Autophagy, Oxidative Stress, and Alcoholic Liver Disease: A Systematic Review and Potential Clinical Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Article Selection and Data Extraction
3. Results and Literature Review
3.1. Studies Selected
3.2. Ethanol, ALD, and Oxidative Stress
3.2.1. Ethanol Metabolism
3.2.2. Protein Modifications
3.2.3. Lipid Peroxidation and DNA Alteration
3.2.4. Mitochondrial Damage
3.2.5. Inflammation
3.2.6. Iron Overload
3.2.7. Protective Mechanisms and the Antioxidant Response
3.3. Autophagy and ALD
3.3.1. The Macroautophagy Pathway
3.3.2. Mitophagy
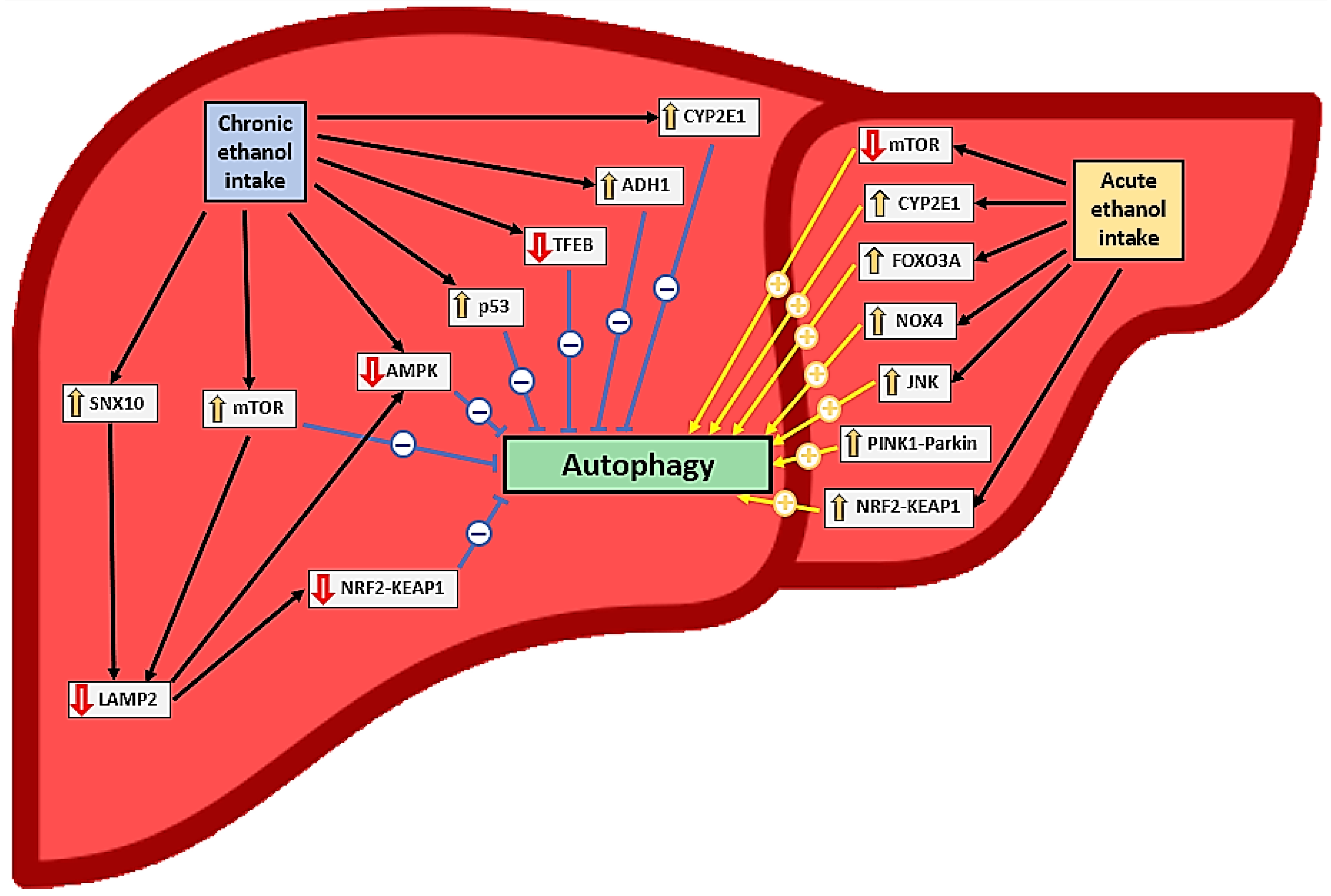
3.3.3. Acute Alcohol Intake and Autophagy in Hepatocytes
3.3.4. Acute Alcohol Intake and Autophagy in Other Cell Types
3.3.5. Chronic Ethanol Intake and Autophagy in Hepatocytes
3.3.6. Chronic Ethanol Intake and Autophagy in Other Cell Types
3.4. Alcohol-Induced Organ–Organ Crosstalk and Autophagy
3.5. Autophagy-Targeting Treatments for ALD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Status Report on Alcohol and Health 2018; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Zakhari, S.; Li, T.-K. Determinants of Alcohol Use and Abuse: Impact of Quantity and Frequency Patterns on Liver Disease. Hepatology 2007, 46, 2032–2039. [Google Scholar] [CrossRef] [PubMed]
- Mathurin, P.; Bataller, R. Trends in the Management and Burden of Alcoholic Liver Disease. J. Hepatol. 2015, 62, S38–S46. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; Mathurin, P.; Cortez-Pinto, H.; Loomba, R. Global Epidemiology of Alcohol-Associated Cirrhosis and HCC: Trends, Projections and Risk Factors. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 37–49. [Google Scholar] [CrossRef]
- Williams, J.A.; Manley, S.; Ding, W.-X. New Advances in Molecular Mechanisms and Emerging Therapeutic Targets in Alcoholic Liver Diseases. World J. Gastroenterol. 2014, 20, 12908–12933. [Google Scholar] [CrossRef]
- Venkatraman, A.; Landar, A.; Davis, A.J.; Chamlee, L.; Sanderson, T.; Kim, H.; Page, G.; Pompilius, M.; Ballinger, S.; Darley-Usmar, V.; et al. Modification of the Mitochondrial Proteome in Response to the Stress of Ethanol-Dependent Hepatotoxicity. J. Biol. Chem. 2004, 279, 22092–22101. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Vasudevan, D.M. Alcohol-Induced Oxidative Stress. Life Sci. 2007, 81, 177–187. [Google Scholar] [CrossRef]
- Manzo-Avalos, S.; Saavedra-Molina, A. Cellular and Mitochondrial Effects of Alcohol Consumption. Int. J. Environ. Res. Public Health 2010, 7, 4281–4304. [Google Scholar] [CrossRef]
- Yang, W.S.; Kim, K.J.; Gaschler, M.M.; Patel, M.; Shchepinov, M.S.; Stockwell, B.R. Peroxidation of Polyunsaturated Fatty Acids by Lipoxygenases Drives Ferroptosis. Proc. Natl. Acad. Sci. USA 2016, 113, E4966–E4975. [Google Scholar] [CrossRef]
- Mueller, S.; Peccerella, T.; Qin, H.; Glassen, K.; Waldherr, R.; Flechtenmacher, C.; Straub, B.K.; Millonig, G.; Stickel, F.; Bruckner, T.; et al. Carcinogenic Etheno DNA Adducts in Alcoholic Liver Disease: Correlation with Cytochrome P-4502E1 and Fibrosis. Alcohol. Clin. Exp. Res. 2018, 42, 252–259. [Google Scholar] [CrossRef]
- Vijayakumar, K.; Cho, G.-W. Autophagy: An Evolutionarily Conserved Process in the Maintenance of Stem Cells and Aging. Cell Biochem. Funct. 2019, 37, 452–458. [Google Scholar] [CrossRef]
- Eid, N.; Ito, Y.; Horibe, A.; Otsuki, Y. Ethanol-Induced Mitophagy in Liver Is Associated with Activation of the PINK1-Parkin Pathway Triggered by Oxidative DNA Damage. Histol. Histopathol. 2016, 31, 1143–1159. [Google Scholar] [CrossRef] [PubMed]
- Rasineni, K.; Donohue, T.M.; Thomes, P.G.; Yang, L.; Tuma, D.J.; McNiven, M.A.; Casey, C.A. Ethanol-induced Steatosis Involves Impairment of Lipophagy, Associated with Reduced Dynamin2 Activity. Hepatol. Commun. 2017, 1, 501–512. [Google Scholar] [CrossRef]
- Ding, W.-X.; Li, M.; Yin, X.-M. Selective Taste of Ethanol-Induced Autophagy for Mitochondria and Lipid Droplets. Autophagy 2011, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Khambu, B.; Wang, L.; Zhang, H.; Yin, X.-M. The Activation and Function of Autophagy in Alcoholic Liver Disease. Curr. Mol. Pharmacol. 2017, 10, 165–171. [Google Scholar] [CrossRef]
- Chao, X.; Ding, W.-X. Role and Mechanisms of Autophagy in Alcohol-Induced Liver Injury. Adv. Pharmacol. 2019, 85, 109–131. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Ding, W.-X. Role of Autophagy in Alcohol and Drug-Induced Liver Injury. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2020, 136, 111075. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, C.; Zhao, W.; He, C.; Ding, J.; Dai, R.; Xu, K.; Xiao, L.; Luo, L.; Liu, S.; et al. Impaired Autophagy in Intestinal Epithelial Cells Alters Gut Microbiota and Host Immune Responses. Appl. Environ. Microbiol. 2018, 84, e00880-18. [Google Scholar] [CrossRef]
- Foerster, E.G.; Mukherjee, T.; Cabral-Fernandes, L.; Rocha, J.D.B.; Girardin, S.E.; Philpott, D.J. How Autophagy Controls the Intestinal Epithelial Barrier. Autophagy 2022, 18, 86–103. [Google Scholar] [CrossRef]
- Li, Y.; Ding, W.-X. Adipose Tissue Autophagy and Homeostasis in Alcohol-Induced Liver Injury. Liver Res. 2017, 1, 54–62. [Google Scholar] [CrossRef]
- Rasineni, K.; Srinivasan, M.P.; Balamurugan, A.N.; Kaphalia, B.S.; Wang, S.; Ding, W.-X.; Pandol, S.J.; Lugea, A.; Simon, L.; Molina, P.E.; et al. Recent Advances in Understanding the Complexity of Alcohol-Induced Pancreatic Dysfunction and Pancreatitis Development. Biomolecules 2020, 10, 669. [Google Scholar] [CrossRef]
- Yang, F.; Luo, J. Endoplasmic Reticulum Stress and Ethanol Neurotoxicity. Biomolecules 2015, 5, 2538–2553. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Qin, X.; Chen, S.; Ceylan, A.F.; Dong, M.; Lin, Z.; Ren, J. Parkin Deficiency Accentuates Chronic Alcohol Intake-Induced Tissue Injury and Autophagy Defects in Brain, Liver and Skeletal Muscle. Acta Biochim. Biophys. Sin. 2020, 52, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, S.; Ni, H.-M.; Huang, H.; Ding, W.-X. Autophagy in Alcohol-Induced Multiorgan Injury: Mechanisms and Potential Therapeutic Targets. BioMed Res. Int. 2014, 2014, 498491. [Google Scholar] [CrossRef] [PubMed]
- Armutcu, F. Organ Crosstalk: The Potent Roles of Inflammation and Fibrotic Changes in the Course of Organ Interactions. Inflamm. Res. 2019, 68, 825–839. [Google Scholar] [CrossRef]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The Gut-Liver Axis in Liver Disease: Pathophysiological Basis for Therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef]
- Ferrell, J.M.; Chiang, J.Y.L. Bile Acid Receptors and Signaling Crosstalk in the Liver, Gut and Brain. Liver Res. 2021, 5, 105–118. [Google Scholar] [CrossRef]
- Poole, L.G.; Dolin, C.E.; Arteel, G.E. Organ-Organ Crosstalk and Alcoholic Liver Disease. Biomolecules 2017, 7, 62. [Google Scholar] [CrossRef]
- Martinez-Lopez, N.; Garcia-Macia, M.; Sahu, S.; Athonvarangkul, D.; Liebling, E.; Merlo, P.; Cecconi, F.; Schwartz, G.J.; Singh, R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016, 23, 113–127. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. Available online: https://www.rayyan.ai/ (accessed on 13 January 2023). [CrossRef]
- Lu, X.; Xuan, W.; Li, J.; Yao, H.; Huang, C.; Li, J. AMPK Protects against Alcohol-Induced Liver Injury through UQCRC2 to up-Regulate Mitophagy. Autophagy 2021, 17, 3622–3643. [Google Scholar] [CrossRef]
- Chen, C.; Wang, S.; Yu, L.; Mueller, J.; Fortunato, F.; Rausch, V.; Mueller, S. H2O2-Mediated Autophagy during Ethanol Metabolism. Redox Biol. 2021, 46, 102081. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lu, J.; Mao, A.; Zhang, R.; Guan, S. Autophagy Inhibition Plays a Protective Role in Ferroptosis Induced by Alcohol via the P62–Keap1–Nrf2 Pathway. J. Agric. Food Chem. 2021, 69, 9671–9683. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, P.; Li, W.; Wu, Y.; An, W. Augmenter of Liver Regeneration Protects against Ethanol-Induced Acute Liver Injury by Promoting Autophagy. Am. J. Pathol. 2019, 189, 552–567. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Li, W.-Z.; Zhang, S.; Hu, B.; Li, Y.-X.; Li, H.-D.; Tang, H.-H.; Li, Q.-W.; Guan, Y.-Y.; Liu, L.-X.; et al. SNX10 Mediates Alcohol-Induced Liver Injury and Steatosis by Regulating the Activation of Chaperone-Mediated Autophagy. J. Hepatol. 2018, 69, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Mahli, A.; Thasler, W.E.; Patsenker, E.; Müller, S.; Stickel, F.; Müller, M.; Seitz, H.K.; Cederbaum, A.I.; Hellerbrand, C. Identification of Cytochrome CYP2E1 as Critical Mediator of Synergistic Effects of Alcohol and Cellular Lipid Accumulation in Hepatocytes in Vitro. Oncotarget 2015, 6, 41464–41478. [Google Scholar] [CrossRef] [PubMed]
- Thomes, P.G.; Ehlers, R.A.; Trambly, C.S.; Clemens, D.L.; Fox, H.S.; Tuma, D.J.; Donohue, T.M. Multilevel Regulation of Autophagosome Content by Ethanol Oxidation in HepG2 Cells. Autophagy 2013, 9, 63–73. [Google Scholar] [CrossRef]
- Ding, W.; Li, M.; Chen, X.; Ni, H.; Lin, C.; Gao, W.; Lu, B.; Stolz, D.B.; Clemens, D.L.; Yin, X. Autophagy Reduces Acute Ethanol-Induced Hepatotoxicity and Steatosis in Mice. Gastroenterology 2010, 139, 1740–1752. [Google Scholar] [CrossRef]
- Guo, R.; Xu, X.; Babcock, S.A.; Zhang, Y.; Ren, J. Aldehyde Dedydrogenase-2 Plays a Beneficial Role in Ameliorating Chronic Alcohol-Induced Hepatic Steatosis and Inflammation through Regulation of Autophagy. J. Hepatol. 2015, 62, 647–656. [Google Scholar] [CrossRef]
- Wu, D.; Wang, X.; Zhou, R.; Yang, L.; Cederbaum, A.I. Alcohol Steatosis and Cytotoxicity: The Role of Cytochrome P4502E1 and Autophagy. Free Radic. Biol. Med. 2012, 53, 1346–1357. [Google Scholar] [CrossRef]
- Wu, D.; Wang, X.; Zhou, R.; Cederbaum, A. CYP2E1 Enhances Ethanol-Induced Lipid Accumulation but Impairs Autophagy in HepG2 E47 Cells. Biochem. Biophys. Res. Commun. 2010, 402, 116–122. [Google Scholar] [CrossRef]
- Samuvel, D.J.; Li, L.; Krishnasamy, Y.; Gooz, M.; Takemoto, K.; Woster, P.M.; Lemasters, J.J.; Zhong, Z. Mitochondrial Depolarization after Acute Ethanol Treatment Drives Mitophagy in Living Mice. Autophagy 2022, 18, 2671–2685. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhou, J.; Chen, X.; Dong, Z.; Yin, X.-M. Diverse Consequences in Liver Injury in Mice with Different Autophagy Functional Status Treated with Alcohol. Am. J. Pathol. 2019, 189, 1744–1762. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Ni, H.-M.; Ding, Y.; Ding, W.-X. Parkin Regulates Mitophagy and Mitochondrial Function to Protect against Alcohol-Induced Liver Injury and Steatosis in Mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G324–G340. [Google Scholar] [CrossRef] [PubMed]
