Application of Multi-Omics Technologies to the Study of Phytochromes in Plants
Abstract
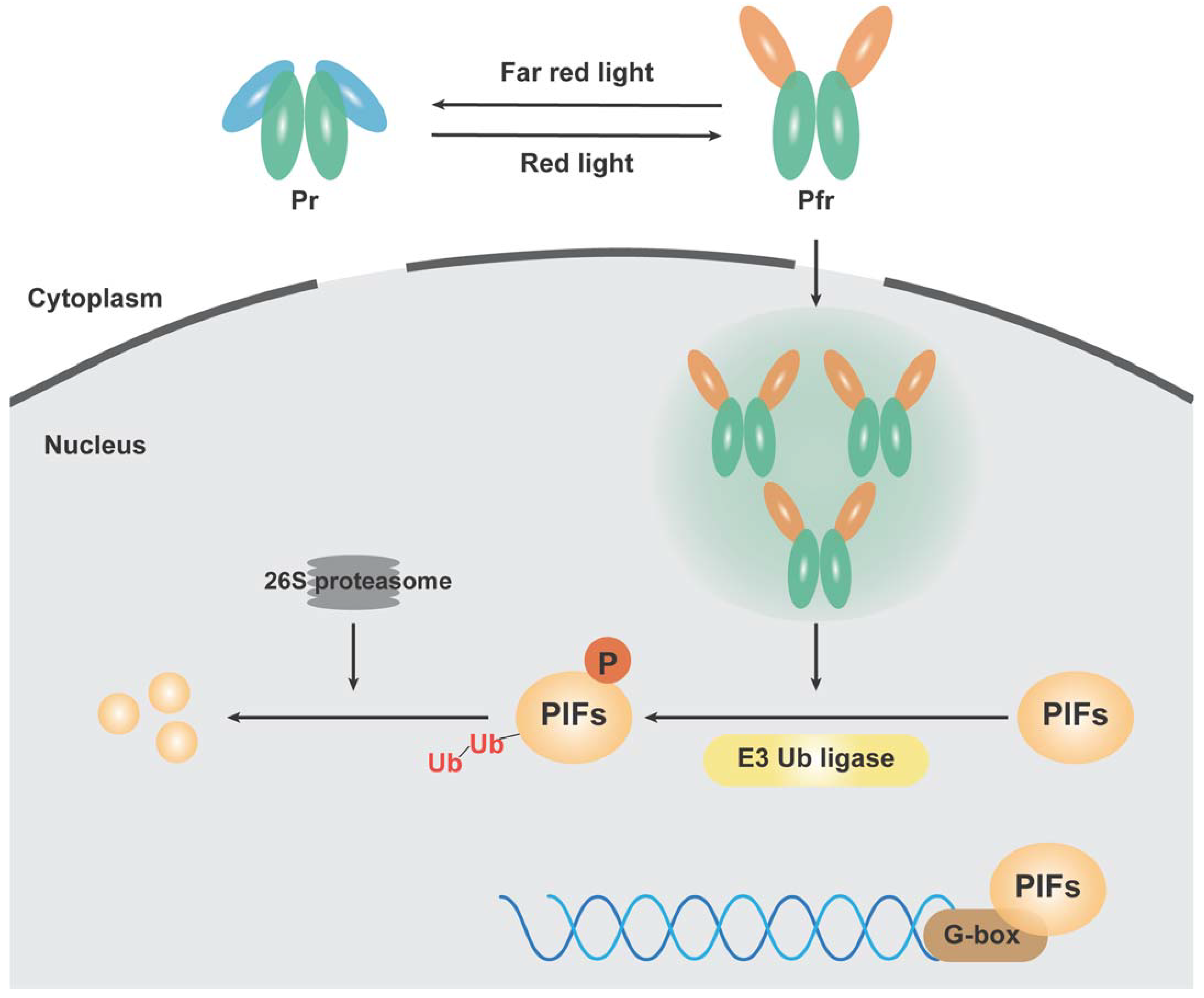
:1. Introduction
2. Analysis of the Mechanism of Phytochrome Action Based on Whole-Genome Transcriptome
3. Application of Proteomics in the Analysis of Phytochrome
4. Metabolomic Analysis to Assess the Effects of Phytochromes on Plants
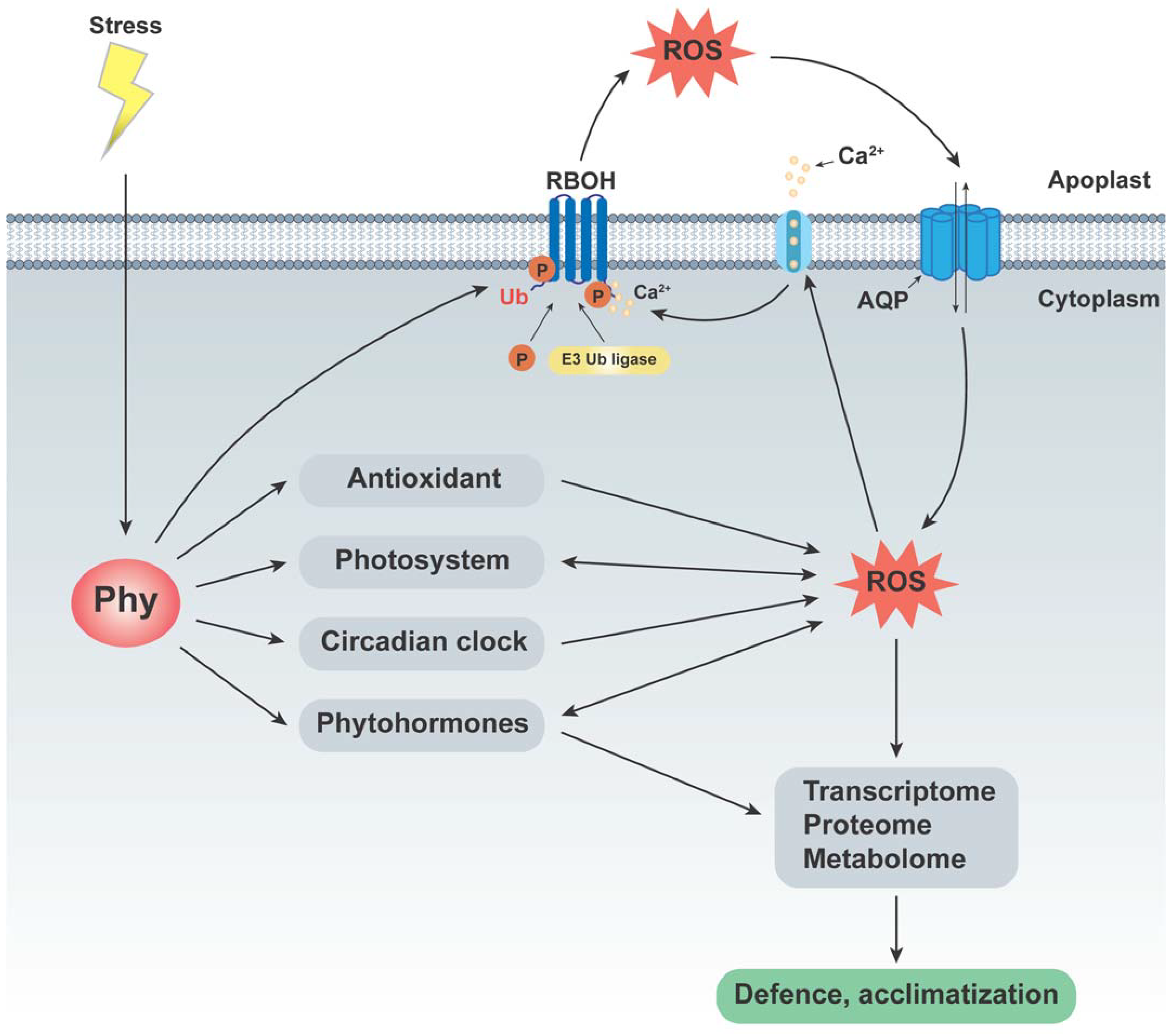
5. Application of Omics to Analyze Epigenetic Changes Associated with Phytochromes
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Liang, T.; Liu, H. How plants coordinate their development in response to light and temperature signals. Plant Cell 2022, 34, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Quail, P.H. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002, 3, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Galvão, V.C.; Fankhauser, C. Sensing the light environment in plants: Photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 2015, 34, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.E.; Schultz, T.F.; Milnamow, M.; Kay, S.A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 2000, 101, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.J.; Liu, D. Cryptochromes: Blue light receptors for plants and animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Wada, M. Evolution of Three LOV Blue Light Receptor Families in Green Plants and Photosynthetic Stramenopiles: Phototropin, ZTL/FKF1/LKP2 and Aureochrome. Plant Cell Physiol. 2013, 54, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Briggs, W.R. Phototropins 1 and 2: Versatile plant blue-light receptors. Plant Sci. 2002, 7, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef]
- Briggs, W.R. Phototropism: Some history, some puzzles, and a look ahead. Plant Physiol. 2014, 164, 13–23. [Google Scholar] [CrossRef]
- Hohm, T.; Preuten, T.; Fankhauser, C. Phototropism: Translating light into directional growth. Am. J. Bot. 2013, 100, 47–59. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Q.; Wang, X.; Zuo, Z.; Oka, Y.; Lin, C. New insights into the mechanisms of phytochrome-cryptochrome coaction. New Phytol. 2018, 217, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Nagatani, A. Phytochrome: Structural basis for its functions. Curr. Opin. Plant Biol. 2010, 13, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Clack, T.; Mathews, S.; Sharrock, R.A. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 1994, 25, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Kagawa, T.; Takano, M. The phytochrome B/phytochrome C heterodimer is necessary for phytochrome C-mediated responses in rice seedlings. PLoS ONE 2014, 9, e97264. [Google Scholar] [CrossRef]
- Whitelam, G.C.; Johnson, E.; Peng, J.; Carol, P.; Anderson, M.L.; Cowl, J.S.; Np, H. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 1993, 5, 757–768. [Google Scholar] [PubMed]
- Tepperman, J.M.; Hwang, Y.S.; Quail, P.H. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006, 48, 728–742. [Google Scholar] [CrossRef]
- Franklin, K.A.; Praekelt, U.; Stoddart, W.M.; Billingham, O.E.; Halliday, K.J.; Whitelam, G.C. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol. 2003, 131, 1340–1346. [Google Scholar] [CrossRef]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and Phytochrome Interacting Factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef]
- Su, L.; Hou, P.; Song, M.; Zheng, X.; Guo, L.; Xiao, Y.; Yan, L.; Li, W.; Yang, J. Synergistic and Antagonistic Action of Phytochrome (Phy) A and PhyB during Seedling De-Etiolation in Arabidopsis thaliana. Int. J. Mol. Sci. 2015, 16, 12199–12212. [Google Scholar] [CrossRef]
- Vierstra, R.D.; Davis, S.J. Bacteriophytochromes: New tools for understanding phytochrome signal transduction. Semin. Cell Dev. Biol. 2000, 11, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-M.; Shim, J.-Y.; Yang, S.-S.; Kang, J.-G.; Kim, J.-L.; Luka, Z.; Song, P.-S. Chromophore-apoprotein interactions in Synechocystis sp. PCC6803 phytochrome Cph1. Biochemistry 2000, 30, 6349–6356. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Chung, Y.H.; Moon, Y.J.; Kim, C.; Watanabe, M.; Song, P.S.; Joe, C.O.; Bogorad, L.; Park, Y.M. Photomovement of the gliding cyanobacterium Synechocystis sp. PCC 6803. Photochem. Photobiol. 1999, 70, 95–102. [Google Scholar] [CrossRef]
- Ng, W.-O.; Grossman, A.R.; Bhaya, D. Multiple Light Inputs Control Phototaxis in Synechocystis sp. Strain PCC6803. J. Bacteriol. 2003, 185, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Brych, A.; Mascarenhas, J.; Jaeger, E.; Charkiewicz, E.; Pokorny, R.; Bölker, M.; Doehlemann, G.; Batschauer, A. White collar 1-induced photolyase expression contributes to UV-tolerance of Ustilago maydis. Microbiologyopen 2016, 5, 224–243. [Google Scholar] [CrossRef] [PubMed]
- Streng, C.; Hartmann, J.; Leister, K.; Krauß, N.; Lamparter, T.; Frankenberg-Dinkel, N.; Weth, F.; Bastmeyer, M.; Yu, Z.; Fischer, R. Fungal phytochrome chromophore biosynthesis at mitochondria. EMBO J. 2021, 40, e108083. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Arreguin, J.A.; Cabrera-Ponce, J.L.; León-Ramírez, C.G.; Camargo-Escalante, M.O.; Ruiz-Herrera, J. Analysis of the photoreceptors involved in the light-depending basidiocarp formation in Ustilago maydis. Arch. Microbiol. 2020, 202, 93–103. [Google Scholar] [CrossRef]
