEBS J. 2011, 278, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.S.; Kelly, C.L.; Mordaka, P.M.; Heap, J.T. Review of NAD(P)H-dependent oxidoreductases: Properties, engineering and application. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Hammerstad, M.; Hersleth, H.P. Overview of structurally homologous flavoprotein oxidoreductases containing the low Mr thioredoxin reductase-like fold—A functionally diverse group. Arch. Biochem. Biophys. 2021, 702, 108826. [Google Scholar] [CrossRef] [PubMed]
- Cotruvo, J.A.; Stubbe, J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 2010, 49, 1297–1309. [Google Scholar] [CrossRef] [Green Version]
- Cotruvo, J.A.; Stubbe, J. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 2011, 50, 1672–1681. [Google Scholar] [CrossRef] [Green Version]
- Boal, A.K.; Cotruvo, J.A.; Stubbe, J.; Rosenzweig, A.C. Structural basis for activation of class Ib ribonucleotide reductase. Science 2010, 329, 1526–1530. [Google Scholar] [CrossRef] [Green Version]
- Lofstad, M.; Gudim, I.; Hammerstad, M.; Røhr, Å.K.; Hersleth, H.-P. Activation of the class Ib ribonucleotide reductase by a flavodoxin reductase in Bacillus cereus. Biochemistry 2016, 55, 4998–5001. [Google Scholar] [CrossRef] [Green Version]
- Gudim, I.; Hammerstad, M.; Lofstad, M.; Hersleth, H.-P. The characterization of different flavodoxin reductase-flavodoxin (FNR-Fld) interactions reveals an efficient FNR-Fld redox pair and identifies a novel FNR subclass. Biochemistry 2018, 57, 5427–5436. [Google Scholar] [CrossRef] [Green Version]
- Hammerstad, M.; Gudim, I.; Hersleth, H.-P. The crystal structures of bacillithiol disulfide reductase Bdr (YpdA) provide structural and functional insight into a new type of FAD-containing NADPH-dependent oxidoreductase. Biochemistry 2020, 59, 4793–4798. [Google Scholar] [CrossRef]
- Linzner, N.; Loi, V.V.; Fritsch, V.N.; Tung, Q.N.; Stenzel, S.; Wirtz, M.; Hell, R.; Hamilton, C.J.; Tedin, K.; Fulde, M.; et al. Staphylococcus aureus uses the bacilliredoxin (BrxAB)/bacillithiol disulfide reductase (YpdA) redox pathway to defend against oxidative stress under infections. Front. Microbiol. 2019, 10, 1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraki, N.; Seo, D.; Shiba, T.; Sakurai, T.; Kurisu, G. Asymmetric dimeric structure of ferredoxin-NAD(P)(+) oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: Implications for binding ferredoxin and NADP(+). J. Mol. Biol. 2010, 401, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Komori, H.; Seo, D.; Sakurai, T.; Higuchi, Y. Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity. Protein Sci. 2010, 19, 2279–2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loutet, S.A.; Kobylarz, M.J.; Chau, C.H.T.; Murphy, M.E.P. IruO Is a reductase for heme degradation by IsdI and IsdG proteins in Staphylococcus aureus. J. Biol. Chem. 2013, 288, 25749–25759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobylarz, M.J.; Heieis, G.A.; Loutet, S.A.; Murphy, M.E.P. Iron uptake oxidoreductase (IruO) uses a flavin adenine dinucleotide semiquinone intermediate for iron-siderophore reduction. ACS Chem. Biol. 2017, 12, 1778–1786. [Google Scholar] [CrossRef] [PubMed]
- Reniere, M.L.; Ukpabi, G.N.; Harry, S.R.; Stec, D.F.; Krull, R.; Wright, D.W.; Bachmann, B.O.; Murphy, M.E.; Skaar, E.P. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 2010, 75, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Gudim, I.; Lofstad, M.; van Beek, W.; Hersleth, H.P. High-resolution crystal structures reveal a mixture of conformers of the Gly61-Asp62 peptide bond in an oxidized flavodoxin from Bacillus cereus. Protein Sci. 2018, 27, 1439–1449. [Google Scholar] [CrossRef] [Green Version]