- Manley, S.; Ni, H.-M.; Williams, J.A.; Kong, B.; DiTacchio, L.; Guo, G.; Ding, W.-X. Farnesoid X Receptor Regulates Forkhead Box O3a Activation in Ethanol-Induced Autophagy and Hepatotoxicity. Redox Biol. 2014, 2, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.-M.; Du, K.; You, M.; Ding, W.-X. Critical Role of FoxO3a in Alcohol-Induced Autophagy and Hepatotoxicity. Am. J. Pathol. 2013, 183, 1815–1825. [Google Scholar] [CrossRef]
- Lin, C.-W.; Zhang, H.; Li, M.; Xiong, X.; Chen, X.; Chen, X.; Dong, X.C.; Yin, X.-M. Pharmacological Promotion of Autophagy Alleviates Steatosis and Injury in Alcoholic and Non-Alcoholic Fatty Liver Conditions in Mice. J. Hepatol. 2013, 58, 993–999. [Google Scholar] [CrossRef]
- Yang, L.; Wu, D.; Wang, X.; Cederbaum, A.I. Cytochrome P4502E1, Oxidative Stress, JNK and Autophagy in Acute Alcohol-Induced Fatty Liver. Free Radic. Biol. Med. 2012, 53, 1170–1180. [Google Scholar] [CrossRef]
- Thomes, P.G.; Trambly, C.S.; Thiele, G.M.; Duryee, M.J.; Fox, H.S.; Haorah, J.; Donohue, T.M. Proteasome Activity and Autophagosome Content in Liver Are Reciprocally Regulated by Ethanol Treatment. Biochem. Biophys. Res. Commun. 2012, 417, 262–267. [Google Scholar] [CrossRef]
- Guo, W.; Zhong, W.; Hao, L.; Sun, X.; Zhou, Z. Activation of MTORC1 by Free Fatty Acids Suppresses LAMP2 and Autophagy Function via ER Stress in Alcohol-Related Liver Disease. Cells 2021, 10, 2730. [Google Scholar] [CrossRef]
- Guo, W.; Zhong, W.; Hao, L.; Dong, H.; Sun, X.; Yue, R.; Li, T.; Zhou, Z. Fatty Acids Inhibit LAMP2-Mediated Autophagy Flux via Activating ER Stress Pathway in Alcohol-Related Liver Disease. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1599–1615. [Google Scholar] [CrossRef] [PubMed]
- Babuta, M.; Furi, I.; Bala, S.; Bukong, T.N.; Lowe, P.; Catalano, D.; Calenda, C.; Kodys, K.; Szabo, G. Dysregulated Autophagy and Lysosome Function Are Linked to Exosome Production by Micro-RNA 155 in Alcoholic Liver Disease. Hepatology 2019, 70, 2123–2141. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhu, P.; Wang, J.; Toan, S.; Ren, J. DNA-PKcs Promotes Alcohol-Related Liver Disease by Activating Drp1-Related Mitochondrial Fission and Repressing FUNDC1-Required Mitophagy. Signal Transduct. Target. Ther. 2019, 4, 56. [Google Scholar] [CrossRef] [PubMed]
- Menk, M.; Graw, J.A.; Poyraz, D.; Möbius, N.; Spies, C.D.; von Haefen, C. Chronic Alcohol Consumption Inhibits Autophagy and Promotes Apoptosis in the Liver. Int. J. Med. Sci. 2018, 15, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Wang, S.; Zhao, K.; Li, Y.; Williams, J.A.; Li, T.; Chavan, H.; Krishnamurthy, P.; He, X.C.; Li, L.; et al. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced Liver Injury and Steatosis in Mice. Gastroenterology 2018, 155, 865–879.e12. [Google Scholar] [CrossRef]
- Kong, X.; Yang, Y.; Ren, L.; Shao, T.; Li, F.; Zhao, C.; Liu, L.; Zhang, H.; McClain, C.J.; Feng, W. Activation of Autophagy Attenuates EtOH-LPS-Induced Hepatic Steatosis and Injury through MD2 Associated TLR4 Signaling. Sci. Rep. 2017, 7, 9292. [Google Scholar] [CrossRef]
- Lu, Y.; Cederbaum, A.I. Autophagy Protects against CYP2E1/Chronic Ethanol-Induced Hepatotoxicity. Biomolecules 2015, 5, 2659–2674. [Google Scholar] [CrossRef]
- King, A.L.; Swain, T.M.; Mao, Z.; Udoh, U.S.; Oliva, C.R.; Betancourt, A.M.; Griguer, C.E.; Crowe, D.R.; Lesort, M.; Bailey, S.M. Involvement of the Mitochondrial Permeability Transition Pore in Chronic Ethanol-Mediated Liver Injury in Mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G265–G277. [Google Scholar] [CrossRef]
- Tan, T.C.H.; Crawford, D.H.G.; Jaskowski, L.A.; Subramaniam, V.N.; Clouston, A.D.; Crane, D.I.; Bridle, K.R.; Anderson, G.J.; Fletcher, L.M. Excess Iron Modulates Endoplasmic Reticulum Stress-Associated Pathways in a Mouse Model of Alcohol and High-Fat Diet-Induced Liver Injury. Lab. Investig. 2013, 93, 1295–1312. [Google Scholar] [CrossRef]
- Liang, S.; Zhong, Z.; Kim, S.Y.; Uchiyama, R.; Roh, Y.S.; Matsushita, H.; Gottlieb, R.A.; Seki, E. Murine Macrophage Autophagy Protects against Alcohol-Induced Liver Injury by Degrading Interferon Regulatory Factor 1 (IRF1) and Removing Damaged Mitochondria. J. Biol. Chem. 2019, 294, 12359–12369. [Google Scholar] [CrossRef]
- Ilyas, G.; Cingolani, F.; Zhao, E.; Tanaka, K.; Czaja, M.J. Decreased Macrophage Autophagy Promotes Liver Injury and Inflammation from Alcohol. Alcohol. Clin. Exp. Res. 2019, 43, 1403–1413. [Google Scholar] [CrossRef]
- Xie, Z.-Y.; Xiao, Z.-H.; Wang, F.-F. Inhibition of Autophagy Reverses Alcohol-Induced Hepatic Stellate Cells Activation through Activation of Nrf2-Keap1-ARE Signaling Pathway. Biochimie 2018, 147, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Denaës, T.; Lodder, J.; Chobert, M.-N.; Ruiz, I.; Pawlotsky, J.-M.; Lotersztajn, S.; Teixeira-Clerc, F. The Cannabinoid Receptor 2 Protects Against Alcoholic Liver Disease Via a Macrophage Autophagy-Dependent Pathway. Sci. Rep. 2016, 6, 28806. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Gea, V.; Hilscher, M.; Rozenfeld, R.; Lim, M.P.; Nieto, N.; Werner, S.; Devi, L.A.; Friedman, S.L. Endoplasmic Reticulum Stress Induces Fibrogenic Activity in Hepatic Stellate Cells through Autophagy. J. Hepatol. 2013, 59, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-S.; Jo, E.; Han, J.-H.; Jung, S.-H.; Lee, D.-H.; Park, I.; Heo, K.-S.; Na, M.; Myung, C.-S. Hepatoprotective Effects of an Acer Tegmentosum Maxim Extract through Antioxidant Activity and the Regulation of Autophagy. J. Ethnopharmacol. 2019, 239, 111912. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Li, L.; Hu, W.; Qu, Y.; Ding, Y.; Meng, L.; Teng, L.; Wang, D. Hepatoprotective Effects of Antrodia cinnamomea: The Modulation of Oxidative Stress Signaling in a Mouse Model of Alcohol-Induced Acute Liver Injury. Oxid. Med. Cell. Longev. 2017, 2017, e7841823. [Google Scholar] [CrossRef]
- Yu, Y.; Tian, Z.; Liang, L.; Yang, X.; Sheng, D.; Zeng, J.; Li, X.; Shi, R.; Han, Z.; Wei, L. Babao Dan Attenuates Acute Ethanol-Induced Liver Injury via Nrf2 Activation and Autophagy. Cell Biosci. 2019, 9, 80. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Hwang, S.H.; Jia, Y.; Seo, W.-D.; Lee, S.-J. Barley Sprout Extracts Reduce Hepatic Lipid Accumulation in Ethanol-Fed Mice by Activating Hepatic AMP-Activated Protein Kinase. Food Res. Int. 2017, 101, 209–217. [Google Scholar] [CrossRef]
- Yuan, F.; Xu, Y.; You, K.; Zhang, J.; Yang, F.; Li, Y. Calcitriol Alleviates Ethanol-Induced Hepatotoxicity via AMPK/MTOR-Mediated Autophagy. Arch. Biochem. Biophys. 2021, 697, 108694. [Google Scholar] [CrossRef]
- Yang, L.; Rozenfeld, R.; Wu, D.; Devi, L.A.; Zhang, Z.; Cederbaum, A. Cannabidiol Protects Liver from Binge Alcohol-Induced Steatosis by Mechanisms Including Inhibition of Oxidative Stress and Increase in Autophagy. Free Radic. Biol. Med. 2014, 68, 260–267. [Google Scholar] [CrossRef]
- Khan, I.; Bhardwaj, M.; Shukla, S.; Min, S.-H.; Choi, D.K.; Bajpai, V.K.; Huh, Y.S.; Kang, S.C. Carvacrol Inhibits Cytochrome P450 and Protects against Binge Alcohol-Induced Liver Toxicity. Food Chem. Toxicol. 2019, 131, 110582. [Google Scholar] [CrossRef]
- Zeng, T.; Zhang, C.-L.; Song, F.-Y.; Zhao, X.-L.; Xie, K.-Q. CMZ Reversed Chronic Ethanol-Induced Disturbance of PPAR-α Possibly by Suppressing Oxidative Stress and PGC-1α Acetylation, and Activating the MAPK and GSK3β Pathway. PLoS ONE 2014, 9, e98658. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-Y.; Zhang, C.-L.; Zhao, X.-L.; Xie, K.-Q.; Zeng, T. Inhibition of Cytochrome P4502E1 by Chlormethiazole Attenuated Acute Ethanol-Induced Fatty Liver. Chem. Biol. Interact. 2014, 222, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Shu, M.-S.; Kim, J.-Y.; Kim, Y.-H.; Sim, K.H.; Sung, W.J.; Eun, J.R. Cilostazol Protects Hepatocytes against Alcohol-Induced Apoptosis via Activation of AMPK Pathway. PLoS ONE 2019, 14, e0211415. [Google Scholar] [CrossRef]
- Guo, X.; Cui, R.; Zhao, J.; Mo, R.; Peng, L.; Yan, M. Corosolic Acid Protects Hepatocytes against Ethanol-Induced Damage by Modulating Mitogen-Activated Protein Kinases and Activating Autophagy. Eur. J. Pharmacol. 2016, 791, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Dong, Y.; Li, B.; Kang, X.; Gu, C.; Zhu, T.; Luo, Y.; Pang, M.; Du, W.; Ge, W. Dihydromyricetin Modulates P62 and Autophagy Crosstalk with the Keap-1/Nrf2 Pathway to Alleviate Ethanol-Induced Hepatic Injury. Toxicol. Lett. 2017, 274, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jin, Q.; Li, X.; Jiang, M.; Cui, B.-W.; Xia, K.-L.; Wu, Y.-L.; Lian, L.-H.; Nan, J.-X. Amelioration of Alcoholic Liver Steatosis by Dihydroquercetin through the Modulation of AMPK-Dependent Lipogenesis Mediated by P2X7R–NLRP3-Inflammasome Activation. J. Agric. Food Chem. 2018, 66, 4862–4871. [Google Scholar] [CrossRef]
- Zhou, Z.-S.; Kong, C.-F.; Sun, J.-R.; Qu, X.-K.; Sun, J.-H.; Sun, A.-T. Fisetin Ameliorates Alcohol-Induced Liver Injury through Regulating SIRT1 and SphK1 Pathway. Am. J. Chin. Med. 2022, 50, 2171–2184. [Google Scholar] [CrossRef]
- Xue, M.; Liang, H.; Zhou, Z.; Liu, Y.; He, X.; Zhang, Z.; Sun, T.; Yang, J.; Qin, Y.; Qin, K. Effect of Fucoidan on Ethanol-Induced Liver Injury and Steatosis in Mice and the Underlying Mechanism. Food Nutr. Res. 2021, 65, 5384. [Google Scholar] [CrossRef]
- Xue, M.; Tian, Y.; Sui, Y.; Zhao, H.; Gao, H.; Liang, H.; Qiu, X.; Sun, Z.; Zhang, Y.; Qin, Y. Protective Effect of Fucoidan against Iron Overload and Ferroptosis-Induced Liver Injury in Rats Exposed to Alcohol. Biomed. Pharmacother. 2022, 153, 113402. [Google Scholar] [CrossRef]
- Song, X.; Yin, S.; Huo, Y.; Liang, M.; Fan, L.; Ye, M.; Hu, H. Glycycoumarin Ameliorates Alcohol-Induced Hepatotoxicity via Activation of Nrf2 and Autophagy. Free Radic. Biol. Med. 2015, 89, 135–146. [Google Scholar] [CrossRef]
