Optimization of Hydrolysis Conditions, Isolation, and Identification of Biologically Active Peptides Derived from Acheta domesticus for Antioxidant and Collagenase Inhibition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Materials
2.2. Chemical Materials
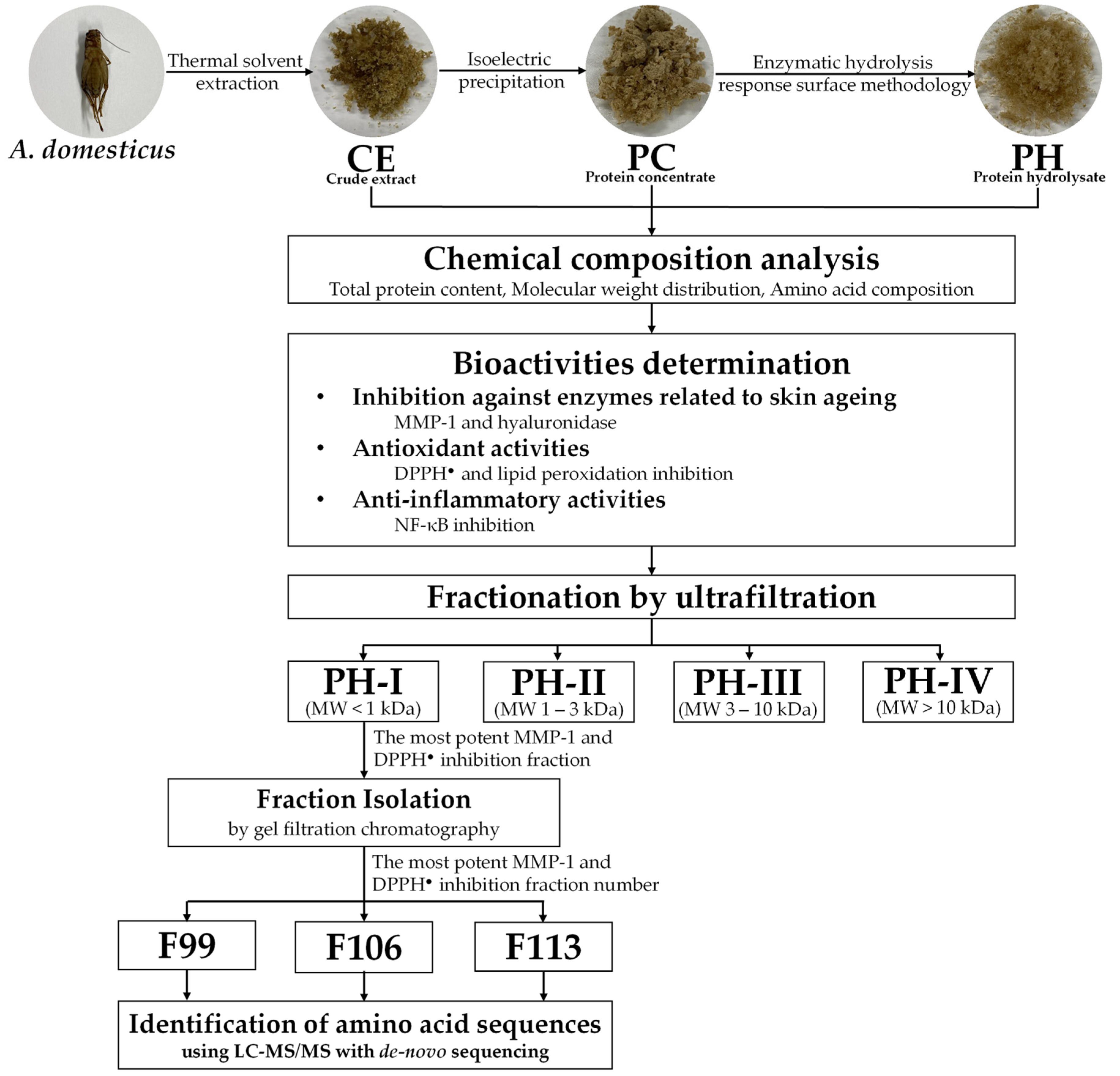
2.3. Preparation of Crude A. domesticus Extract (CE) by Thermal Solvent Extraction