- Brych, A.; Haas, F.B.; Parzefall, K.; Panzer, S.; Schermuly, J.; Altmüller, J.; Engelsdorf, T.; Terpitz, U.; Rensing, S.A.; Kiontke, S.; et al. Coregulation of gene expression by White collar 1 and phytochrome in Ustilago maydis. Fungal Genet. Biol. 2021, 152, 103570. [Google Scholar] [CrossRef]
- Xue, P.; Bai, Y.; Rottwinkel, G.; Averbukh, E.; Ma, Y.; Roeder, T.; Scheerer, P.; Krauß, N.; Lamparter, T. Phytochrome Mediated Responses in Agrobacterium fabrum: Growth, Motility and Plant Infection. Curr. Microbiol. 2021, 78, 2708–2719. [Google Scholar] [CrossRef]
- Bae, G.; Choi, G. Decoding of Light Signals by Plant Phytochromes and Their Interacting Proteins. Annu. Rev. Plant Biol. 2008, 59, 281–311. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Nagatani, A. Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 2004, 7, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, A.M.; Figueiredo, D.D.; Tepperman, J.; Borba, A.R.; Lourenço, T.; Abreu, I.A.; Ouwerkerk, P.B.; Quail, P.H.; Margarida Oliveira, M.; Saibo, N.J. Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim. Biophys. Acta 2016, 1859, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Tepperman, J.M.; Cohn, M.M.; Monte, E.; Al-Sady, B.; Erickson, E.; Quail, P.H. Dynamic Antagonism between Phytochromes and PIF Family Basic Helix-Loop-Helix Factors Induces Selective Reciprocal Responses to Light and Shade in a Rapidly Responsive Transcriptional Network in Arabidopsis. Plant Cell 2012, 24, 1398–1419. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Zhu, L.; Huq, E. Multiple kinases promote light-induced degradation of PIF1. Plant Signal. Behav. 2014, 6, 1119–1121. [Google Scholar] [CrossRef]
- Legris, M.; Ince, Y.; Fankhauser, C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 2019, 10, 5219. [Google Scholar] [CrossRef]
- Lee, N.; Choi, G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr. Opin. Plant Biol. 2017, 35, 54–60. [Google Scholar] [CrossRef]
- Liang, S.; Gao, X.; Wang, Y.; Zhang, H.; Yin, K.; Chen, S.; Zhang, M.; Zhao, R. Phytochrome-interacting factors regulate seedling growth through ABA signaling. Biochem. Biophys. Res. Commun. 2020, 526, 1100–1105. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodríguez-Concepción, M.; Halliday, K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef]
- Song, J.; Liu, Q.; Hu, B.; Wu, W. Photoreceptor PhyB Involved in Arabidopsis Temperature Perception and Heat-Tolerance Formation. Int. J. Mol. Sci. 2017, 18, 1194. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Martin, S.S.; Feoktistova, K.; Lagarias, J.C. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. USA 2011, 108, 11854–11859. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Bussell, A.N.; Lye, S.H.; Wang, T.; Hu, W.; McLoughlin, K.E.; Weber, E.L.; Li, H.; Vierstra, R.D. Photosensing and Thermosensing by Phytochrome B Require Both Proximal and Distal Allosteric Features within the Dimeric Photoreceptor. Sci. Rep. 2017, 7, 13648. [Google Scholar] [CrossRef] [PubMed]
- Bianchetti, R.; De Luca, B.; de Haro, L.A.; Rosado, D.; Demarco, D.; Conte, M.; Bermudez, L.; Freschi, L.; Fernie, A.R.; Michaelson, L.V.; et al. Phytochrome-Dependent Temperature Perception Modulates Isoprenoid Metabolism. Plant Physiol. 2020, 183, 869–882. [Google Scholar] [CrossRef]
- Carlson, K.D.; Bhogale, S.; Anderson, D.; Tomanek, L.; Madlung, A. Phytochrome A Regulates Carbon Flux in Dark Grown Tomato Seedlings. Front. Plant Sci. 2019, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Ushijima, T.; Hanada, K.; Gotoh, E.; Yamori, W.; Kodama, Y.; Tanaka, H.; Kusano, M.; Fukushima, A.; Tokizawa, M.; Yamamoto, Y.Y.; et al. Light Controls Protein Localization through Phytochrome-Mediated Alternative Promoter Selection. Cell 2017, 171, 1316–1325.e12. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, B.; Li, Y.; André, D.; Nilsson, O. Phytochrome B and PHYTOCHROME INTERACTING FACTOR8 modulate seasonal growth in trees. New Phytol. 2021, 232, 2339–2352. [Google Scholar] [CrossRef]
- Carlson, K.D.; Bhogale, S.; Anderson, D.; Zaragoza-Mendoza, A.; Madlung, A. Subfunctionalization of phytochrome B1/B2 leads to differential auxin and photosynthetic responses. Plant Direct 2020, 4, e00205. [Google Scholar] [CrossRef]
- Gramegna, G.; Rosado, D.; Sánchez Carranza, A.P.; Cruz, A.B.; Simon-Moya, M.; Llorente, B.; Rodríguez-Concepcíon, M.; Freschi, L.; Rossi, M. PHYTOCHROME-INTERACTING FACTOR 3 mediates light-dependent induction of tocopherol biosynthesis during tomato fruit ripening. Plant Cell Environ. 2019, 42, 1328–1339. [Google Scholar] [CrossRef]
- Liao, J.; Deng, B.; Cai, X.; Yang, Q.; Hu, B.; Cong, J.; Zhang, Y.; Wang, G.; Xin, G.; Li, Y.; et al. Time-course transcriptome analysis reveals regulation of Arabidopsis seed dormancy by the transcription factors WOX11/12. J. Exp. Bot. 2023, 74, 1090–1106. [Google Scholar] [CrossRef]