- Hammerstad, M.; Røhr, Å.K.; Hersleth, H.P. A Research-inspired biochemistry laboratory module-combining expression, purification, crystallization, structure-solving, and characterization of a flavodoxin-like protein. Biochem. Mol. Biol. Educ. 2019, 47, 318–332. [Google Scholar] [CrossRef] [Green Version]
- Skråmo, S.; Hersleth, H.P.; Hammerstad, M.; Andersson, K.K.; Røhr, Å.K. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of a ferredoxin/flavodoxin-NADP(H) oxidoreductase (Bc0385) from Bacillus cereus. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 777–780. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Q.; Lawson, R.J.; Buddha, M.R.; Wei, C.C.; Crane, B.R.; Munro, A.W.; Stuehr, D.J. Bacterial flavodoxins support nitric oxide production by Bacillus subtilis nitric-oxide synthase. J. Biol. Chem. 2007, 282, 2196–2202. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaar, E.P.; Gaspar, A.H.; Schneewind, O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J. Bacteriol. 2006, 188, 1071–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vonrhein, C.; Flensburg, C.; Keller, P.; Sharff, A.; Smart, O.; Paciorek, W.; Womack, T.; Bricogne, G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 293–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Joosten, R.P.; Long, F.; Murshudov, G.N.; Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 2014, 1, 213–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeldin, O.B.; Gerstel, M.; Garman, E.F. RADDOSE-3D: Time- and space-resolved modelling of dose in macromolecular crystallography. J. Appl. Crystallogr. 2013, 46, 1225–1230. [Google Scholar] [CrossRef]
- Zallot, R.; Oberg, N.; Gerlt, J.A. The EFI web resource for genomic enzymology tools: Leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry 2019, 58, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.Z.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Novichkov, P.S.; Kazakov, A.E.; Ravcheev, D.A.; Leyn, S.A.; Kovaleva, G.Y.; Sutormin, R.A.; Kazanov, M.D.; Riehl, W.; Arkin, A.P.; Dubchak, I.; et al. RegPrecise 3.0-A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 2013, 14, 745. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins 2017, 85, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Christoffer, C.; Chen, S.Y.; Bharadwaj, V.; Aderinwale, T.; Kumar, V.; Hormati, M.; Kihara, D. LZerD webserver for pairwise and multiple protein-protein docking. Nucleic Acids Res. 2021, 49, W359–W365. [Google Scholar] [CrossRef]
- Christoffer, C.; Bharadwaj, V.; Luu, R.; Kihara, D. LZerD protein-protein docking webserver enhanced with de novo structure prediction. Front. Mol. Biosci. 2021, 8, 724947I. [Google Scholar] [CrossRef]
- Wu, R.Y.; Skaar, E.P.; Zhang, R.G.; Joachimiak, G.; Gornicki, P.; Schneewind, O.; Joachimiak, A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J. Biol. Chem. 2005, 280, 2840–2846. [Google Scholar] [CrossRef] [Green Version]
- Schuelke-Sanchez, A.E.; Cornetta, A.R.; Kocian, T.A.J.; Conger, M.A.; Liptak, M.D. Ruffling is essential for Staphylococcus aureus IsdG-catalyzed degradation of heme to staphylobilin. J. Inorg. Biochem. 2022, 230, 111775. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Reniere, M.L.; Skaar, E.P.; Murphy, M.E.P. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 2008, 283, 30957–30963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meschi, F.; Wiertz, F.; Klauss, L.; Blok, A.; Ludwig, B.; Merli, A.; Heering, H.A.; Rossi, G.L.; Ubbink, M. Efficient electron transfer in a protein network lacking specific interactions. J. Am. Chem. Soc. 2011, 133, 16861–16867. [Google Scholar] [CrossRef]
- Pinochet-Barros, A.; Helmann, J.D. Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid. Redox Signal. 2018, 29, 1858–1871. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Rensing, C.; Helmann, J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017, 15, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baichoo, N.; Wang, T.; Ye, R.; Helmann, J.D. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 2002, 45, 1613–1629. [Google Scholar] [CrossRef] [Green Version]