2.4. Preparation of Protein Concentrate (PC) by Isoelectric Precipitation
2.5. Optimization of PH Preparation by Enzymatic Hydrolysis
2.6. Degree of Hydrolysis Determination
2.7. Determination of Inhibitory Activities against Enzymes Related to Skin Aging
2.7.1. MMP-1 Inhibitory Activity
2.7.2. Hyaluronidase Inhibitory Activity
2.8. Determination of Antioxidant Activities
2.8.1. DPPH Assay
2.8.2. Inhibition of Lipid Peroxidation by Ferric Thiocyanate (FTC) Assay
2.9. Chemical Composition Analysis
2.9.1. Total Protein Content
2.9.2. Protein MW Distribution Profile
2.9.3. Amino Acid Composition
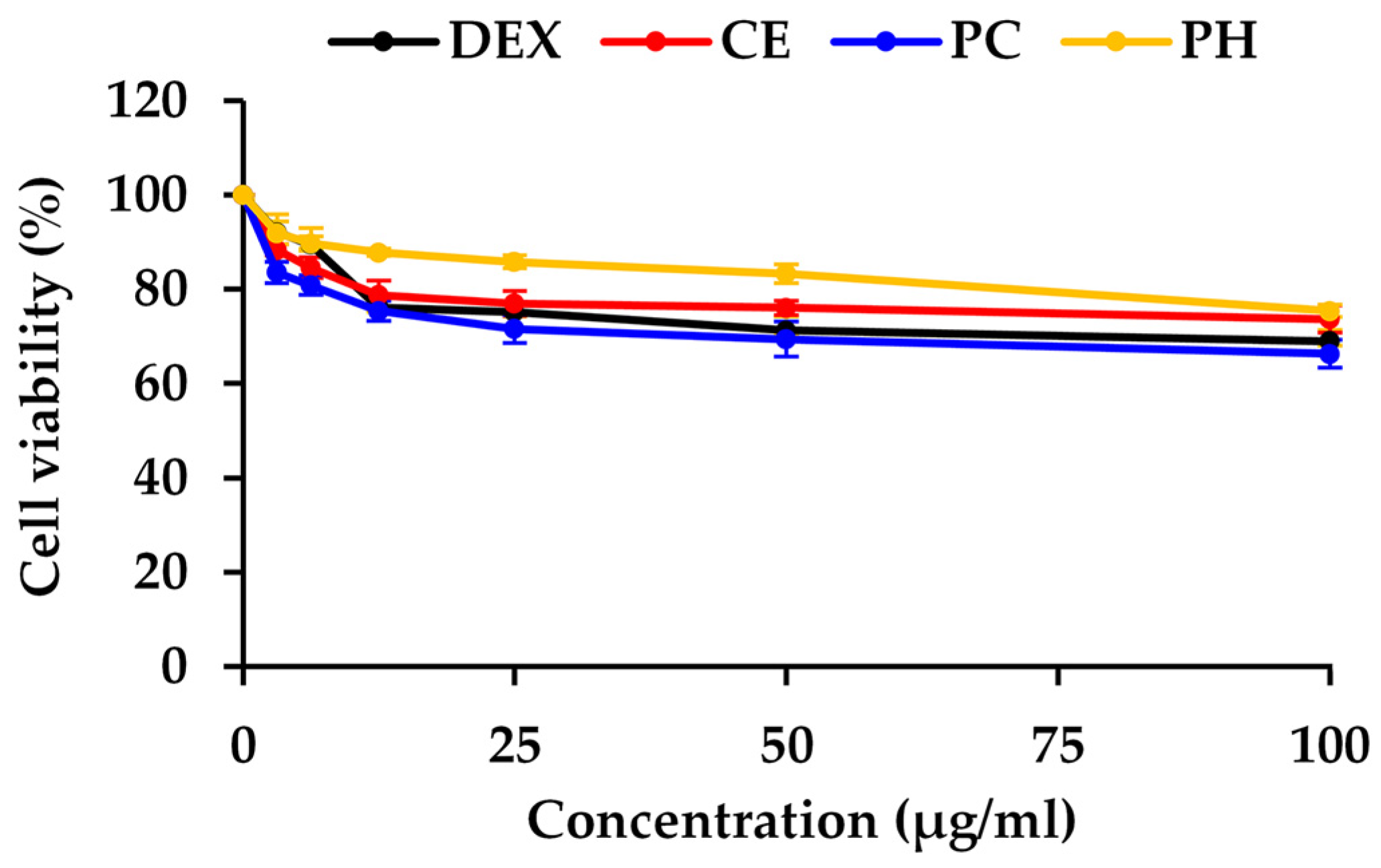
2.10. Determination of Cytotoxicity by MTT Assay
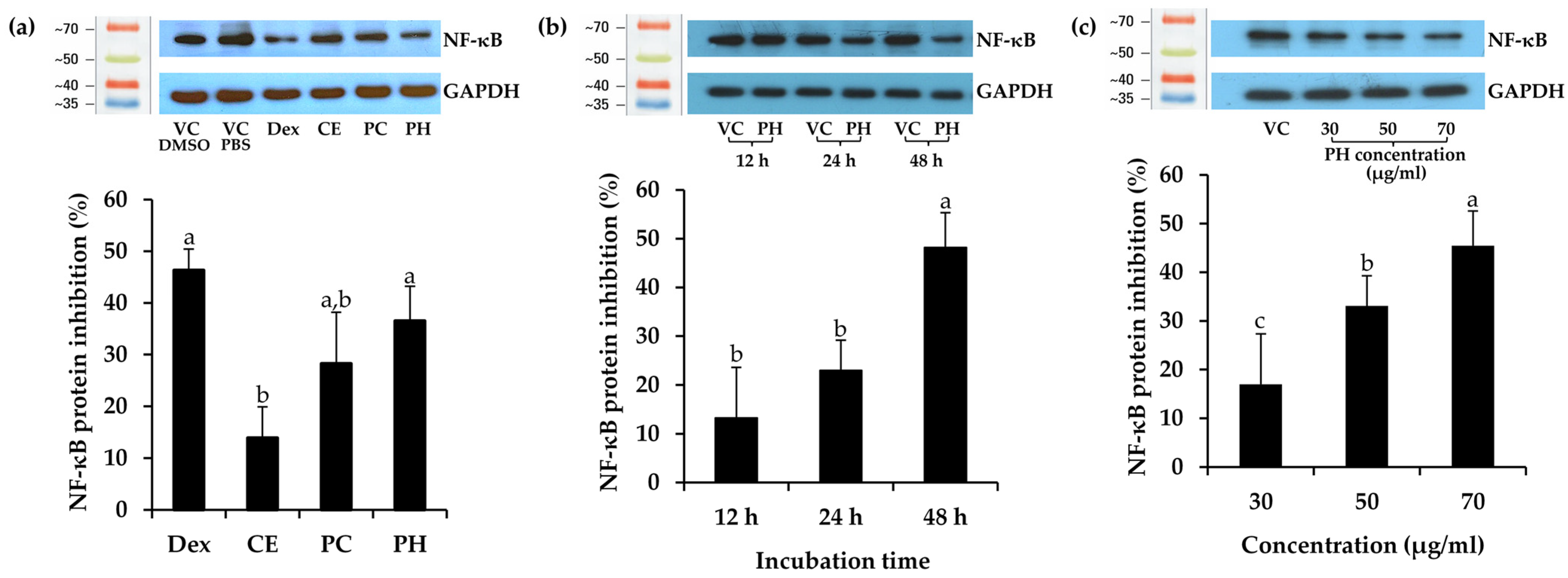