- Yoo, Y.H.; Nalini Chandran, A.K.; Park, J.C.; Gho, Y.S.; Lee, S.W.; An, G.; Jung, K.H. OsPhyB-Mediating Novel Regulatory Pathway for Drought Tolerance in Rice Root Identified by a Global RNA-Seq Transcriptome Analysis of Rice Genes in Response to Water Deficiencies. Front. Plant Sci. 2017, 8, 580. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Park, J.-H.; Jung, S.; Hwang, D.; Nam, H.G.; Hong, S. Antagonistic Roles of PhyA and PhyB in Far-Red Light-Dependent Leaf Senescence in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, W.; Xu, P.; Pan, J.; Zhang, T.; Li, Y.; Li, G.; Yang, H.; Lian, H. phyB Interacts with BES1 to Regulate Brassinosteroid Signaling in Arabidopsis. Plant Cell Physiol. 2019, 60, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Dash, L.; McEwan, R.E.; Montes, C.; Mejia, L.; Walley, J.W.; Dilkes, B.P.; Kelley, D.R. slim shady is a novel allele of PHYTOCHROME B present in the T-DNA line SALK_015201. Plant Direct 2021, 5, e00326. [Google Scholar] [CrossRef] [PubMed]
- Cagnola, J.I.; Cerdan, P.D.; Pacin, M.; Andrade, A.; Rodriguez, V.; Zurbriggen, M.D.; Legris, M.; Buchovsky, S.; Carrillo, N.; Chory, J.; et al. Long-Day Photoperiod Enhances Jasmonic Acid-Related Plant Defense. Plant Physiol. 2018, 178, 163–173. [Google Scholar] [CrossRef]
- Kippes, N.; VanGessel, C.; Hamilton, J.; Akpinar, A.; Budak, H.; Dubcovsky, J.; Pearce, S. Effect of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods. BMC Plant Biol. 2020, 20, 297. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, H.; Li, X.; Shen, H.; Gao, J.; Hou, S.; Zhang, B.; Mayes, S.; Bennett, M.; Ma, J.; et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C(4) model system. Nat. Plants 2020, 6, 1167–1178. [Google Scholar] [CrossRef]
- Wang, E.; Zhou, T.; Jing, S.; Dong, L.; Sun, X.; Fan, Y.; Shen, Y.; Liu, T.; Song, B. Leaves and stolons transcriptomic analysis provide insight into the role of phytochrome F in potato flowering and tuberization. Plant J. 2023, 113, 402–415. [Google Scholar] [CrossRef]
- Calderon, R.H.; Dalton, J.; Zhang, Y.; Quail, P.H. Shade triggers posttranscriptional PHYTOCHROME-INTERACTING FACTOR-dependent increases in H3K4 trimethylation. Plant Physiol. 2022, 190, 1915–1926. [Google Scholar] [CrossRef]
- Bianchetti, R.; Bellora, N.; de Haro, L.A.; Zuccarelli, R.; Rosado, D.; Freschi, L.; Rossi, M.; Bermudez, L. Phytochrome-Mediated Light Perception Affects Fruit Development and Ripening Through Epigenetic Mechanisms. Front. Plant Sci. 2022, 13, 870974. [Google Scholar] [CrossRef]
- Thomas, S.; Kumar, R.; Sharma, K.; Barpanda, A.; Sreelakshmi, Y.; Sharma, R.; Srivastava, S. iTRAQ-based proteome profiling revealed the role of Phytochrome A in regulating primary metabolism in tomato seedling. Sci. Rep. 2021, 11, 7540. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Li, Y.; Wang, J.; Xiao, X.; Guo, X.; Tang, D.; Liu, X. Protein identification and mRNA analysis of phytochrome-regulated genes in Arabidopsis under red light. Sci. China C Life Sci. 2009, 52, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Cho, D.S.; Park, W.M.; Na, H.J.; Nam, H.G. Proteomic pattern-based analyses of light responses in Arabidopsis thaliana wild-type and photoreceptor mutants. Proteomics 2006, 6, 3040–3049. [Google Scholar] [CrossRef] [PubMed]
- Phee, B.K.; Shin, D.H.; Cho, J.H.; Kim, S.H.; Kim, J.I.; Lee, Y.H.; Jeon, J.S.; Bhoo, S.H.; Hahn, T.R. Identification of phytochrome-interacting protein candidates in Arabidopsis thaliana by co-immunoprecipitation coupled with MALDI-TOF MS. Proteomics 2006, 6, 3671–3680. [Google Scholar] [CrossRef] [PubMed]
- Luklová, M.; Novák, J.; Kopecká, R.; Kameniarová, M.; Gibasová, V.; Brzobohatý, B.; Černý, M. Phytochromes and Their Role in Diurnal Variations of ROS Metabolism and Plant Proteome. Int. J. Mol. Sci. 2022, 23, 14134. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; McKenzie, S.D.; Chung, J.; Aryal, U.K.; Leon-Salas, W.D.; Puthiyaveetil, S. Photosystem stoichiometry adjustment is a photoreceptor-mediated process in Arabidopsis. Sci. Rep. 2022, 12, 10982. [Google Scholar] [CrossRef] [PubMed]
- Singiri, J.R.; Priyanka, G.; Trishla, V.S.; Adler-Agmon, Z.; Grafi, G. Moonlight Is Perceived as a Signal Promoting Genome Reorganization, Changes in Protein and Metabolite Profiles and Plant Growth. Plants 2023, 12, 1121. [Google Scholar] [CrossRef]
- Ghassemian, M.; Lutes, J.; Tepperman, J.M.; Chang, H.S.; Zhu, T.; Wang, X.; Quail, P.H.; Lange, B.M. Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Arch. Biochem. Biophys. 2006, 448, 45–59. [Google Scholar] [CrossRef]
- Han, X.; Tohge, T.; Lalor, P.; Dockery, P.; Devaney, N.; Esteves-Ferreira, A.A.; Fernie, A.R.; Sulpice, R. Phytochrome A and B Regulate Primary Metabolism in Arabidopsis Leaves in Response to Light. Front. Plant Sci. 2017, 8, 1394. [Google Scholar] [CrossRef]
- Yang, D.; Seaton, D.D.; Krahmer, J.; Halliday, K.J. Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc. Natl. Acad. Sci. USA 2016, 113, 7667–7672. [Google Scholar] [CrossRef]
- Swathy, P.S.; Kiran, K.R.; Joshi, M.B.; Mahato, K.K.; Muthusamy, A. He-Ne laser accelerates seed germination by modulating growth hormones and reprogramming metabolism in brinjal. Sci. Rep. 2021, 11, 7948. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, T.; Sawada, Y.; Imai, H.; Kanai, M.; Hirai, M.Y.; Mano, S.; Uemura, M.; Nishimura, M.; Kusaba, M.; Nagatani, A. Regulation of Sugar and Storage Oil Metabolism by Phytochrome during De-etiolation. Plant Physiol. 2020, 182, 1114–1129. [Google Scholar] [CrossRef]
- Li, F.; Liu, Y.; Zhang, X.; Liu, L.; Yan, Y.; Ji, X.; Kong, F.; Zhao, Y.; Li, J.; Peng, T.; et al. Transcriptome and Metabolome Analyses Reveals the Pathway and Metabolites of Grain Quality under Phytochrome B in Rice (Oryza sativa L.). Rice 2022, 15, 52. [Google Scholar] [CrossRef]