- Zhang, Y.-P.; Yang, X.-Q.; Yu, D.-K.; Xiao, H.-Y.; Du, J.-R. Nrf2 Signalling Pathway and Autophagy Impact on the Preventive Effect of Green Tea Extract against Alcohol-Induced Liver Injury. J. Pharm. Pharmacol. 2021, 73, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zhuo, H.; Yang, L.; Ouyang, H.; Chen, J.; Liu, B.; Huang, H. A Peptide HEPFYGNEGALR from Apostichopus Japonicus Alleviates Acute Alcoholic Liver Injury by Enhancing Antioxidant Response in Male C57BL/6J Mice. Molecules 2022, 27, 5839. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Chen, X.; Fu, Z.; Yin, J.; Wang, Y.; Sun, W.; Ren, H.; Zhang, Y. Kinsenoside Alleviates Alcoholic Liver Injury by Reducing Oxidative Stress, Inhibiting Endoplasmic Reticulum Stress, and Regulating AMPK-Dependent Autophagy. Front. Pharmacol. 2022, 12, 747325. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, L.; Qin, Y.; Zhou, Y.; Liu, W.; Li, Y.; Chen, Y.; Xu, Y. Protective Effects of Rare Earth Lanthanum on Acute Ethanol-Induced Oxidative Stress in Mice via Keap 1/Nrf2/P62 Activation. Sci. Total Environ. 2021, 758, 143626. [Google Scholar] [CrossRef] [PubMed]
- Kurhaluk, N.; Tkachenko, H.; Lukash, O. Melatonin Modulates Oxidative Phosphorylation, Hepatic and Kidney Autophagy-Caused Subclinical Endotoxemia and Acute Ethanol-Induced Oxidative Stress. Chronobiol. Int. 2020, 37, 1709–1724. [Google Scholar] [CrossRef]
- Lin, G.-S.; Zhao, M.-M.; Fu, Q.-C.; Zhao, S.-Y.; Ba, T.-T.; Yu, H.-X. Palmatine Attenuates Hepatocyte Injury by Promoting Autophagy via the AMPK/MTOR Pathway after Alcoholic Liver Disease. Drug Dev. Res. 2022, 83, 1613–1622. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, M.; Deng, Y.; Guo, C.; Liao, L.; He, L.; Peng, C.; Li, Y. Functional Teas from Penthorum Chinense Pursh Alleviates Ethanol-Induced Hepatic Oxidative Stress and Autophagy Impairment in Zebrafish via Modulating the AMPK/P62/Nrf2/MTOR Signaling Axis. Plant Foods Hum. Nutr. 2022, 77, 514–520. [Google Scholar] [CrossRef]
- Cao, P.; Zhang, Y.; Huang, Z.; Sullivan, M.A.; He, Z.; Wang, J.; Chen, Z.; Hu, H.; Wang, K. The Preventative Effects of Procyanidin on Binge Ethanol-Induced Lipid Accumulation and ROS Overproduction via the Promotion of Hepatic Autophagy. Mol. Nutr. Food Res. 2019, 63, 1801255. [Google Scholar] [CrossRef]
- Yu, X.; Xu, Y.; Zhang, S.; Sun, J.; Liu, P.; Xiao, L.; Tang, Y.; Liu, L.; Yao, P. Quercetin Attenuates Chronic Ethanol-Induced Hepatic Mitochondrial Damage through Enhanced Mitophagy. Nutrients 2016, 8, 27. [Google Scholar] [CrossRef]
- Zhao, X.; Xue, X.; Wang, J.; Dai, S.; Peng, C.; Li, Y. Quercetin Alleviates Ethanol-Induced Hepatic Steatosis in L02 Cells by Activating TFEB Translocation to Compensate for Inadequate Autophagy. Phytother. Res. 2023, 37, 62–76. [Google Scholar] [CrossRef]
- Lin, H.; Guo, X.; Liu, J.; Liu, P.; Mei, G.; Li, H.; Li, D.; Chen, H.; Chen, L.; Zhao, Y.; et al. Improving Lipophagy by Restoring Rab7 Cycle: Protective Effects of Quercetin on Ethanol-Induced Liver Steatosis. Nutrients 2022, 14, 658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Zhou, X.; Zhao, J. Quercetin Prevents Alcohol-induced Liver Injury through Targeting of PI3K/Akt/Nuclear Factor-κB and STAT3 Signaling Pathway. Exp. Ther. Med. 2017, 14, 6169–6175. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Guo, X.; Zhou, F.; Xiao, L.; Liu, J.; Jiang, C.; Xing, M.; Yao, P. Quercetin Alleviates Ethanol-Induced Liver Steatosis Associated with Improvement of Lipophagy. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 125, 21–28. [Google Scholar] [CrossRef]
- Zhao, X.; Gong, L.; Wang, C.; Liu, M.; Hu, N.; Dai, X.; Peng, C.; Li, Y. Quercetin Mitigates Ethanol-Induced Hepatic Steatosis in Zebrafish via P2X7R-Mediated PI3K/ Keap1/Nrf2 Signaling Pathway. J. Ethnopharmacol. 2021, 268, 113569. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, F.; Fang, Z.; Hu, C. Resveratrol Ameliorates Alcoholic Fatty Liver by Inducing Autophagy. Am. J. Chin. Med. 2016, 44, 1207–1220. [Google Scholar] [CrossRef]
- You, M.; Liang, X.; Ajmo, J.M.; Ness, G.C. Involvement of Mammalian Sirtuin 1 in the Action of Ethanol in the Liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G892–G898. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, R.; Zhao, Y.; Fu, R.; Wang, R.; Zhao, H.; Wang, Z.; Tang, F.; Zhang, N.; Tian, X.; et al. Promotion of Autophagosome–Lysosome Fusion via Salvianolic Acid A-Mediated SIRT1 up-Regulation Ameliorates Alcoholic Liver Disease. RSC Adv. 2018, 8, 20411–20422. [Google Scholar] [CrossRef]
- Fu, X.; Zhong, Z.; Hu, F.; Zhang, Y.; Li, C.; Yan, P.; Feng, L.; Shen, J.; Huang, B. The Protective Effects of Selenium-Enriched Spirulina Platensis on Chronic Alcohol-Induced Liver Injury in Mice. Food Funct. 2018, 9, 3155–3165. [Google Scholar] [CrossRef]
- Song, X.-Y.; Liu, P.-C.; Liu, W.-W.; Zhou, J.; Hayashi, T.; Mizuno, K.; Hattori, S.; Fujisaki, H.; Ikejima, T. Silibinin Inhibits Ethanol- or Acetaldehyde-Induced Ferroptosis in Liver Cell Lines. Toxicol. Vitro Int. J. Publ. Assoc. BIBRA 2022, 82, 105388. [Google Scholar] [CrossRef]
- Atef, M.M.; Hafez, Y.M.; Alshenawy, H.A.; Emam, M.N. Ameliorative Effects of Autophagy Inducer, Simvastatin on Alcohol-Induced Liver Disease in a Rat Model. J. Cell. Biochem. 2019, 120, 7679–7688. [Google Scholar] [CrossRef]
- Zhou, R.; Lin, J.; Wu, D. Sulforaphane Induces Nrf2 and Protects against CYP2E1-Dependent Binge Alcohol-Induced Liver Steatosis. Biochim. Biophys. Acta 2014, 1840, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, Y.; Yuan, F.; Peng, L.; Qiu, C. Tangeretin Protects Mice from Alcohol-Induced Fatty Liver by Activating Mitophagy through the AMPK–ULK1 Pathway. J. Agric. Food Chem. 2022, 70, 11236–11244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, R.; Wang, X.; Jiang, Y.; Xu, W.; Shao, Y.; Yue, C.; Shi, W.; Jin, H.; Ge, T.; et al. Activation of UQCRC2-Dependent Mitophagy by Tetramethylpyrazine Inhibits MLKL-Mediated Hepatocyte Necroptosis in Alcoholic Liver Disease. Free Radic. Biol. Med. 2022, 179, 301–316. [Google Scholar] [CrossRef]
- Thoreen, C.C.; Kang, S.A.; Chang, J.W.; Liu, Q.; Zhang, J.; Gao, Y.; Reichling, L.J.; Sim, T.; Sabatini, D.M.; Gray, N.S. An ATP-Competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-Resistant Functions of MTORC1. J. Biol. Chem. 2009, 284, 8023–8032. [Google Scholar] [CrossRef]
- Wu, W.-B.; Chen, Y.-Y.; Zhu, B.; Peng, X.-M.; Zhang, S.-W.; Zhou, M.-L. Excessive Bile Acid Activated NF-Kappa B and Promoted the Development of Alcoholic Steatohepatitis in Farnesoid X Receptor Deficient Mice. Biochimie 2015, 115, 86–92. [Google Scholar] [CrossRef]
- Liuzzi, J.P.; Yoo, C. Role of Zinc in the Regulation of Autophagy During Ethanol Exposure in Human Hepatoma Cells. Biol. Trace Elem. Res. 2013, 156, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tan, H.-Y.; Wang, N.; Zhang, Z.-J.; Lao, L.; Wong, C.-W.; Feng, Y. The Role of Oxidative Stress and Antioxidants in Liver Diseases. Int. J. Mol. Sci. 2015, 16, 26087–26124. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cederbaum, A.I. Oxidative Stress and Alcoholic Liver Disease. Semin. Liver Dis. 2009, 29, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jia, Z.; Misra, H.; Li, Y.R. Oxidative Stress and Redox Signaling Mechanisms of Alcoholic Liver Disease: Updated Experimental and Clinical Evidence. J. Dig. Dis. 2012, 13, 133–142. [Google Scholar] [CrossRef]
- Balbo, S.; Hashibe, M.; Gundy, S.; Brennan, P.; Canova, C.; Simonato, L.; Merletti, F.; Richiardi, L.; Agudo, A.; Castellsagué, X.; et al. N2-Ethyldeoxyguanosine as a Potential Biomarker for Assessing Effects of Alcohol Consumption on DNA. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2008, 17, 3026–3032. [Google Scholar] [CrossRef]
- Lucey, M.R.; Mathurin, P.; Morgan, T.R. Alcoholic Hepatitis. N. Engl. J. Med. 2009, 360, 2758–2769. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.K.; Yates, E.; Lilly, K.; Dhanda, A.D. Oxidative Stress in Alcohol-Related Liver Disease. World J. Hepatol. 2020, 12, 332–349. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Cho, Y.E.; Hwang, S. Crosstalk between Oxidative Stress and Inflammatory Liver Injury in the Pathogenesis of Alcoholic Liver Disease. Int. J. Mol. Sci. 2022, 23, 774. [Google Scholar] [CrossRef]
- Cederbaum, A.I. Alcohol Metabolism. Clin. Liver Dis. 2012, 16, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Cederbaum, A.I.; Lu, Y.; Wu, D. Role of Oxidative Stress in Alcohol-Induced Liver Injury. Arch. Toxicol. 2009, 83, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, T.; Kusumanchi, P.; Han, S.; Yang, Z.; Liangpunsakul, S. Alcohol Metabolizing Enzymes, Microsomal Ethanol Oxidizing System, Cytochrome P450 2E1, Catalase, and Aldehyde Dehydrogenase in Alcohol-Associated Liver Disease. Biomedicines 2020, 8, 50. [Google Scholar] [CrossRef]
- Galli, A.; Pinaire, J.; Fischer, M.; Dorris, R.; Crabb, D.W. The Transcriptional and DNA Binding Activity of Peroxisome Proliferator-Activated Receptor Alpha Is Inhibited by Ethanol Metabolism. A Novel Mechanism for the Development of Ethanol-Induced Fatty Liver. J. Biol. Chem. 2001, 276, 68–75. [Google Scholar] [CrossRef]
- You, M.; Fischer, M.; Deeg, M.A.; Crabb, D.W. Ethanol Induces Fatty Acid Synthesis Pathways by Activation of Sterol Regulatory Element-Binding Protein (SREBP). J. Biol. Chem. 2002, 277, 29342–29347. [Google Scholar] [CrossRef]
- Donohue, T.M.; Osna, N.A.; Trambly, C.S.; Whitaker, N.P.; Thomes, P.G.; Todero, S.L.; Davis, J.S. Early Growth Response-1 Contributes to Steatosis Development after Acute Ethanol Administration. Alcohol. Clin. Exp. Res. 2012, 36, 759–767. [Google Scholar] [CrossRef]
- Sacitharan, P.K.; Bou-Gharios, G.; Edwards, J.R. SIRT1 Directly Activates Autophagy in Human Chondrocytes. Cell Death Discov. 2020, 6, 41. [Google Scholar] [CrossRef]
- Zhao, R.-Z.; Jiang, S.; Zhang, L.; Yu, Z.-B. Mitochondrial Electron Transport Chain, ROS Generation and Uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Stickel, F. Risk Factors and Mechanisms of Hepatocarcinogenesis with Special Emphasis on Alcohol and Oxidative Stress. Biol. Chem. 2006, 387, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Cederbaum, A.I. CYP2E1 and Oxidative Liver Injury by Alcohol. Free Radic. Biol. Med. 2008, 44, 723–738. [Google Scholar] [CrossRef]
- Ambade, A.; Mandrekar, P. Oxidative Stress and Inflammation: Essential Partners in Alcoholic Liver Disease. Int. J. Hepatol. 2012, 2012, 853175. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Chen, Y.; Wang, J.; Hao, L.; Huang, C.; Griffiths, A.; Sun, Z.; Zhou, Z.; Song, Z. ER Stress-Induced Upregulation of NNMT Contributes to Alcohol-Related Fatty Liver Development. J. Hepatol. 2020, 73, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Ma, Y.; Lai, S.; Dou, X.; Li, S. NNMT Aggravates Hepatic Steatosis, but Alleviates Liver Injury in Alcoholic Liver Disease. J. Hepatol. 2021, 74, 1248–1250. [Google Scholar] [CrossRef]