- Baichoo, N.; Helmann, J.D. Recognition of DNA by Fur: A reinterpretation of the Fur box consensus sequence. J. Bacteriol. 2002, 184, 5826–5832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayrapetyan, H.; Siezen, R.; Abee, T.; Groot, M.N. Comparative genomics of iron-transporting systems in Bacillus cereus strains and impact of iron sources on growth and biofilm formation. Front. Microbiol. 2016, 7, 842. [Google Scholar] [CrossRef] [PubMed]
- Pi, H.L.; Helmann, J.D. Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2017, 114, 12785–12790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaar, E.P.; Gaspar, A.H.; Schneewind, O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 2004, 279, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Skaar, E.P.; Schneewind, O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: Stealing iron from heme. Microbes Infect. 2004, 6, 390–397. [Google Scholar] [CrossRef]
- Sevilla, E.; Bes, M.T.; Peleato, M.L.; Fillat, M.F. Fur-like proteins: Beyond the ferric uptake regulator (Fur) paralog. Arch. Biochem. Biophys. 2021, 701, 108770. [Google Scholar] [CrossRef]
- Seo, D.; Muraki, N.; Kurisu, G. Kinetic and structural insight into a role of the re-face Tyr328 residue of the homodimer type ferredoxin-NADP(+) oxidoreductase from Rhodopseudomonas palustris in the reaction with NADP(+)/NADPH. Biochim. Biophys. Acta Bioenergetics. 2020, 1861, 148140. [Google Scholar] [CrossRef]
- Bashir, Q.; Scanu, S.; Ubbink, M. Dynamics in electron transfer protein complexes. FEBS J. 2011, 278, 1391–1400. [Google Scholar] [CrossRef] [Green Version]
- Seo, D.; Asano, T. C-terminal residues of ferredoxin-NAD(P)(+) reductase from Chlorobaculum tepidum are responsible for reaction dynamics in the hydride transfer and redox equilibria with NADP(+)/NADPH. Photosynth. Res. 2018, 136, 275–290. [Google Scholar] [CrossRef] [Green Version]
- Seo, D.; Asano, T.; Komori, H.; Sakurai, T. Role of the C-terminal extension stacked on the re-face of the isoalloxazine ring moiety of the flavin adenine dinucleotide prosthetic group in ferredoxin-NADP(+) oxidoreductase from Bacillus subtilis. Plant Physiol. Biochem. 2014, 81, 143–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waksman, G.; Krishna, T.S.R.; Williams, C.H.; Kuriyan, J. Crystal structure of Escherichia-coli thioredoxin reductase refined at 2-Ångström resolution—Implications for a large conformational change during catalysis. J. Mol. Biol. 1994, 236, 800–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoor, M.; Gudim, I.; Hersleth, H.P.; Hammerstad, M. Thioredoxin reductase from Bacillus cereus exhibits distinct reduction and NADPH-binding properties. FEBS Open Bio 2021, 11, 3019–3031. [Google Scholar] [CrossRef] [PubMed]


| BcIsdGapo | BcIsdGheme | BcFNR1V329H | BcFNR2H326V | |
|---|---|---|---|---|
| Data collection | ||||
| X-ray source | MAXIV BioMAX | MAXIV BioMAX | ID30B | ID30B |
| Detector | Eiger 16M | Eiger 16M | Pilatus 6M | Pilatus 6M |
| Wavelength (Å) | 0.9700 | 0.9700 | 0.9763 | 0.9763 |
| Space group | P212121 | P212121 | P21212 | P212121 |
| a, b, c (Å) | 46.4, 48.9, 101.7 | 46.7, 48.9, 100.9 | 164.6, 56.9, 94.4 | 71.9, 105.1, 216.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Rotation | Standard | Standard | Standard | Standard |
| Rotation range per image (°) | 0.20 | 0.20 | 0.05 | 0.10 |
| Total rotation range (°) | 180 | 180 | 100 | 110 |
| Exposure time per image (s) | 0.011 | 0.011 | 0.020 | 0.020 |
| Flux (ph/s)/Transmission (%) | 1.9 × 1012/100 | 2.4 × 1012/100 | 3.7 × 1011/1.7 | 1.4 × 1012/6.1 |
| Beam size (µm2) | 50 × 50 | 50 × 50 | 30 × 30 | 30 × 30 |