- Wang, X.; Hou, J.; Quedenau, C.; Chen, W. Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals. Mol. Syst. Biol. 2016, 12, 875. [Google Scholar] [CrossRef]
- Qiu, Y.; Sun, G.; Fen, L.; Wei, M. Functions of Plant Phytochrome Signaling Pathways in Adaptation to Diverse Stresses. Int. J. Mol. Sci. 2023, 24, 13201. [Google Scholar] [CrossRef]
- Schippers, J.H. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhao, X.; Chory, J. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019, 29, 4186–4199.e3. [Google Scholar] [CrossRef] [PubMed]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 15190. [Google Scholar] [CrossRef]
- Murcia, G.; Enderle, B.; Hiltbrunner, A.; Casal, J.J. Phytochrome B and PCH1 protein dynamics store night temperature information. Plant J. 2021, 105, 22–33. [Google Scholar] [CrossRef]
- Li, N.; Bo, C.; Zhang, Y.; Wang, L. PHYTOCHROME INTERACTING FACTORS PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 4577–4589. [Google Scholar] [CrossRef]
- Yang, J.; Qu, X.; Li, T.; Gao, Y.; Du, H.; Zheng, L.; Ji, M.; Zhang, P.; Zhang, Y.; Hu, J.; et al. HY5–HDA9 orchestrates the transcription of HsfA2 to modulate salt stress response in Arabidopsis. J. Integr. Plant Biol. 2022, 65, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Bajracharya, A.; Xi, J.; Grace, K.F.; Bayer, E.E.; Grant, C.A.; Clutton, C.H.; Baerson, S.R.; Agarwal, A.K.; Qiu, Y. PHYTOCHROME-INTERACTING FACTOR 4/HEMERA-mediated thermosensory growth requires the Mediator subunit MED14. Plant Physiol. 2022, 190, 2706–2721. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Ramakrishnan, M.; Khanna, K.; Landi, M.; Prasad, R.; Bhardwaj, R.; Zheng, B. Brassinosteroids and metalloids: Regulation of plant biology. J. Hazard. Mater. 2022, 424 Pt C, 127518. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, B.; Gardestrom, P.; Keech, O. In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J. Exp. Bot. 2014, 65, 4037–4049. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xu, G.; Jing, Y.; Tang, W.; Lin, R. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef]
- Yuan, H.Y.; Saha, S.; Vandenberg, A.; Bett, K.E. Flowering and Growth Responses of Cultivated Lentil and Wild Lens Germplasm toward the Differences in Red to Far-Red Ratio and Photosynthetically Active Radiation. Front. Plant Sci. 2017, 8, 386. [Google Scholar] [CrossRef]
- de Wit, M.; Spoel, S.H.; Sanchez-Perez, G.F.; Gommers, C.M.M.; Pieterse, C.M.J.; Voesenek, L.A.C.J.; Pierik, R. Perception of low red:far–red ratio compromises both salicylic acid– and jasmonic acid–dependent pathogen defences in Arabidopsis. Plant J. 2013, 75, 90–103. [Google Scholar] [CrossRef]
- Liu, Y.; Wei, H.; Ma, M.; Li, Q.; Kong, D.; Sun, J.; Ma, X.; Wang, B.; Chen, C.; Xie, Y.; et al. Arabidopsis FHY3 and FAR1 Regulate the Balance between Growth and Defense Responses under Shade Conditions. Plant Cell 2019, 31, 2089–2106. [Google Scholar] [CrossRef]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, A.; Valverde, F.; Pineiro, M.; Jarillo, J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 2012, 24, 982–999. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Lee, H.-J.; Jung, J.-H.; Lee, S.; Park, C.-M. HOS1 Facilitates the Phytochrome B-Mediated Inhibition of PIF4 Function during Hypocotyl Growth in Arabidopsis. Mol. Plant 2017, 10, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef]
- Pazos-Navarro, M.; Ribalta, F.M.; Hurgobin, B.; Croser, J.S.; Kaur, P. Gene networks underlying faster flowering induction in response to far-red light. Cold Spring Harb. Lab. 2018, 2017, 234161. [Google Scholar]
- Lorenzo, C.D.; Alonso Iserte, J.; Sanchez Lamas, M.; Antonietti, M.S.; Garcia Gagliardi, P.; Hernando, C.E.; Dezar, C.A.A.; Vazquez, M.; Casal, J.J.; Yanovsky, M.J.; et al. Shade delays flowering in Medicago sativa. Plant J. 2019, 99, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.; Jiang, P.; Sun, X.; Yu, L.; Wang, E.; Qin, J.; Zhang, F.; Prat, S.; Song, B. Long-distance control of potato storage organ formation by SELF PRUNING 3D and FLOWERING LOCUS T-like 1. Plant Commun. 2023, 4, 100547. [Google Scholar] [CrossRef]
- Li, Q.; Wu, G.; Zhao, Y.; Wang, B.; Zhao, B.; Kong, D.; Wei, H.; Chen, C.; Wang, H. CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol. J. 2020, 18, 2520–2532. [Google Scholar] [CrossRef]
- Sun, G.; Yang, L.; Zhan, W.; Chen, S.; Song, M.; Wang, L.; Jiang, L.; Guo, L.; Wang, K.; Ye, X.; et al. HFR1, a bHLH Transcriptional Regulator from Arabidopsis thaliana, Improves Grain Yield, Shade and Osmotic Stress Tolerances in Common Wheat. Int. J. Mol. Sci. 2022, 23, 12057. [Google Scholar] [CrossRef]