- Campagna, R.; Vignini, A. NAD+ Homeostasis and NAD+-Consuming Enzymes: Implications for Vascular Health. Antioxidants 2023, 12, 376. [Google Scholar] [CrossRef]
- Shin, J.H.; Park, C.W.; Yoon, G.; Hong, S.M.; Choi, K.Y. NNMT Depletion Contributes to Liver Cancer Cell Survival by Enhancing Autophagy under Nutrient Starvation. Oncogenesis 2018, 7, 58. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, X.; Wang, Y.; Huang, X.; Yang, J.; Zeng, J.; Li, G.; Xie, X.; Zhang, J. Nicotinamide N-Methyltransferase Inhibits Autophagy Induced by Oxidative Stress through Suppressing the AMPK Pathway in Breast Cancer Cells. Cancer Cell Int. 2020, 20, 191. [Google Scholar] [CrossRef]
- Campagna, R.; Mateuszuk, Ł.; Wojnar-Lason, K.; Kaczara, P.; Tworzydło, A.; Kij, A.; Bujok, R.; Mlynarski, J.; Wang, Y.; Sartini, D.; et al. Nicotinamide N-Methyltransferase in Endothelium Protects against Oxidant Stress-Induced Endothelial Injury. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119082. [Google Scholar] [CrossRef]
- Guarino, M.; Dufour, J.-F. Nicotinamide and NAFLD: Is There Nothing New Under the Sun? Metabolites 2019, 9, 180. [Google Scholar] [CrossRef] [PubMed]
- Filomeni, G.; Rotilio, G.; Ciriolo, M.R. Disulfide Relays and Phosphorylative Cascades: Partners in Redox-Mediated Signaling Pathways. Cell Death Differ. 2005, 12, 1555–1563. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Jiang, J.; Zhan, M.; Zhang, H.; Wang, Q.-T.; Sun, S.-N.; Guo, X.-K.; Yin, H.; Wei, Y.; Liu, J.O.; et al. Targeting Neoantigens in Hepatocellular Carcinoma for Immunotherapy: A Futile Strategy? Hepatology 2021, 73, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Zapotoczny, B.; Braet, F.; Kus, E.; Ginda-Mäkelä, K.; Klejevskaja, B.; Campagna, R.; Chlopicki, S.; Szymonski, M. Actin-spectrin Scaffold Supports Open Fenestrae in Liver Sinusoidal Endothelial Cells. Traffic Cph. Den. 2019, 20, 932–942. [Google Scholar] [CrossRef]
- Kimata, Y.; Kohno, K. Endoplasmic Reticulum Stress-Sensing Mechanisms in Yeast and Mammalian Cells. Curr. Opin. Cell Biol. 2011, 23, 135–142. [Google Scholar] [CrossRef]
- Bailey, S.M. A Review of the Role of Reactive Oxygen and Nitrogen Species in Alcohol-Induced Mitochondrial Dysfunction. Free Radic. Res. 2003, 37, 585–596. [Google Scholar] [CrossRef]
- Haldar, S.M.; Stamler, J.S. S-Nitrosylation at the Interface of Autophagy and Disease. Mol. Cell 2011, 43, 1–3. [Google Scholar] [CrossRef]
- Rizza, S.; Cardaci, S.; Montagna, C.; Di Giacomo, G.; De Zio, D.; Bordi, M.; Maiani, E.; Campello, S.; Borreca, A.; Puca, A.A.; et al. S-Nitrosylation Drives Cell Senescence and Aging in Mammals by Controlling Mitochondrial Dynamics and Mitophagy. Proc. Natl. Acad. Sci. USA 2018, 115, E3388–E3397. [Google Scholar] [CrossRef]
- Dalleau, S.; Baradat, M.; Guéraud, F.; Huc, L. Cell Death and Diseases Related to Oxidative Stress: 4-Hydroxynonenal (HNE) in the Balance. Cell Death Differ. 2013, 20, 1615–1630. [Google Scholar] [CrossRef]
- Linhart, K.; Bartsch, H.; Seitz, H.K. The Role of Reactive Oxygen Species (ROS) and Cytochrome P-450 2E1 in the Generation of Carcinogenic Etheno-DNA Adducts. Redox Biol. 2014, 3, 56–62. [Google Scholar] [CrossRef]
- Abdelmegeed, M.A.; Choi, Y.; Ha, S.-K.; Song, B.-J. Cytochrome P450-2E1 Promotes Aging-Related Hepatic Steatosis, Apoptosis and Fibrosis through Increased Nitroxidative Stress. Free Radic. Biol. Med. 2016, 91, 188–202. [Google Scholar] [CrossRef]
- Nair, J.; Srivatanakul, P.; Haas, C.; Jedpiyawongse, A.; Khuhaprema, T.; Seitz, H.K.; Bartsch, H. High Urinary Excretion of Lipid Peroxidation-Derived DNA Damage in Patients with Cancer-Prone Liver Diseases. Mutat. Res. 2010, 683, 23–28. [Google Scholar] [CrossRef]
- Neeley, W.L.; Essigmann, J.M. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem. Res. Toxicol. 2006, 19, 491–505. [Google Scholar] [CrossRef]
- Nassir, F.; Ibdah, J.A. Role of Mitochondria in Alcoholic Liver Disease. World J. Gastroenterol. 2014, 20, 2136–2142. [Google Scholar] [CrossRef]
- Grattagliano, I.; Russmann, S.; Diogo, C.; Bonfrate, L.; Oliveira, P.J.; Wang, D.Q.-H.; Portincasa, P. Mitochondria in Chronic Liver Disease. Curr. Drug Targets 2011, 12, 879–893. [Google Scholar] [CrossRef]
- García-Ruiz, C.; Kaplowitz, N.; Fernandez-Checa, J.C. Role of Mitochondria in Alcoholic Liver Disease. Curr. Pathobiol. Rep. 2013, 1, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Bratton, S.B.; Salvesen, G.S. Regulation of the Apaf-1–Caspase-9 Apoptosome. J. Cell Sci. 2010, 123, 3209–3214. [Google Scholar] [CrossRef] [PubMed]
- Harijith, A.; Ebenezer, D.L.; Natarajan, V. Reactive Oxygen Species at the Crossroads of Inflammasome and Inflammation. Front. Physiol. 2014, 5, 352. [Google Scholar] [CrossRef] [PubMed]
- Seki, E.; Brenner, D.A.; Karin, M. A Liver Full of JNK: Signaling in Regulation of Cell Function and Disease Pathogenesis, and Clinical Approaches. Gastroenterology 2012, 143, 307–320. [Google Scholar] [CrossRef]
- Pastorino, J.G.; Shulga, N.; Hoek, J.B. TNF-α-Induced Cell Death in Ethanol-Exposed Cells Depends on P38 MAPK Signaling but Is Independent of Bid and Caspase-8. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G503–G516. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondrial DNA Mutations in Disease and Aging. Environ. Mol. Mutagen. 2010, 51, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Lenaz, G. Mitochondria and Reactive Oxygen Species. Which Role in Physiology and Pathology? Adv. Exp. Med. Biol. 2012, 942, 93–136. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Lemasters, J.J. Mitophagy Selectively Degrades Individual Damaged Mitochondria after Photoirradiation. Antioxid. Redox Signal. 2011, 14, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, E.; Cavallini, G.; Donati, A.; Gori, Z. The Anti-Ageing Effects of Caloric Restriction May Involve Stimulation of Macroautophagy and Lysosomal Degradation, and Can Be Intensified Pharmacologically. Biomed. Pharmacother. 2003, 57, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Dolganiuc, A.; Thomes, P.G.; Ding, W.-X.; Lemasters, J.J.; Donohue, T.M. Autophagy in Alcohol-Induced Liver Diseases. Alcohol. Clin. Exp. Res. 2012, 36, 1301–1308. [Google Scholar] [CrossRef]
- Wu, D.; Cederbaum, A. Glutathione Depletion in CYP2E1-Expressing Liver Cells Induces Toxicity Due to the Activation of P38 Mitogen-Activated Protein Kinase and Reduction of Nuclear Factor-ΚB DNA Binding Activity. Mol. Pharmacol. 2004, 66, 749–760. [Google Scholar] [CrossRef]
- Albano, E. Oxidative Mechanisms in the Pathogenesis of Alcoholic Liver Disease. Mol. Aspects Med. 2008, 29, 9–16. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Morgan, M.J.; Choksi, S.; Liu, Z.-G. TNF-Induced Activation of the Nox1 NADPH Oxidase and Its Role in the Induction of Necrotic Cell Death. Mol. Cell 2007, 26, 675–687. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Wang, H.; Xie, Q.; Xu, Y. Notch1-Nuclear Factor ΚB Involves in Oxidative Stress-Induced Alcoholic Steatohepatitis. Alcohol Alcohol. 2014, 49, 10–16. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R.; Szabo, G. Interleukin-1 and Inflammasomes in Alcoholic Liver Disease/Acute Alcoholic Hepatitis and Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis. Hepatology 2016, 64, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, J.; Schroder, K. NLRP3 Inflammasome Activation: The Convergence of Multiple Signalling Pathways on ROS Production? Nat. Rev. Immunol. 2010, 10, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, W.; Zhong, W.; Sun, X.; Zhou, Z. Pharmacological Inhibition of NOX4 Ameliorates Alcohol-Induced Liver Injury in Mice through Improving Oxidative Stress and Mitochondrial Function. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Rusyn, I.; Yin, M.; Gäbele, E.; Yamashina, S.; Dikalova, A.; Kadiiska, M.B.; Connor, H.D.; Mason, R.P.; Segal, B.H.; et al. NADPH Oxidase-Derived Free Radicals Are Key Oxidants in Alcohol-Induced Liver Disease. J. Clin. Investig. 2000, 106, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Pritchard, M.T.; McMullen, M.R.; Wang, Q.; Nagy, L.E. Chronic Ethanol Feeding Increases Activation of NADPH Oxidase by Lipopolysaccharide in Rat Kupffer Cells: Role of Increased Reactive Oxygen in LPS-Stimulated ERK1/2 Activation and TNF-Alpha Production. J. Leukoc. Biol. 2006, 79, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Milic, S.; Mikolasevic, I.; Orlic, L.; Devcic, E.; Starcevic-Cizmarevic, N.; Stimac, D.; Kapovic, M.; Ristic, S. The Role of Iron and Iron Overload in Chronic Liver Disease. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 2144–2151. [Google Scholar] [CrossRef]
- Purohit, V.; Russo, D.; Salin, M. Role of Iron in Alcoholic Liver Disease: Introduction and Summary of the Symposium. Alcohol Fayettev. N 2003, 30, 93–97. [Google Scholar] [CrossRef]
- Wen, Q.; Liu, J.; Kang, R.; Zhou, B.; Tang, D. The Release and Activity of HMGB1 in Ferroptosis. Biochem. Biophys. Res. Commun. 2019, 510, 278–283. [Google Scholar] [CrossRef]
- Filomeni, G.; De Zio, D.; Cecconi, F. Oxidative Stress and Autophagy: The Clash between Damage and Metabolic Needs. Cell Death Differ. 2015, 22, 377–388. [Google Scholar] [CrossRef]
- Choi, D.W.; Kim, S.Y.; Kim, S.K.; Kim, Y.C. Factors Involved in Hepatic Glutathione Depletion Induced by Acute Ethanol Administration. J. Toxicol. Environ. Health A 2000, 60, 459–469. [Google Scholar] [CrossRef]
- Mantena, S.K.; King, A.L.; Andringa, K.K.; Landar, A.; Darley-Usmar, V.; Bailey, S.M. Novel Interactions of Mitochondria and Reactive Oxygen/Nitrogen Species in Alcohol Mediated Liver Disease. World J. Gastroenterol. 2007, 13, 4967–4973. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, V.; Hay, N. Molecular Pathways: Reactive Oxygen Species Homeostasis in Cancer Cells and Implications for Cancer Therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 4309–4314. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS Signaling under Metabolic Stress: Cross-Talk between AMPK and AKT Pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef]