| Crystal size (µm3) | 200 × 50 × 30 | 150 × 70 × 30 | 50 × 30 × 20 | 30 × 25 × 25 |
| Absorbed X-ray dose (MGy) | ||||
| - Av. diffraction weighted dose; | 1.0 | 1.4 | 1.3 | 6.4 |
| - Average dose (exposed regions); | 1.2 | 2.0 | 2.5 | 12.9 |
| - Max. dose. | 2.5 | 3.5 | 3.1 | 13.2 |
| Mosaicity (°) | 0.22 | 0.27 | 0.06 | 0.22 |
| Resolution range (Å) | 42.22–1.90 (1.94–1.90) | 44.04–2.00 (2.05–2.00) | 53.81–2.60 (2.72–2.60) | 68.21–4.20 (4.70–4.20) |
| Total no. of reflections | 119,246 | 105,677 | 98,595 | 47,888 |
| No. of unique reflections | 18,723 | 16,227 | 26,835 | 11,894 |
| Rmeas | 0.070 (1.003) | 0.071 (1.313) | 0.100 (0.575) | 0.202 (1.092) |
| Rmerge | 0.064 (0.901) | 0.065 (1.213) | 0.087 (0.498) | 0.177 (0.962) |
| Completeness (%) | 99.1 (95.2) | 99.7 (100.0) | 96.1 (97.6) | 95.8 (96.8) |
| Multiplicity | 6.4 (5.2) | 6.5 (6.7) | 3.7 (3.6) | 4.0 (4.2) |
| <I/σ(I)> | 12.7 (1.8) | 12.2 (1.6) | 8.7 (2.2) | 5.6 (1.7) |
| CC1/2 | 0.997 (0.556) | 0.997 (0.579) | 0.996 (0.848) | 0.994 (0.552) |
| Refinement statistics | ||||
| Rwork/Rfree (%) | 19.7/23.6 | 20.1/23.9 | 18.8/23.8 | 22.7/31.1 |
| Mean protein/cofactor/waters isotropic B factor (Å2) | 46.4/-/46.5 | 59.9/87.9/56.6 | 81.8/53.8/53.2 | 186.2/168.8/- |
| Wilson B-factor (Å2) | 37.8 | 47.7 | 55.8 | 157.8 |
| Protein assembly in asymmetric unit (AU) | Homodimer | Homodimer | Homodimer | Asymmetric homodimer |
| Protein residues in gene | 107 | 107 | 349 | 331 |
| Total modelled residues in AU | ||||
| - Protein residues by chain; | A: 1–106, B: 1–106 | A: 1–106, B: 1–106 | A: 4–349, B: 4–349 | A: 6–319, B:5–331, C: 6–318, D: 6–331 |
| - Cofactors; | - | A: 1 heme, B: 1 heme | A: 1 FAD, B: 1 FAD | A: 1 FAD, C: 1 FAD |
| - Added waters. | 82 | 42 | 59 | 0 |
| Matthews coefficient (Å3/Da) | 2.4 | 2.3 | 2.8 | 2.8 |
| Solvent content (%) | 48.5 | 45.9 | 55.9 | 56.1 |
| Ramachandran favored/allowed/outliers (%) | 97.1/2.9/0.0 | 97.1/2.4/0.5 | 96.7/3.2/0.2 | 90.6/8.8/0.6 |
| RMSD bond lengths (Å) | 0.007 | 0.008 | 0.008 | 0.004 |
| RMSD bond angles (°) | 0.78 | 0.94 | 0.98 | 0.75 |
| Estimated overall coordinate error based on Luzzati plot (Å) | 0.25 | 0.28 | 0.37 | 1.33 |
| PDB ID | 8AVH | 8AVI | 8C3M | 8C16 |
| FNR1 a,b | FNR1V329H | FNR2 a,b | FNR2H326V | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (min−1) | KM (μM) | kcat/KM (μM−1 min−1) | kcat (min−1) | KM (μM) | kcat/KM (μM−1 min−1) | kcat (min−1) | KM (μM) | kcat/KM (μM−1 min−1) | kcat (min−1) | KM (μM) | kcat/KM (μM−1 min−1) | |
| NrdI | 8.0 ± 0.1 | 2.7 ± 0.2 | 3.0 ± 0.2 | 1.8 ± 0.2 | 4.0 ± 1.3 | 0.5 ± 0.2 | 100 ± 4 | 61 ± 5 | 1.6 ± 0.2 | 36 ± 3 | 64 ± 9 | 0.6 ± 0.1 |
| Fld1 | 7.3 ± 0.5 | 4.7 ± 0.8 | 1.5 ± 0.3 | n.a. | n.a. | n.a. | 2778 ± 401 | 25 ± 7 | 111 ± 47 | 183 ± 31 | 43 ± 11 | 4 ± 2 |
| Fld2 | 42 ± 8 | 60 ± 22 | 0.7 ± 0.4 | 19 ± 6 | 12 ± 7 | 1.5 ± 1.0 | 9125 ± 1450 | 13 ± 5 | 701 ± 360 | n.a. | n.a. | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammerstad, M.; Rugtveit, A.K.; Dahlen, S.; Andersen, H.K.; Hersleth, H.-P. Functional Diversity of Homologous Oxidoreductases—Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction. Antioxidants 2023, 12, 1224. https://doi.org/10.3390/antiox12061224
Hammerstad M, Rugtveit AK, Dahlen S, Andersen HK, Hersleth H-P. Functional Diversity of Homologous Oxidoreductases—Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction. Antioxidants. 2023; 12(6):1224. https://doi.org/10.3390/antiox12061224
Chicago/Turabian StyleHammerstad, Marta, Anne Kristine Rugtveit, Sondov Dahlen, Hilde Kristin Andersen, and Hans-Petter Hersleth. 2023. "Functional Diversity of Homologous Oxidoreductases—Tuning of Substrate Specificity by a FAD-Stacking Residue for Iron Acquisition and Flavodoxin Reduction" Antioxidants 12, no. 6: 1224. https://doi.org/10.3390/antiox12061224