- Lempiäinen, T.; Rintamäki, E.; Aro, E.M.; Tikkanen, M. Plants acclimate to Photosystem I photoinhibition by readjusting the photosynthetic machinery. Plant Cell Environ. 2022, 45, 2954–2971. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. ASCORBATE AND GLUTATHIONE: Keeping Active Oxygen Under Control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Spicher, L.; Almeida, J.; Gutbrod, K.; Pipitone, R.; Dörmann, P.; Glauser, G.; Rossi, M.; Kessler, F. Essential role for phytol kinase and tocopherol in tolerance to combined light and temperature stress in tomato. J. Exp. Bot. 2017, 68, 5845–5856. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Maruta, T.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Transcriptional control of vitamin C defective 2 and tocopherol cyclase genes by light and plastid-derived signals: The partial involvement of GENOMES UNCOUPLED 1. Plant Sci. 2015, 231, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Chen, L.; Thao, N.P.; Fujiwara, M.; Wong, H.L.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, I.M.Y.; Yuan, S.; Yoshihara, S.; Suzaki, T.; Ezura, H.; Miura, K. Stimulation of Tomato Drought Tolerance by PHYTOCHROME A and B1B2 Mutations. Int. J. Mol. Sci. 2023, 24, 1560. [Google Scholar] [CrossRef]
- Abdellatif, I.M.Y.; Yuan, S.; Na, R.; Yoshihara, S.; Hamada, H.; Suzaki, T.; Ezura, H.; Miura, K. Functional Characterization of Tomato Phytochrome A and B1B2 Mutants in Response to Heat Stress. Int. J. Mol. Sci. 2022, 23, 1681. [Google Scholar] [CrossRef] [PubMed]
- Kreslavski, V.D.; Khudyakova, A.Y.; Kosobryukhov, A.A.; Balakhnina, T.I.; Shirshikova, G.N.; Alharby, H.F.; Allakhverdiev, S.I. The Effect of Short-Term Heating on Photosynthetic Activity, Pigment Content, and Pro-/Antioxidant Balance of A. thaliana Phytochrome Mutants. Plants 2023, 12, 867. [Google Scholar] [CrossRef]
- Willems, P.; Mhamdi, A.; Stael, S.; Storme, V.; Kerchev, P.; Noctor, G.; Gevaert, K.; Van Breusegem, F. The ROS Wheel: Refining ROS Transcriptional Footprints. Plant Physiol. 2016, 171, 1720–1733. [Google Scholar] [CrossRef]
- Alba, R.; Kelmenson, P.M.; Cordonnier-Pratt, M.M.; Pratt, L.H. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 2000, 17, 362–373. [Google Scholar] [CrossRef]
- Hauser, B.A.; Cordonnier-Pratt, M.M.; Daniel-Vedele, F.; Pratt, L.H. The phytochrome gene family in tomato includes a novel subfamily. Plant Mol. Biol. 1995, 29, 1143–1155. [Google Scholar] [CrossRef]
- Whippo, C.W.; Hangarter, R.P. Phytochrome modulation of blue-light-induced phototropism. Plant Cell Environ. 2010, 27, 1223–1228. [Google Scholar] [CrossRef]
- Correll, M.J.; Kiss, J.Z. The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol. 2005, 46, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Karve, A.A.; Jawdy, S.S.; Gunter, L.E.; Allen, S.M.; Yang, X.; Tuskan, G.A.; Wullschleger, S.D.; Weston, D.J. Initial characterization of shade avoidance response suggests functional diversity between Populus phytochrome B genes. New Phytol. 2012, 196, 726–737. [Google Scholar] [CrossRef]
- Shinomura, T.; Uchida, K.; Furuya, M. Elementary processes of photoperception by phytochrome A for high irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000, 122, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, L.; Duan, J.; Cheng, J.; Shen, Y.; Wang, X.; Han, R.; Li, H.; Li, Z.; Wang, L.; et al. Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc. Natl. Acad. Sci. USA 2018, 115, e11864–e11873. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, Z.; Feng, S.; Li, J.; Tan-Wilson, A.; Qu, L.J.; Wang, H.; Deng, X.W. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 2009, 21, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xie, F.; Jiang, Y.; Li, Z.; Huang, X.; Li, L. Phytochrome A Negatively Regulates the Shade Avoidance Response by Increasing Auxin/Indole Acidic Acid Protein Stability. Dev. Cell 2018, 44, 29–41.e4. [Google Scholar] [CrossRef]
- Tuinen, A.V.; Kerckhoffs, L.H.J.; Nagatani, A.; Koornneef, R.E.K.M. Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol. Genet. Genom. 1995, 246, 133–141. [Google Scholar] [CrossRef]
- Wei, X.; Wang, W.; Xu, P.; Wang, W.; Guo, T.; Kou, S.; Liu, M.; Niu, Y.; Yang, H.Q.; Mao, Z. Phytochrome B interacts with SWC6 and ARP6 to regulate H2A.Z deposition and photomorphogensis in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1133–1146. [Google Scholar] [CrossRef]
- Casal, J.J. Phytochrome A Enhances the Promotion of Hypocotyl Growth Caused by Reductions in Levels of Phytochrome B in Its Far-Red-Light-Absorbing Form in Light-Grown Arabidopsis thaliana. Plant Physiol. 1996, 112, 965–973. [Google Scholar] [CrossRef]
- Raven, J.A.; Cockell, C.S. Influence on photosynthesis of starlight, moonlight, planetlight, and light pollution (reflections on photosynthetically active radiation in the universe). Astrobiology 2006, 6, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Breitler, J.C.; Djerrab, D.; Leran, S.; Toniutti, L.; Guittin, C.; Severac, D.; Pratlong, M.; Dereeper, A.; Etienne, H.; Bertrand, B. Full moonlight-induced circadian clock entrainment in Coffea arabica. BMC Plant Biol. 2020, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Lamas, M.; Lorenzo, C.D.; Cerdán, P.D. Bottom-up Assembly of the Phytochrome Network. PLoS Genet. 2016, 12, e1006413. [Google Scholar] [CrossRef] [PubMed]
- Kugan, H.M.; Rejab, N.A.; Sahruzaini, N.A.; Harikrishna, J.A.; Baisakh, N.; Cheng, A. Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore? Int. J. Mol. Sci. 2021, 22, 4588. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Mas, P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 2018, 9, 567. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, Y.; Yao, H.; Zheng, Y.; Cao, S.; Wang, H. Arabidopsis Circadian Clock Repress Phytochrome a Signaling. Front. Plant Sci. 2022, 13, 809563. [Google Scholar] [CrossRef]