- Meijer, A.J.; Codogno, P. Regulation and Role of Autophagy in Mammalian Cells. Int. J. Biochem. Cell Biol. 2004, 36, 2445–2462. [Google Scholar] [CrossRef]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in Mammalian Cells: Revisiting a 40-Year-Old Conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Chaperone-Mediated Autophagy: A Unique Way to Enter the Lysosome World. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Ding, W.-X. Mechanisms, Pathophysiological Roles and Methods for Analyzing Mitophagy—Recent Insights. Biol. Chem. 2018, 399, 147–178. [Google Scholar] [CrossRef] [PubMed]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Garcia-Macia, M.; Santos-Ledo, A.; Leslie, J.; Paish, H.L.; Collins, A.L.; Scott, R.S.; Watson, A.; Burgoyne, R.A.; White, S.; French, J.; et al. A Mammalian Target of Rapamycin-Perilipin 3 (MTORC1-Plin3) Pathway Is Essential to Activate Lipophagy and Protects Against Hepatosteatosis. Hepatology 2021, 74, 3441–3459. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Levine, B. To Be or Not to Be? How Selective Autophagy and Cell Death Govern Cell Fate. Cell 2014, 157, 65–75. [Google Scholar] [CrossRef]
- Kumar, A.V.; Mills, J.; Lapierre, L.R. Selective Autophagy Receptor P62/SQSTM1, a Pivotal Player in Stress and Aging. Front. Cell Dev. Biol. 2022, 10, 793328. [Google Scholar] [CrossRef]
- Mizushima, N. The Role of the Atg1/ULK1 Complex in Autophagy Regulation. Curr. Opin. Cell Biol. 2010, 22, 132–139. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Høyer-Hansen, M.; Jäättelä, M. AMP-Activated Protein Kinase: A Universal Regulator of Autophagy? Autophagy 2007, 3, 381–383. [Google Scholar] [CrossRef]
- Liang, J.; Shao, S.H.; Xu, Z.-X.; Hennessy, B.; Ding, Z.; Larrea, M.; Kondo, S.; Dumont, D.J.; Gutterman, J.U.; Walker, C.L.; et al. The Energy Sensing LKB1-AMPK Pathway Regulates P27(Kip1) Phosphorylation Mediating the Decision to Enter Autophagy or Apoptosis. Nat. Cell Biol. 2007, 9, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes Form at ER–Mitochondria Contact Sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-Binding Proteins, Atg14L and Rubicon, Reciprocally Regulate Autophagy at Different Stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.-Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.-L. ULK1 Induces Autophagy by Phosphorylating Beclin-1 and Activating VPS34 Lipid Kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef]
- Obara, K.; Ohsumi, Y. Dynamics and Function of PtdIns(3)P in Autophagy. Autophagy 2008, 4, 952–954. [Google Scholar] [CrossRef] [PubMed]
- Polson, H.E.J.; de Lartigue, J.; Rigden, D.J.; Reedijk, M.; Urbé, S.; Clague, M.J.; Tooze, S.A. Mammalian Atg18 (WIPI2) Localizes to Omegasome-Anchored Phagophores and Positively Regulates LC3 Lipidation. Autophagy 2010, 6, 506–522. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, S.R.; Mizushima, N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Molecular Dissection of Autophagy: Two Ubiquitin-like Systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The Autophagosome: Origins Unknown, Biogenesis Complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- Tan, C.T.; Soh, N.J.H.; Chang, H.-C.; Yu, V.C. P62/SQSTM1 in Liver Diseases: The Usual Suspect with Multifarious Identities. FEBS J. 2021, 290, 892–912. [Google Scholar] [CrossRef]
- Takahashi, Y.; He, H.; Tang, Z.; Hattori, T.; Liu, Y.; Young, M.M.; Serfass, J.M.; Chen, L.; Gebru, M.; Chen, C.; et al. An Autophagy Assay Reveals the ESCRT-III Component CHMP2A as a Regulator of Phagophore Closure. Nat. Commun. 2018, 9, 2855. [Google Scholar] [CrossRef]
- Takahashi, Y.; Liang, X.; Hattori, T.; Tang, Z.; He, H.; Chen, H.; Liu, X.; Abraham, T.; Imamura-Kawasawa, Y.; Buchkovich, N.J.; et al. VPS37A Directs ESCRT Recruitment for Phagophore Closure. J. Cell Biol. 2019, 218, 3336–3354. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The Hairpin-Type Tail-Anchored SNARE Syntaxin 17 Targets to Autophagosomes for Fusion with Endosomes/Lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef]
- Shen, Q.; Shi, Y.; Liu, J.; Su, H.; Huang, J.; Zhang, Y.; Peng, C.; Zhou, T.; Sun, Q.; Wan, W.; et al. Acetylation of STX17 (Syntaxin 17) Controls Autophagosome Maturation. Autophagy 2021, 17, 1157–1169. [Google Scholar] [CrossRef]
- Hubert, V.; Peschel, A.; Langer, B.; Gröger, M.; Rees, A.; Kain, R. LAMP-2 Is Required for Incorporating Syntaxin-17 into Autophagosomes and for Their Fusion with Lysosomes. Biol. Open 2016, 5, 1516–1529. [Google Scholar] [CrossRef]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Iorio, R.; Celenza, G.; Petricca, S. Mitophagy: Molecular Mechanisms, New Concepts on Parkin Activation and the Emerging Role of AMPK/ULK1 Axis. Cells 2021, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Holmström, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-Mediated Mitophagy Is Dependent on VDAC1 and P62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Narendra, D.P. Mechanisms of Mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial Membrane Potential Regulates PINK1 Import and Proteolytic Destabilization by PARL. J. Cell Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, M.; Kujuro, Y.; Okatsu, K.; Koyano, F.; Kosako, H.; Kimura, M.; Suzuki, N.; Uchiyama, S.; Tanaka, K.; Matsuda, N. Parkin-Catalyzed Ubiquitin-Ester Transfer Is Triggered by PINK1-Dependent Phosphorylation. J. Biol. Chem. 2013, 288, 22019–22032. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Johansen, T. P62/SQSTM1: A Missing Link between Protein Aggregates and the Autophagy Machinery. Autophagy 2006, 2, 138–139. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kominami, E.; Tanaka, K.; Komatsu, M. Selective Turnover of P62/A170/SQSTM1 by Autophagy. Autophagy 2008, 4, 1063–1066. [Google Scholar] [CrossRef]
- Rodriguez, Y.; Dunfield, J.; Roderique, T.; Ni, H.-M. Liver-Adipose Tissue Crosstalk in Alcohol-Associated Liver Disease: The Role of MTOR. Liver Res. 2022, 6, 227–237. [Google Scholar] [CrossRef]
- Li, Y.; Chao, X.; Wang, S.; Williams, J.A.; Ni, H.-M.; Ding, W.-X. Role of Mechanistic Target of Rapamycin and Autophagy in Alcohol-Induced Adipose Atrophy and Liver Injury. Am. J. Pathol. 2020, 190, 158–175. [Google Scholar] [CrossRef]
- Sakane, S.; Hikita, H.; Shirai, K.; Myojin, Y.; Sasaki, Y.; Kudo, S.; Fukumoto, K.; Mizutani, N.; Tahata, Y.; Makino, Y.; et al. White Adipose Tissue Autophagy and Adipose-Liver Crosstalk Exacerbate Nonalcoholic Fatty Liver Disease in Mice. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1683–1699. [Google Scholar] [CrossRef] [PubMed]
- Sid, B.; Verrax, J.; Calderon, P.B. Role of AMPK Activation in Oxidative Cell Damage: Implications for Alcohol-Induced Liver Disease. Biochem. Pharmacol. 2013, 86, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Zhu, M.; Zhou, S.; Feng, W.; Chen, H. Rapamycin-Loaded MPEG-PLGA Nanoparticles Ameliorate Hepatic Steatosis and Liver Injury in Non-Alcoholic Fatty Liver Disease. Front. Chem. 2020, 8, 407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xue, X.; Wang, C.; Wang, J.; Peng, C.; Li, Y. Emerging Roles of Sirtuins in Alleviating Alcoholic Liver Disease: A Comprehensive Review. Int. Immunopharmacol. 2022, 108, 108712. [Google Scholar] [CrossRef]
- Rafiee, S.; Mohammadi, H.; Ghavami, A.; Sadeghi, E.; Safari, Z.; Askari, G. Efficacy of Resveratrol Supplementation in Patients with Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis of Clinical Trials. Complement. Ther. Clin. Pract. 2021, 42, 101281. [Google Scholar] [CrossRef]
- Nguyen-Khac, E.; Thevenot, T.; Piquet, M.-A.; Benferhat, S.; Goria, O.; Chatelain, D.; Tramier, B.; Dewaele, F.; Ghrib, S.; Rudler, M.; et al. Glucocorticoids plus N -Acetylcysteine in Severe Alcoholic Hepatitis. N. Engl. J. Med. 2011, 365, 1781–1789. [Google Scholar] [CrossRef]
- Morley, K.C.; Baillie, A.; Van Den Brink, W.; Chitty, K.E.; Brady, K.; Back, S.E.; Seth, D.; Sutherland, G.; Leggio, L.; Haber, P.S. N-Acetyl Cysteine in the Treatment of Alcohol Use Disorder in Patients with Liver Disease: Rationale for Further Research. Expert Opin. Investig. Drugs 2018, 27, 667–675. [Google Scholar] [CrossRef]
- Ni, Y.-H.; Huo, L.-J.; Li, T.-T. Antioxidant Axis Nrf2-Keap1-ARE in Inhibition of Alcoholic Liver Fibrosis by IL-22. World J. Gastroenterol. 2017, 23, 2002–2011. [Google Scholar] [CrossRef]
- Lu, C.; Xu, W.; Zhang, F.; Shao, J.; Zheng, S. Nrf2 Knockdown Disrupts the Protective Effect of Curcumin on Alcohol-Induced Hepatocyte Necroptosis. Mol. Pharm. 2016, 13, 4043–4053. [Google Scholar] [CrossRef]
- Ospina, R.; Marmolejo-Ramos, F. Performance of Some Estimators of Relative Variability. Front. Appl. Math. Stat. 2019, 5, 43. [Google Scholar] [CrossRef]

| First Author, Year | Models and Methods | Summary of Effects |
|---|---|---|
| Lu et al., 2021 [31] | AML-12 cells treated with 200 mM ethanol over 24 h. | AMPK enhances mitophagy in hepatocytes, and the AMPK–NFE2L2–UQCRC2 axis regulates liver mitophagy. |
| Chen et al., 2021 [32] | Huh7 cells and murine primary hepatocytes treated with 50–100 mM ethanol over 24 h. | Acute ethanol exposure induces NOX4 and CYP2E1 overexpression and significantly increases autophagy. Antioxidants efficiently block CYP2E1- and NOX4-mediated autophagy induction. |
| Zhao et al., 2021 [33] | HepG2 cells treated with 0, 100, 200, 300, 400, and 500 mM alcohol over 12 h. | Alcohol induces reversible ferroptosis, which is significantly reduced by ferrostatin-1. Inhibiting autophagy protects HepG2 cells against alcohol-induced ferroptosis by activating the p62−Keap1−Nrf2 pathway. |
| Liu et al., 2019 [34] | HepG2 cells treated with 200 mmol/L ethanol for 12 h. | Autophagy is inhibited in ethanol-treated HepG2 cells. ALR-expressing HepG2 cells have increased survival rates, improved mitochondrial membrane potential, and increased ATP levels after ethanol treatment. This protection is associated with the upregulation of autophagy markers and downregulation of p62 and mTOR phosphorylation. |
| You et al., 2018 [35] | Mouse hepatocytes treated with 80 mM ethanol for 6 h. | SNX10 deficiency upregulates LAMP2A expression and CMA activation via Nrf2 and AMPK signaling in vitro, significantly ameliorating ethanol-induced liver damage and hepatic steatosis. |
| Mahli et al., 2015 [36] | Primary human hepatocytes and HepG2 cells treated with 50 mM alcohol over 16–24 h. | Alcohol and steatosis increase CYP2E1 levels and activity, lipid peroxidation, oxidative stress, pro-inflammatory gene expression, and autophagy via the CYP2E1 and JNK pathways. Autophagy improves the effects of alcohol on lipid accumulation and inflammatory gene expression in liver cells. |