- Bernula, P.; Pettkó-Szandtner, A.; Hajdu, A.; Kozma-Bognár, L.; Josse, E.M.; Ádám, É.; Nagy, F.; Viczián, A. SUMOylation of PHYTOCHROME INTERACTING FACTOR 3 promotes photomorphogenesis in Arabidopsis thaliana. New Phytol. 2021, 229, 2050–2061. [Google Scholar] [CrossRef]
- Shen, Y.; Kim, J.I.; Song, P.S. NDPK2 as a signal transducer in the phytochrome-mediated light signaling. J. Biol. Chem. 2005, 280, 5740–5749. [Google Scholar] [CrossRef]
- Fankhauser, C.; Yeh, K.C.; Lagarias, J.C.; Zhang, H.; Elich, T.D.; Chory, J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 1999, 284, 1539–1541. [Google Scholar] [CrossRef]
- Ma, A.; Qi, X. Mining plant metabolomes: Methods, applications, and perspectives. Plant Commun. 2021, 2, 100238. [Google Scholar] [CrossRef] [PubMed]
- Melis, A. Carbon partitioning in photosynthesis. Curr. Opin. Chem. Biol. 2013, 17, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Cortés, L.E.; Weldegergis, B.T.; Boccalandro, H.E.; Dicke, M.; Ballaré, C.L. Trading direct for indirect defense? Phytochrome B inactivation in tomato attenuates direct anti–herbivore defenses whilst enhancing volatile–mediated attraction of predators. New Phytol. 2016, 212, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Kendrick, J.W.C.A.R.E. Fluence-response curves and action spectra for promotion and inhibition of seed germination in wildtype and long-hypocotyl mutants of Arabidopsis thaliana L. Planta 1985, 163, 43–54. [Google Scholar]
- Su, Y.-S.; Lagarias, J.C. Light-Independent Phytochrome Signaling Mediated by Dominant GAF Domain Tyrosine Mutants ofArabidopsisPhytochromes in Transgenic Plants. Plant Cell 2007, 19, 2124–2139. [Google Scholar] [CrossRef]
- Herbert, A.H.-D.A.S.J. Intensifying Plant Density Response of Corn with Artificial Shade. Agron. J. 1992, 84, 547–551. [Google Scholar]
- Ren, M.; Ma, J.; Lu, D.; Wu, C.; Zhu, S.; Chen, X.; Wu, Y.; Shen, Y. STAY-GREEN Accelerates Chlorophyll Degradation in Magnolia sinostellata under the Condition of Light Deficiency. Int. J. Mol. Sci. 2023, 24, 8510. [Google Scholar] [CrossRef]
- Zhou, D.; Li, T.; Yang, Y.; Qu, Z.; Ouyang, L.; Jiang, Z.; Lin, X.; Zhu, C.; Peng, L.; Fu, J.; et al. OsPLS4 Is Involved in Cuticular Wax Biosynthesis and Affects Leaf Senescence in Rice. Front. Plant Sci. 2020, 11, 782. [Google Scholar] [CrossRef]
- Graham, I.A. Seed Storage Oil Mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef]
- Patel, D.; Basu, M.; Hayes, S.; Majláth, I.; Hetherington, F.M.; Tschaplinski, T.J.; Franklin, K.A. Temperature-dependent shade avoidance involves the receptor-like kinase ERECTA. Plant J. 2013, 73, 980–992. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Y.; Wang, Y.; Du, Y.; Song, C.; Song, P.; Yang, Q.; He, F.; Bai, X.; Huang, L.; et al. Glycine-serine-rich effector PstGSRE4 in Puccinia striiformis f. sp. tritici inhibits the activity of copper zinc superoxide dismutase to modulate immunity in wheat. PLoS Pathog. 2022, 18, e1010702. [Google Scholar] [CrossRef] [PubMed]
- Jumtee, K.; Bamba, T.; Okazawa, A.; Fukusaki, E.; Kobayashi, A. Integrated metabolite and gene expression profiling revealing phytochrome A regulation of polyamine biosynthesis of Arabidopsis thaliana. J. Exp. Bot. 2008, 59, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.R., 2nd; Frost, E.; Vidi, P.A.; Kessler, F.; Staehelin, L.A. Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 2006, 18, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M.; et al. Characterization of the ABA–regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Kempa, S.; Krasensky, J.; Dal Santo, S.; Kopka, J.; Jonak, C. A Central Role of Abscisic Acid in Stress-Regulated Carbohydrate Metabolism. PLoS ONE 2008, 3, e3935. [Google Scholar]
- Tisné, S.; Barbier, F.; Granier, C. The ERECTA gene controls spatial and temporal patterns of epidermal cell number and size in successive developing leaves of Arabidopsis thaliana. Ann. Bot. 2011, 108, 159–168. [Google Scholar] [CrossRef]
- Sun, W.; Hui Xu, X.; Lu, X.; Xie, L.; Bai, B.; Zheng, C.; Sun, H.; He, Y.; Xie, X.Z. The Rice Phytochrome Genes, PHYA and PHYB, Have Synergistic Effects on Anther Development and Pollen Viability. Sci. Rep. 2017, 7, 6439. [Google Scholar] [CrossRef]
- Yang, W.; Xu, P.; Zhang, J.; Zhang, S.; Li, Z.; Yang, K.; Chang, X.; Li, Y. OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. J. Genet. Genom. 2022, 49, 414–426. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, A.; Liu, X.; Chen, J. Grain Size Associated Genes and the Molecular Regulatory Mechanism in Rice. Int. J. Mol. Sci. 2022, 23, 3169. [Google Scholar] [CrossRef]
- Appenroth, K.J.; Lenk, G.; Goldau, L.; Sharma, R. Tomato seed germination: Regulation of different response modes by phytochrome B2 and phytochrome A. Plant Cell Environ. 2006, 29, 701–709. [Google Scholar] [CrossRef]
- Mardani Korrani, F.; Amooaghaie, R.; Ahadi, A. He-Ne Laser Enhances Seed Germination and Salt Acclimation in Salvia officinalis Seedlings in a Manner Dependent on Phytochrome and H(2)O(2). Protoplasma 2023, 260, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Zander, M.; Yoo, C.Y.; Phan, A.; Garza, R.M.; Wanamaker, S.A.; He, Y.; Nery, J.R.; Chen, H.; Chen, M.; et al. PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 2021, 53, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bordiya, Y.; Kathare, P.K.; Zhao, B.; Zong, W.; Huq, E.; Sung, S. Phytochrome B triggers light-dependent chromatin remodelling through the PRC2-associated PHD finger protein VIL1. Nat. Plants 2021, 7, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Sofo, A.; Scopa, A.; Roychoudhury, A.; Gill, S.S.; Iqbal, M.; Lukatkin, A.S.; Pereira, E.; Duarte, A.C.; Ahmad, I. Lipids and proteins—Major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. Pollut. Res. 2014, 22, 4099–4121. [Google Scholar] [CrossRef] [PubMed]