| Thomes et al., 2013 [37] | Hep G2 cells treated with 50 mM ethanol over 24 h. | Ethanol treatment increases LC3-II expression and decreases its degradation in a dose-dependent manner depending on ADH and CYP2E1 expression. Blocking ethanol oxidation and ROS production prevents the enhancement of LC3-II expression. Direct exposure to acetaldehyde enhances LC3-II content. |
| Ding et al., 2010 [38] | Murine hepatocytes and HepG2 cells treated with 40, 80, and 160 mM ethanol for 24 h. | Ethanol-induced autophagy requires ethanol metabolism, ROS generation, and mTOR signaling inhibition in vitro. It is selective for cells with damaged mitochondria and accumulated lipid droplets (but not long-lived proteins) and protects cells from ethanol’s toxic effects. Increasing autophagy reduces acute ethanol hepatotoxicity and steatosis. |
| First Author, Year | Models and Methods | Summary of Effects |
|---|---|---|
| Guo et al., 2015 [39] | HepG2 cells treated with 100 mM ethanol for 4 days. | Ethanol and acetaldehyde increase IL-6 and IFN-γ levels and suppress autophagy in ADH1-expressing HepG2 cells. Lysosomal inhibitors mimic ethanol-induced p62 accumulation. |
| Wu et al., 2012 [40] | HepG2 cells treated with 100 mM ethanol for 8 days. | Inhibiting autophagy enhances ethanol hepatotoxicity, steatosis, and oxidative stress in CYP2E1-expressing HepG2 cells. These cells show increased fat accumulation and oxidative stress but decreased autophagy. The antioxidant N-acetylcysteine and CYP2E1 inhibition blunt these effects. |
| Wu et al., 2010 [41] | HepG2 cells treated with 50, 100, and 150 mM ethanol for 4–5 days. | CYP2E1-expressing HepG2 cells have exacerbated lipid and TG accumulation and increased p62 levels. HepG2 cells show increased autophagy. Inhibiting autophagy increases lipid accumulation and TG levels in HepG2 cells and, to a lesser extent, in CYP2E1-expressing HepG2 cells. Ethanol induces CYP2E1 activity and oxidative stress in CYP2E1-expressing HepG2 cells. |
| First Author, Year | Models and Methods | Summary of Effects |
|---|---|---|
| Samuvel et al., 2022 [42] | Male C57BL/6 mice gavaged one dose ethanol (2–6 g/kg in normal saline, 20 µL/g body weight). | Acute ethanol treatment induces dose-dependent mitochondrial depolarization, leading to type 2 mitophagy sequestration (probably through the PINK1–Parkin pathway) and subsequent lysosomal processing. |
| Chen et al., 2021 [32] | Male C57BL/6 mice gavaged a total of 4.5 g/kg (body weight) of ethanol over 24 h. | Acute ethanol exposure induces autophagy and ROS-generating CYP2E1 and NOX4 enzymes. NOX4 and CYP2E1 overexpression significantly increases autophagy. Ethanol and H2O2 (but not acetaldehyde) induce autophagy in primary mouse hepatocytes. Antioxidants efficiently block CYP2E1- and NOX4-mediated autophagy induction. |
| Yan et al., 2019 [43] | Male/female C57BL/6 mice; ethanol (5 g/kg body weight) gavage for 16 h. | Atg5-KO mice are more susceptible to acute alcohol treatment, but liver damage is unexpectedly improved with the chronic plus binge model. |
| Liu et al., 2019 [34] | Male C57BL/6 mice gavaged a total of 10 mL/kg 55% ethanol over 12 h. | Mice overexpressing ALR have less liver damage with alcohol exposure, associated with the upregulation of autophagy markers and downregulation of p62 and mTOR phosphorylation. Autophagy is inhibited in ALR-KO mice. |
| Eid et al., 2016 [12] | Adult male Wistar rats given one intraperitoneal ethanol dose (40% v/v, 5 g/kg) over 24 h. | Ethanol induces a low level of hepatocyte apoptosis but enhances mitophagic vacuole formation (increased LC3 puncta formation and co-localization of Parkin and LC3). PINK1 and Parkin are located around damaged mitochondria in the hepatocytes of ethanol-treated rats with an enhanced formation of mitochondrial spheroids. |
| Williams et al., 2015 [44] | Male C57BL/6J mice gavaged with a total of 4.5 g/kg of ethanol per kg of body weight over 16 h. | Parkin prevents liver damage in an acute alcoholic model. Ethanol-fed Parkin-KO mice exhibit severe mitochondrial damage; reduced mitophagy, β-oxidation, mitochondrial respiration; and cytochrome C oxidase activity. |
| Manley et al., 2014 [45] | Male C57BL mice gavaged with a total of 4.5 g/kg ethanol per kg of body weight ethanol over 16 h. | FXR-KO mice have exacerbated hepatotoxicity, steatosis, decreased essential autophagy-related gene expression, increased Akt activation, and decreased FOXO3a activity. Ethanol treatment induces hepatic mitochondrial spheroid formation in FXR-KO, but not WT, mice. |
| Ni et al., 2013 [46] | Male C57BL/6 mice gavaged with a total of 4.5 g/kg ethanol per kg of body weight ethanol over 6, 12, and 16 h. | Ethanol-fed mice have increased mRNA and protein levels of autophagy-related genes in hepatocytes and FOXO3a activity. Suppressing FOXO3a activity in hepatocytes inhibits autophagy-related gene expression and enhances cell death, steatosis, and liver damage. A SIRT1 agonist enhances ethanol-induced autophagy-related gene expression by increasing FOXO3a deacetylation. |
| Lin et al., 2013 [47] | Intraperitoneal ethanol (33%, v/v, 1.2 g/kg body weight) injection over 10 min. | Macroautophagy is activated under acute conditions. Hepatic steatosis and liver damage are exacerbated by autophagy inhibition and alleviated by autophagy activation. |
| Yang et al., 2012 [48] | Male SV/129 and C57BL/6J mice intraperitoneally injected with ethanol (0.93 g/kg body weight) and later gavaged three times (total: 3.75 g/kg body weight) over 18 h. | WT mice show CYP2E1 activation; increased oxidative stress, JNK signaling, and SREBP expression; and decreased autophagy. Acute alcohol-induced fatty liver and oxidative stress are blunted in CYP2E1-KO and JNK inhibitor-treated mice. N-acetylcysteine decreases acute alcohol-induced oxidative stress, JNK activation, and steatosis but not CYP2E1 activation. Acute alcohol-induced fatty liver is the same in JNK1 and JNK2-KO mice as in WT mice. |
| Thomes et al., 2012 [49] | Female C57Bl/6 mice gavaged a total of 6 g/kg body weight ethanol over 12 h; precision-cut liver slices treated with 50 mM ethanol over 12–24 h. | Acute ethanol administration elevates autophagosomes without affecting hepatic proteasome activity. Liver slices show inhibited proteasome activity and enhanced autophagosome expression depending on ethanol oxidation. |
| Ding et al., 2010 [38] | Male C57BL/6 mice gavaged with a total of 4.5 g/kg ethanol per kg of body weight over 16 h. | Ethanol-induced autophagy in vivo requires ethanol metabolism, ROS generation, and mTOR signaling inhibition. Increasing autophagy reduces acute ethanol hepatotoxicity and steatosis. |
| First Author, Year | Models and Methods | Summary of Effects |
|---|---|---|
| Lu et al., 2021 [31] | NIAAA model with male C57BL/6J mice. | Alcohol induces low UQCRC2 expression, which is alleviated by AMPK. The AMPK–NFE2L2–UQCRC2 axis regulates liver mitophagy. |
| Guo et al., 2021 [50] | Lieber–DeCarli model with male C57BL/6J mice. Liver tissues from patients with alcoholic hepatitis were examined. | Palmitic acid in alcohol-fed mice induces ER stress and mTORC1-dependent LAMP2 suppression. mTORC1 signaling induction and CHOP were detected in patient livers. |
| Guo et al., 2021 [51] | Lieber–DeCarli model with male C57BL/6J mice. Liver tissues from patients with alcoholic hepatitis were examined. | The pathogenesis of ALD is mediated by hepatic free fatty acid accumulation, which suppresses the LAMP2–autophagy flux pathway through ER stress signaling. |
| Babuta et al., 2019 [52] | Lieber–DeCarli and NIAAA models with female C57BL/6 mice. Liver tissues from patients with alcoholic hepatitis were examined. | Chronic alcohol intake impairs autophagy in livers, decreasing mTOR and Rheb and increasing Beclin-l and Atg7 expression, disrupting autophagy at the lysosomal level by decreasing LAMP1 and LAMP2 expression. Alcohol increases miR-155 targeting of mTOR, Rheb, LAMP1, and LAMP2. miR-155-deficient mice are protected from alcohol-induced autophagy disruption and have attenuated exosome production. LAMP1/2 downregulation increases exosome release in hepatocytes in the presence and absence of alcohol. |
| Zhou et al., 2019 [53] | Female C57BL mice fed liquid diet with 4% (vol/vol) alcohol for 16 weeks. | Chronic alcohol consumption increases DNA-PKcs in the liver, leading to liver damage and mitochondrial dysfunction through p53 activation and defective mitophagy. |
| Yan et al., 2019 [43] | NIAAA model with male/female C57BL/6 mice. | Mice lacking the Atg7 gene had more liver damage from alcohol and were more susceptible to chronic plus binge drinking. Long-term autophagy deficiency worsened the liver’s response to alcohol. |
| Menk et al., 2018 [54] | Lieber–DeCarli model with male Wistar rats for 12 weeks. | Chronic alcohol consumption causes stress in the liver, impairs autophagy-related gene expression, disrupts autophagic flux, and increases apoptosis in the liver. |
| You et al., 2018 [35] | Lieber–DeCarli model with male FVB and C57BL/6J mice for 4 weeks. | SNX10 deficiency increases LAMP2A expression and CMA activation, improving liver damage and fat accumulation caused by alcohol through the activation of Nrf2 and AMPK signaling. |
| Chao et al., 2018 [55] | NIAAA model with male C57BL/6N mice. Liver tissues from patients with alcoholic hepatitis were examined. | Alcohol-fed mice had lower levels of TFEB, decreased lysosome and autophagy activity, and increased mTOR activation. Activating the TFEB pathway reversed these effects. Mice lacking TFEB or both TFEB and TFE3 had more severe liver damage from alcohol. Patient liver tissues had lower levels of nuclear TFEB than control tissues. |
| Kong et al., 2017 [56] | Lieber–DeCarli model with male C57BL6 mice for 16 days with intraperitoneal LPS injection (10 mg/kg) on the final day. | Alcohol-fed mice experienced fat accumulation, liver damage, and increased inflammation. LPS worsened alcohol-induced oxidative stress and reduced autophagy activity. |
| Williams et al., 2015 [44] | NIAAA model with male C57BL/6J mice. | Parkin prevented liver damage in chronic alcohol-fed mice. Mice lacking Parkin had severe mitochondrial damage, reduced mitophagy and mitochondrial function, and an impaired ability to adapt to alcohol treatment. |
| Guo et al., 2015 [39] | Female FVB mice fed a 4% (vol/vol) alcohol liquid diet for 6 weeks. | Chronic alcohol intake causes liver damage, disturbed fat metabolism, increased inflammation and oxidative stress, and decreased autophagy. Expressing the ALDH2 gene reduced these effects. Lysosomal inhibitors had the same effects as alcohol on p62 accumulation. |
| Lu and Cederbaum, 2015 [57] | Lieber–DeCarli model with male SV/129 mice for 4 weeks. | Inhibiting autophagy increased liver damage and fat accumulation in mice with normal or increased CYP2E1 levels but not in mice lacking CYP2E1. Autophagy did not affect CYP2E1 activity or induction by alcohol. Mice with normal or increased CYP2E1 levels had decreased autophagy-related gene expression and increased p62 levels. |
| King et al., 2014 [58] | Lieber–DeCarli model with male C57BL/6J mice. | Alcohol-treated mice experienced fat accumulation, increased autophagy, decreased mitochondrial function, and increased CypD levels. Their mitochondria were more sensitive to damage than those of mice lacking CypD. CypD deficiency impaired autophagy but did not prevent fat accumulation caused by alcohol. |
| Tan et al., 2013 [59] | Male C57/BL6 mice fed 5–20% (vol/vol) ethanol and a high-fat diet for 8 weeks. | Mice lacking the HFE gene had liver damage, fibrosis, and increased cell death. Iron overload in these mice caused stress responses and impaired autophagy-related gene expression and activity. |
| Lin et al., 2013 [47] | Lieber–DeCarli model with C57BL/6 mice for 4 weeks. | Macroautophagy was activated during chronic alcohol consumption. Inhibiting autophagy worsened liver damage and fat accumulation, while activating autophagy improved these conditions. |
| Wu et al., 2012 [40] | Male SV129 mice gavaged with a total of 3 g/kg body weight ethanol over 4 days. | Alcohol treatment caused liver damage, increased CYP2E1 levels, and oxidative stress in mice with normal or increased CYP2E1 levels but not in mice lacking CYP2E1. Alcohol impaired autophagy in mice with increased CYP2E1 levels. Inhibiting autophagy worsened alcohol-induced liver damage, fat accumulation, and oxidative stress in these mice. |
| Thomes et al., 2012 [49] | Lieber–DeCarli model with GFP-LC3 tg mice for 4–6 weeks. | Chronic alcohol-fed mice had reduced proteasome activity and increased autophagy markers in liver cells. Inhibiting the proteasome further increased autophagy markers. |
| First Author, Year | Models and Methods | Summary of Effect |
|---|---|---|
| Liang et al., 2019 [60] | Lieber–DeCarli model with female C57BL/6 mice and intraperitoneal LPS injection (1 mg/kg) on the final day. | Chronic alcohol feeding increased liver damage and inflammation in mice lacking the Atg7 gene in immune cells and increased inflammatory gene expression in normal mice. Mice lacking Atg7 experienced impaired mitochondrial function, increased oxidative stress, and increased inflammation. Silencing p62 or deleting Atg7 caused the accumulation of IRF1 and increased inflammatory gene expression. |
| Ilyas et al., 2019 [61] | Female C57BL/6J mice fed 5% ethanol liquid diet for 21 days with intraperitoneal LPS injection (7.5 mg/kg) on the final day. | Mice lacking the Atg5 gene in immune cells had similar fat accumulation compared to normal mice when fed alcohol but had increased liver damage, inflammation, and cell death. Blocking the IL-1 receptor reduced alcohol-induced inflammation. |
| Xie et al., 2018 [62] | HSC-T6 cells treated with 100 mmol/L alcohol. | The treatment increased autophagy and oxidative stress and activated HSCs. Inhibiting autophagy reversed HSC activation and reduced oxidative stress in HSCs. The Nrf2-Keap1-ARE pathway was involved in regulating HSC activation and oxidative stress through autophagy. |
| Kong et al., 2017 [56]. | RAW 264.7 cells treated with various alcohol doses for 48h plus LPS (100 ng/mL) for 6h. | The protective effects of autophagy are associated with decreased cellular MD2/TLR4 expression in RAW 264.7 cells. |
| Denaës et al., 2016 [63] | NIAAA model with C57BL/6N mice. | Mice lacking the CB2 gene in immune cells experienced worsened alcohol-induced inflammation and fat accumulation. Activating the CB2 receptor reduced alcohol-induced liver inflammation and fat accumulation in normal mice but not in mice lacking the ATG5 gene in immune cells. Macrophage autophagy mediated the protective effects of the CB2 receptor. |
| Hernández-Gea et al., 2013 [64] | Lieber–DeCarli model with C57/BL6 mice. | An increase in the UPR, as indicated by XBP1 mRNA splicing, triggered autophagy. The Nrf2-mediated antioxidant response was activated during ER stress. Blocking the IRE1α pathway in HSCs reduced their activation and autophagy activity, reducing fibrosis through a p38 MAPK-dependent mechanism. |
| Drug | Pharmacological Classes | Experimental Model | Main Pathways Involved | Disease Prevention or Potential Benefits |
|---|---|---|---|---|
| AT extract [65] | Flavonoids, phenolic compounds, steroidal glycosides, coumarins. | Intragastric administration of ethanol (5 g/kg b.d., 7 days) or carbon tetrachloride ± AT extract (50 and 150 mg/kg/d) to mice, HepG2 and SK-Hep-1 cells exposed to ethanol. | Induction of autophagy through the activation of Nrf2 and MAPK and increased HO-1 levels. | Reduced liver damage and histopathological changes via increased antioxidant activity. |
| ACE [66] | Basidiomycete triterpenoids, flavonoids, fatty acids, amino acids. | Administration of white wine (9.52 g/kg, 56°, 2 weeks) and ACE (75, 225, and 675 mg/kg, 2 weeks) to mice. | Reduced Akt/ NF-κB signalling. | Reduced alcohol-induced hepatotoxicity, oxidative stress, and regulation of AST, ALT, oxidation-related enzyme, inflammatory cytokine, and caspase levels. |
| BBD [67] | Traditional Chinese medicine. | Mice gavaged with ethanol (50%, 5 g/kg), pretreated with BBD (0.125, 0.25, and 0.5 g/kg). | Induction of autophagy through increased NRF2 expression and suppression of CYP450 2E1 induction. | BBD reduced alcohol-induced steatosis, hepatic lipid peroxidation, antioxidant depletion, and oxidative stress. |
| BSE [68] | High levels of flavonoids and polyphenols. | Lieber–DeCarli model for 10 days with intraperitoneal injection of 31.5% ethanol on the last day and BSE (100 and 200 mg/kg/d) gavage, cultured hepatocytes. | Autophagy induction via AMPK activation. | BSE decreased hepatic lipid accumulation and inflammatory macrophage infiltration; in vitro, it induced hepatic β-oxidation and reduced fatty acid synthesis. |
| Calcitriol [69] | Active form of vitamin D. | In vitro, human L02 hepatocytes were pretreated with 100 nM calcitriol, then stimulated acutely with 300 nM ethanol. | Induction of autophagy through the AMPK/mTOR signaling pathway and upregulation of ATG16L1. | Calcitriol alleviated ethanol-induced cytotoxicity and apoptosis caused by oxidative stress and mitochondrial damage in hepatocytes. |
| CBD [70] | Antagonist of CB1/CB2 receptor agonists (negative allosteric modulator of CB1, inverse agonist of CB2). | Mouse acute binge drinking model with intraperitoneal CBD injection (5 mg/kg, q 12 h), HepG2 (E47) cells exposed to ethanol ± CBD. | Induction of autophagy through the blunted activation of the JNK/MAPK pathway. | CBD prevented ethanol-induced autophagy reduction and reduced oxidative stress and acute alcohol-induced liver steatosis in mice. |
| CBZ [47] | Antiepileptic. | Lieber–DeCarli model ± intraperitoneal CBZ (25 mg/kg), chloroquine (60 mg/kg), or rapamycin (2 mg/kg) injection. | Enhanced mTOR-independent autophagy. | CBZ alleviated hepatic steatosis and liver damage and improved insulin sensitivity. |
| Carvacrol [71] | Monoterpenoid phenol. | Mouse model of acute ethanol intake with carvacrol pretreatment (10 mL/kg). | Induction of autophagy, likely through the inactivation of p38, and inhibition of cytochrome p450. | Carvacrol reduced the TG content and ethanol-induced liver histopathological changes. |
| CMZ [40,72,73] | Thiazole derivative. | Chronic ethanol intake mouse model with CMZ (50 mg/kg), acute ethanol intake mouse model ± CMZ (50 mg/kg). | Induction of autophagy through the activation of the AMPK, MAPK, and PI3K/Akt/GSK3β pathways, and inhibition of CYP2E1. | CMZ suppressed chronic ethanol-induced oxidative stress and pro-inflammatory cytokine production, attenuated acute ethanol-induced fatty liver. |
| Cilostazol [74] | Selective phosphodiesterase III inhibitor. | Acute alcohol intake rat model ± intraperitoneal cilostazol (10 mg/kg/d for 4 days; primary rat hepatocytes were examined. | Autophagy induction via AMPK pathway activation. | Cilostazol protected hepatocytes from apoptosis in vivo and in vitro. |
| Corosolic acid [75] | Pentacyclic triterpene acid extracted from Lagerstroemia speciosa. | Chronic ethanol intake mouse model (intragastric, 60%; 4.5, 6.5, and 9 g/kg/d for 4 weeks) ± corosolic acid (20%, 4 mL b.d., 5–12 weeks). HepG2 cells and BRL-3A liver cells were examined. | Induction of autophagy through the activation of the AMPK pathway and reduction of ROS levels. | Corosolic acid ameliorated alcoholic liver damage, reduced histopathological changes in vivo, and decreased ethanol-induced ROS elevation. |
| DMY [76] | Bioactive flavonoid from Ampelopsis grossedentata. | Lieber–DeCarli mouse model (1% 2 d, 2% 2 d, 4% 7 d, and 4% 6 weeks) ± oral DMY (75 and 150 mg/kg/d). | Induction of autophagy through the activation of the Keap-1/Nrf2 pathway and upregulation of p62. | DMY attenuated ethanol induced hepatic enzyme release, lipid peroxidation, TG accumulation, proinflammatory cytokine elevation, and histopathological changes while alleviating IL-1β and IL-6 elevation and pathological changes. |
| TAX [77] | Dihydroflavone. | Acute ethanol intake mouse model (intragastric) ± TAX (1, 5, and 25 mg/kg), HepG2 cells exposed to ethanol and TAX. | Induction of autophagy via AMPK activation and upregulated SIRT1 expression. | TAX reduced liver damage and inhibited alcohol-induced lipid accumulation in mouse livers. |
| Fisetin [78] | Plant polyphenol flavonoid. | Lieber–DeCarli mouse model ± fisetin; human primary HSCs co-cultured with ethanol. | Activation of autophagy through the activation of SIRT1 and inhibition of Sphk1-mediated ER stress. | Fisetin inhibited ER stress and improved alcohol-induced liver damage and fibrosis through the suppression of HSC activation. |