- Pallavi, S.; Bhushan, J.A.; Shanker, D.R.; Mohammad, P. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct Regulation of the NADPH Oxidase RBOHD by the PRR-Associated Kinase BIK1 during Plant Immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef]
- Lee, D.; Lal, N.K.; Lin, Z.-J.D.; Ma, S.; Liu, J.; Castro, B.; Toruño, T.; Dinesh-Kumar, S.P.; Coaker, G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat. Commun. 2020, 11, 1838. [Google Scholar] [CrossRef]
- Fichman, Y.; Xiong, H.; Sengupta, S.; Morrow, J.; Loog, H.; Azad, R.K.; Hibberd, J.M.; Liscum, E.; Mittler, R. Phytochrome B regulates reactive oxygen signaling during abiotic and biotic stress in plants. New Phytol. 2022, 237, 1711–1727. [Google Scholar] [CrossRef]
- Xiong, H.; Hua, L.; Reyna-Llorens, I.; Shi, Y.; Chen, K.-M.; Smirnoff, N.; Kromdijk, J.; Hibberd, J.M. Photosynthesis-independent production of reactive oxygen species in the rice bundle sheath during high light is mediated by NADPH oxidase. Proc. Natl. Acad. Sci. USA 2021, 118, e2022702118. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.H.; Kim, J.H.; Kim, S.G.; Sim, H.J.; Lee, G.; Halitschke, R.; Baldwin, I.T.; Kim, J.I.; Park, C.M. Shoot phytochrome B modulates reactive oxygen species homeostasis in roots via abscisic acid signaling in Arabidopsis. Plant J. 2018, 94, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Chen, L.; Yang, T.; Zou, W.; Lin, H.; Xi, D. The role of phytochromes in Nicotiana tabacum against Chilli veinal mottle virus. Plant Physiol. Biochem. 2019, 139, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.G.; Miller, G.; Wallace, I.; Harper, J.; Mittler, R.; Gilroy, S. Orchestrating rapid long–distance signaling in plants with Ca2+, ROS and electrical signals. Plant J. 2017, 90, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Ha, J.H.; Park, C.M. Abscisic acid-mediated phytochrome B signaling promotes primary root growth in Arabidopsis. Plant Signal. Behav. 2018, 13, e1473684. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for Enhanced Defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. Pathogen–associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef]
- Lindermayr, C.; Rudolf, E.E.; Durner, J.; Groth, M. Interactions between metabolism and chromatin in plant models. Mol. Metab. 2020, 38, 100951. [Google Scholar] [CrossRef]
- Tang, M.; Li, R.; Chen, P. Exogenous glutathione can alleviate chromium toxicity in kenaf by activating antioxidant system and regulating DNA methylation. Chemosphere 2023, 337, 139305. [Google Scholar] [CrossRef]
| Transcriptomics | ||||
|---|---|---|---|---|
| Species/Tissue | Experimental Condition | Platform | Key Points of Interest | Ref. |
| A. thaliana seedling | Heat stress treatment | Microarray | Phytochrome (phy) B is an essential thermal sensor in plants. | [41] |
| Tomato fruit | Normal growth conditions | RNA-seq | The effect of phyb1b2 on tomato fruit ripening is greater than that of phyA. | [44] |
| Tomato seedling | Seedlings grown in the dark and exposed to red light (R) treatment | RNA-seq | PhyA helps tomato seedlings maintain growth in the dark. | [45] |
| A. thaliana leaf | High R: far-red light (FR) and low R: FR | RNA-seq | The phytochrome-dependent selective promoter is involved in genome-wide regulation with changes in light wavelength. | [46] |
| Hybrid aspen Populus tremula × tremuloides bud | After 12 weeks of exposure to 14 h light/10 h dark, the temperature dropped to 4 °C for eight weeks, and then plants were exposed to 21 °C and 18 h light/6 h dark | RNA-seq | PhyB in poplar regulates the seasonal growth of trees. | [47] |
| Tomato seedling | Dark environment and fast harvest under green light and rapid R | RNA-seq | The subfunctionalization of phyB is mainly related to auxin and photosynthetic responses. | [48] |
| Tomato fruit | Normal growth conditions | RNA-seq | The activation of PIF3 by phytochrome increases the content of tocopherols in fruits. | [49] |
| A. thaliana seed | Normal growth conditions | RNA-seq | A new phyB downstream transcription factor, WUSCHEL-RELATED HO-MEOBOX 11/12 (WOX11/12), was identified as a precise regulator of seed dormancy. | [50] |
| Rice seedling | Drought stress | RNA-seq | Twenty-nine drought-tolerance genes were identified, and the scavenging effect of phyB on reactive oxygen species (ROS) in rice was demonstrated. | [51] |
| A. thaliana leaf | R or FR | Microarray | There are antagonistic effects of phyA and phyB on leaf senescence. | [52] |
| A. thaliana seedling | R | RNA-seq | PhyB negatively regulates the BR signaling pathway. | [53] |
| A. thaliana seedling | indole-3-acetic acid (IAA) treatment | RNA-seq | Slim shady is a mutant allele of phyB. | [54] |
| A. thaliana leaf | Short days (SD) or long days (LD) | Microarray | Long days improve plant resistance. | [55] |
| Triticum turgidum L. leaf | SD or LD | RNA-seq | PhyB and phyC are essential for wheat flowering. | [56] |
| Setaria italica seedling | Normal growth conditions | RNA-seq | C4 model plants were successfully developed. | [57] |
| Solanum tuberosum L. leaf | Normal growth conditions | RNA-seq | PhyF has regulatory effects on flowering and stem fragmentation in potato. | [58] |
| Epigenetics | ||||
| Species/tissue | Experimental condition | Platform | Key points of interest | Ref. |
| A. thaliana seedling | Simulated shade (30 mmol m−2 s−1, R/FR ~ 0.3) | RNA-seq and ChIP-seq | Histone 3 lysine 4 trimethylation (H3K4me3) buffers light fluctuations in plants. | [59] |
| Tomato fruit | Normal growth conditions | MethylC-seq, RNA-seq, and sRNAome | The changes in the mRNA profile of maturation-related genes induced by phyB1B2 involve DNA methylase/demethylase, histone-modifying enzymes, remodeling factors, and transcriptional regulatory factors. | [60] |
| Proteomics | ||||
| Species/tissue | Experimental condition | Platform | Key points of interest | Ref. |
| Tomato seedling | In the dark or under continuous FR | iTRAQ | PhyA is the primary regulator of tomato under FR. | [61] |
| A. thaliana seedling | Culture under 4 °C in darkness for 4 days, and then transfer to continuous R at 23–25 °C for growth | MALDI-TOF-TOF MS | Double mutants under R shorten their hypocotyl and regulate proteins involved in stress and defense, proteins with binding function or cofactor requirements, storage proteins, energy proteins, protein-fate proteins, and featureless functional proteins, especially essential photorespiratory proteins. | [62] |
| A. thaliana leaf | R, FR, and blue light | MALDI-TOF MS | PhyB mutants retain physiological responses to R, but phyA mutants do not respond to FR, and the wild type under FR has little effect on chloroplast-associated proteins. | [63] |
| A. thaliana seedling | After four days of dark growth, apply R for 20 min | MALDI-TOF MS | Protein phosphatase type 2C (PP2C) and a 66 kDa protein are new proteins discovered to interact with phyB. | [64] |
| A. thaliana seedling | 12 h photoperiod, with harvest in the middle of the light or dark period | RPLC-MS | Three hundred novel oscillations were identified in the four mutants, and most of the proteins were enriched in the ROS metabolic pathway. | [65] |
| A. thaliana leaf | Plant’s photosystem I (PSI) light or plant’s photosystem II (PSII) light, with 16 h light/8 h dark or 8 h light/16 h dark | LC-MS/MS | The light quality response of PSI gene transcripts and proteins is closely related to phyB. | [66] |
| Tomato seedling | Dark environment, with fast harvest under green, safe light, and rapid R | MALDI-TOF-TOF MS | PhyA enhances the enzymatic upregulation of glycolysis, β-oxidation, and the tricarboxylic acid (TCA) cycle that accelerates the breakdown of glucose and fat. It also increases the abundance of storage proteins and controls the distribution of sucrose in seedlings by regulating the expression of sucrose transporter to ensure the development and growth of seedlings. | [45] |
| A. thaliana leaf | Continuous R after 4 days of dark growth | MALDI-TOF-TOF MS | The abundance of the α subunit of heterotrimer g protein (Gα) in phyAB double mutants is upregulated, and sensitivity to light is enhanced. | [62] |
| Nicotiana tabacum seedling | full moonlight | GC-MS | Moonlight is also a critical light signal for plants. | [67] |
| Metabolomics | ||||
| Species/tissue | Experimental condition | Platform | Key points of interest | Ref. |
| A. thaliana seedling | R or FR | GC-MS | In condition F, phyB increases the Calvin cycle and the biosynthesis of chlorophylls, carotenoids, isoprenoid quinones, thylakoid lipids, sterols, and amino acids. | [68] |
| A. thaliana leaf | High light or low light | GC-MS | The number of plastid particles in the phyAB double mutant is reduced, and the oxidation reaction is inhibited. | [69] |
| A. thaliana seedling | Normal growth conditions | GC-MS | Phytochrome changes the diurnal growth ratio of plants. | [70] |
| Tomato seedling | In darkness or under continuous FR | GC-MS | PhyA is the primary regulator of tomato under FR. | [61] |
| Solanum melongena L. seed | He–Ne laser irradiation | LC-MS | He-Ne laser irradiation can break the dormant period of seeds in advance. | [71] |
| A. thaliana seedling | R or FR | LC-MS | Primary and secondary metabolites, such as niacin, alkaloids, phenylpropanoids, glucosinolates (GSLs), and flavonoids, are all affected in yellowing seedlings. | [72] |
| Oryza sativa L. seed | Normal growth conditions | UPLC-MS/MS | The absence of phyB in rice promotes the improvement in rice seed quality. | [73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Gao, Y.; Zhang, Q.; Liu, F.; Hu, W. Application of Multi-Omics Technologies to the Study of Phytochromes in Plants. Antioxidants 2024, 13, 99. https://doi.org/10.3390/antiox13010099
Wu S, Gao Y, Zhang Q, Liu F, Hu W. Application of Multi-Omics Technologies to the Study of Phytochromes in Plants. Antioxidants. 2024; 13(1):99. https://doi.org/10.3390/antiox13010099
Chicago/Turabian StyleWu, Shumei, Yue Gao, Qi Zhang, Fen Liu, and Weiming Hu. 2024. "Application of Multi-Omics Technologies to the Study of Phytochromes in Plants" Antioxidants 13, no. 1: 99. https://doi.org/10.3390/antiox13010099