| Fucoidan [79,80] | Long-chain sulfated polysaccharide from various brown algae species. | Mice gavaged with ethanol (56%: 6 [7] mL/kg for 4 weeks then 8 [9] mL/kg for 12 [16] weeks) with daily intragastric fucoidan (100 and 200 [150 and 300] mg/kg). | Induction of autophagy via AMPKα1, SIRT1, and p62/Nrf2/Keap1/SLC7A11 pathway upregulation. | Fucoidan inhibited alcohol-induced steatosis, inflammation, oxidative stress, and histopathological changes; reduced the serum ferritin level; and alleviated liver iron deposition. |
| GMC [81] | Coumarin extracted from licorice. | Chronic and acute ethanol gavage mouse models ± GMC. | Induction of autophagy through the activation of Nrf2 and p38. | GCM prevented acute and chronic ethanol-induced hepatic steatosis in vivo and alleviated oxidative stress. |
| Green tea extract [82] | Tea polyphenols. | Chronic ethanol intake mouse model (50%, 15 mL/kg, intragastric) ± three doses extract (50, 120, and 300 mg tea polyphenols/kg body weight) q.d. for 4 weeks. | Induction of autophagy through increased Nrf2 activation and decreased Keap1 expression. | Dose-dependent improvement of functional and histopathological changes in hepatocytes after ethanol intake. |
| HEPFYGNEGALR (P03) [83] | Peptide isolated from Apostichopus japonicus. | Mice were given one intragastric dose of 50% ethanol (12 mL/kg) after oral P03 (20 mg/kg/d) or spermidine for 35 days and compared with controls without ethanol. | Induction of autophagy through the activation of the Nrf2/HO-1 pathway and blockade of NF-κB nuclear translocation. | Reduced hepatomegaly, liver inflammation, lipid droplet accumulation and increased antioxidant enzyme activities. |
| KD [84] | Major active ingredient extracted from Anoectochilus roxburghii. | Lieber–DeCarli mouse model ± 5% carbon tetrachloride in olive oil (intraperitoneal injection) and KD (20 40 mg/kg) or silymarin (80 mg/kg); control without ethanol. | Autophagy induction through AMPK activation. | KD alleviated alcoholic liver damage by reducing oxidative stress and lipid accumulation. |
| Lanthanum nitrate [85] | Rare earth element. | Acute ethanol intake mouse model (50%, 12 mL/kg, intragastric) after lanthanum nitrate (0.1, 0.2, 1.0, 2.0, and 20.0 mg/kg) administration for 30 days. | Induction of autophagy through the activation of the Keap1/Nrf2/p62 pathway. | Improved redox homeostasis and histopathological changes. |
| Melatonin [86] | Pineal gland hormone. | Acute ethanol intake mouse model (0.75 g/kg, intraperitoneal) ± melatonin (10 mg/kg, intraperitoneal) for 10 days. | Improved mitochondrial oxidation of NADH and decreased mitochondrial ability to oxidize FAD. | Prevented lysosomal destruction of liver tissue by limiting the increased activity of lysosomal enzymes and the resulting oxidative stress. |
| NAC [38] | Amino acid modified from L-cysteine. | Acute ethanol intake mouse model. Murine hepatocytes were exposed to ethanol ± NAC. | Reduction of autophagy via mTOR activation and reversed ROS levels. | NAC reduced TG and TBARS contents and ROS stress and reversed ethanol-induced mTOR inhibition. |
| PLT [87] | Protoberberine alkaloid. | Mouse hepatocytes were exposed to 75% alcohol for 2–3 weeks and PLT (0, 20, 50, 100, 150 and 200 μg/mL). | Induction of autophagy via AMPK/mTOR pathway activation. | PLT reduced ethanol-induced liver cell damage by inhibiting hepatocyte apoptosis through autophagy promotion. |
| PCP [88] | Gallic acid, lutein, quercetin, luteolin, apigenin, among others. | Acute ethanol intake (350 mM for 32 h) and/or PCP (100, 50 and 25 μg/mL) with zebrafish larvae. | Induction of autophagy through activating the AMPK/p62/Nrf2/mTOR signaling pathways and reduced oxidative stress. | PCP ameliorated ethanol-induced liver function damage and fat accumulation. |
| Procyanidin [89] | Polyphenol flavonoid. | Acute ethanol intake mouse model ± procyanidin (50 mg/kg for 11 days). | Induction of autophagy through increased LC3-II and reduced p62 levels, reduced ROS levels, and elevated GSH content. | Procyanidin eliminated lipid droplets and damaged mitochondria, thereby reducing hepatic lipid deposition and ROS overproduction. |
| Quercetin [90,91,92,93,94,95] | Plant-derived flavonoid. | Chronic and chronic plus single binge ethanol mouse models ± quercetin (control group without ethanol), L02 cells exposed to 3% ethanol for 24 h plus quercetin (20, 40, and 80 μM) for 24 h (control group without ethanol); transgenic zebrafish larvae were given quercetin (25, 50, and 100 μM for 48 h 3 days after fertilization) and ethanol for 32 h. | Induction of autophagy through FOXO3a activation and reversal of ethanol’s effects on AMPK and ERK2. | Quercetin inhibited inflammation and alleviated chronic ethanol-induced hepatic mitochondrial damage via mitophagy activation. |
| Rapamycin (sirolimus) [15,40,47] | Selective immunosuppressant (mTOR inhibitor). | Chronic and acute ethanol intake mouse models ± rapamycin. | Induction of autophagy via inhibition of mTOR signaling. | Rapamycin reduced ethanol-induced steatosis. |
| Resveratrol [46,96,97] | Dietary polyphenol. | Lieber–DeCarli mouse model plus acute ethanol binge, HepG2 cells exposed to oleic acid and alcohol. | Induction of autophagy via increased sirtuin-1 signaling. | Resveratrol increased the number of autophagosomes, reduced hepatic lipid accumulation, and protected against alcoholic liver steatosis. |
| SaIA [98] | Phenolic carboxylic acid extracted from Salvia miltiorrhiza. | Lieber–DeCarli mouse model with intragastric SaIA (8 and 16 mg/kg/d); AML-12 hepatocytes were examined. | Induction of autophagy via SIRT1 activation. | SaIA restored autophagosome-lysosome fusion, protected the liver from chronic ethanol exposure, decreased transaminase levels, attenuated histopathological liver damage, and prevented ethanol-induced liver cell damage in vitro. |
| Se-SP [99] | Microalga of the cyanobacterial class with chemical element enrichment. | Chronic ethanol intake mouse model (30%, 10 mL/kg by gavage for 15 days) and intragastric Se-SP (100, 200, and 400 mg/kg/d for 42 days). | Reduction of autophagy via decreased LC3 and increased p70s6k expression, and decreased p53, caspase 1, and 3 expression. | Se-SP protected against alcoholic liver damage by increasing antioxidant enzyme levels, inhibiting DNA damage and apoptosis, and inducing pyrosis. |
| Silibinin [100] | Flavonoid glycoside. | HepG2 and HL7702 cells exposed to ethanol or acetaldehyde and silibinin. | Induction of autophagy via PINK1 and Parkin activation. | Silibinin inhibited ethanol-induced ferroptosis, resolved oxidative stress, and reduced iron levels. |
| Simvastatin [101] | Statin. | Chronic ethanol intake rat model ± simvastatin (10 mg/kg/d). | Induction os autophagy, selectively inhibited HMG-CoA reductase. | Simvastatin ameliorated alcohol-induced liver histopathological changes, transaminase elevation, attenuated oxidative stress, and inflammation. |
| Sulforaphane [102] | Isothiocyanate derived from glucoraphanin present in Brassica. | Acute binge drinking mouse model ± sulforaphane (0.05 g/kg for 5 days), HepG2 (E47) cells treated with or without 100 mM ethanol ± 6 uM sulforaphane. | Induction of autophagy via Nrf2 activation. | Sulforaphane prevented binge ethanol–induced oxidative stress and steatosis in CYP2E1 KI mice and lipid accumulation in HepG2 (E47) cells. |
| Tangeretin [103] | Flavonoid derived from citrus peel. | Chronic binge drinking mouse model ± tangeretin (20 and 40 mg/kg). | Induction of autophagy via AMPK/Ulk1 signalling pathway activation. | Tangeretin dose-dependently normalized serum ALT and AST levels, liver weight, and serum and liver triacylglycerol contents; restored mitochondrial respiratory function; and suppressed steatosis. |
| TMP [104] | Alkylpyrazine extracted from Ligusticum wallichii. | Chronic ethanol intake mouse model ± TMP, LO2 cells exposed to ethanol (100 mM) and/or TMP (40 μM for 24 h). | Induction of autophagy via increased UQCRC2 expression and RIPK1/RIPK3 necrosome activation. | Reduced necroptosis and leakage of damage-associated molecular patterns and promoted the clearance of impaired mitochondria. |
| Torin 1 [55,105] | Pyridoquinoline (ATP-competitive mTOR kinase inhibitor). | Chronic plus binge ethanol intake mouse model ± torin 1. | Induction of autophagy via inhibition of mTORC1 and mTORC2 and increased hepatic TFEB levels. | Torin 1 reduced steatosis and liver damage induced by ethanol. |
| UDCA [106] | Hydrophilic bile acid (non–FXR agonistic). | Lieber–DeCarli mouse model ± UDCA. | Attenuated NF-κB activation. | UDCA attenuated and prevented the progression of alcoholic hepatic cholestasis. |
| Zinc (Zn) [107] | Chemical element. | VL-17A cells exposed to 100 mM ethanol for 24 h and 0, 10, 20, and 40 μM Zn for 48 h. | Induction of autophagy via ERK1/2 activation. | Zn depletion significantly suppressed autophagy, Zn exposure stimulated autophagy, cotreatment with ethanol, and 40 μM Zn had an additive effect on autophagy induction. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salete-Granado, D.; Carbonell, C.; Puertas-Miranda, D.; Vega-Rodríguez, V.-J.; García-Macia, M.; Herrero, A.B.; Marcos, M. Autophagy, Oxidative Stress, and Alcoholic Liver Disease: A Systematic Review and Potential Clinical Applications. Antioxidants 2023, 12, 1425. https://doi.org/10.3390/antiox12071425
Salete-Granado D, Carbonell C, Puertas-Miranda D, Vega-Rodríguez V-J, García-Macia M, Herrero AB, Marcos M. Autophagy, Oxidative Stress, and Alcoholic Liver Disease: A Systematic Review and Potential Clinical Applications. Antioxidants. 2023; 12(7):1425. https://doi.org/10.3390/antiox12071425
Chicago/Turabian StyleSalete-Granado, Daniel, Cristina Carbonell, David Puertas-Miranda, Víctor-José Vega-Rodríguez, Marina García-Macia, Ana Belén Herrero, and Miguel Marcos. 2023. "Autophagy, Oxidative Stress, and Alcoholic Liver Disease: A Systematic Review and Potential Clinical Applications" Antioxidants 12, no. 7: 1425. https://doi.org/10.3390/antiox12071425
APA StyleSalete-Granado, D., Carbonell, C., Puertas-Miranda, D., Vega-Rodríguez, V.-J., García-Macia, M., Herrero, A. B., & Marcos, M. (2023). Autophagy, Oxidative Stress, and Alcoholic Liver Disease: A Systematic Review and Potential Clinical Applications. Antioxidants, 12(7), 1425. https://doi.org/10.3390/antiox12071425