2.11. Determination of Anti-Inflammatory Activity
2.12. Fractionation and Isolation of Bioactive Peptides from PH
2.12.1. Fractionation of PH by Ultrafiltration
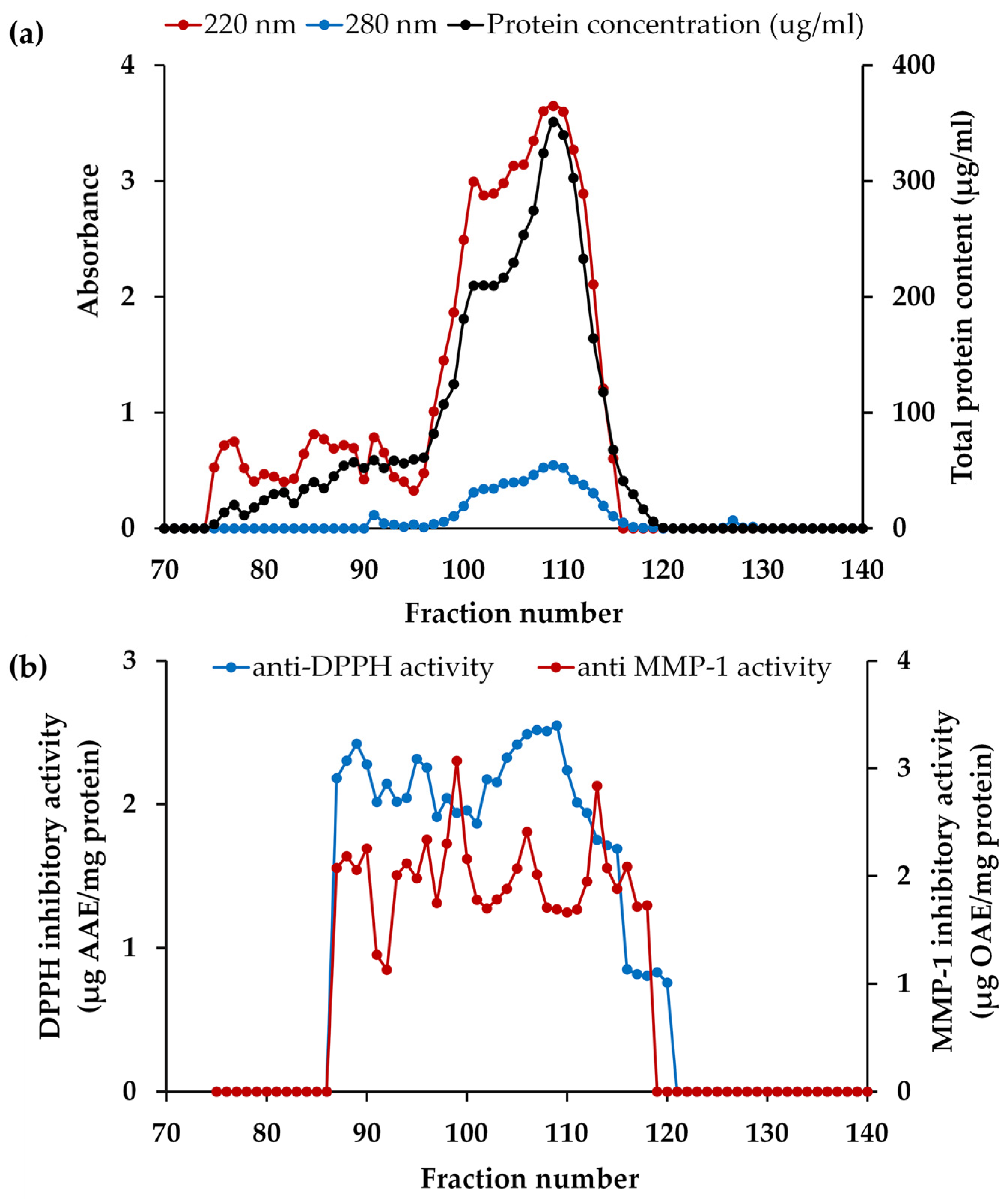
2.12.2. Fraction Isolation by Gel Filtration Chromatography
2.13. Identification of Bioactive Peptide from PH
2.14. Statistical Analysis
3. Results and Discussions
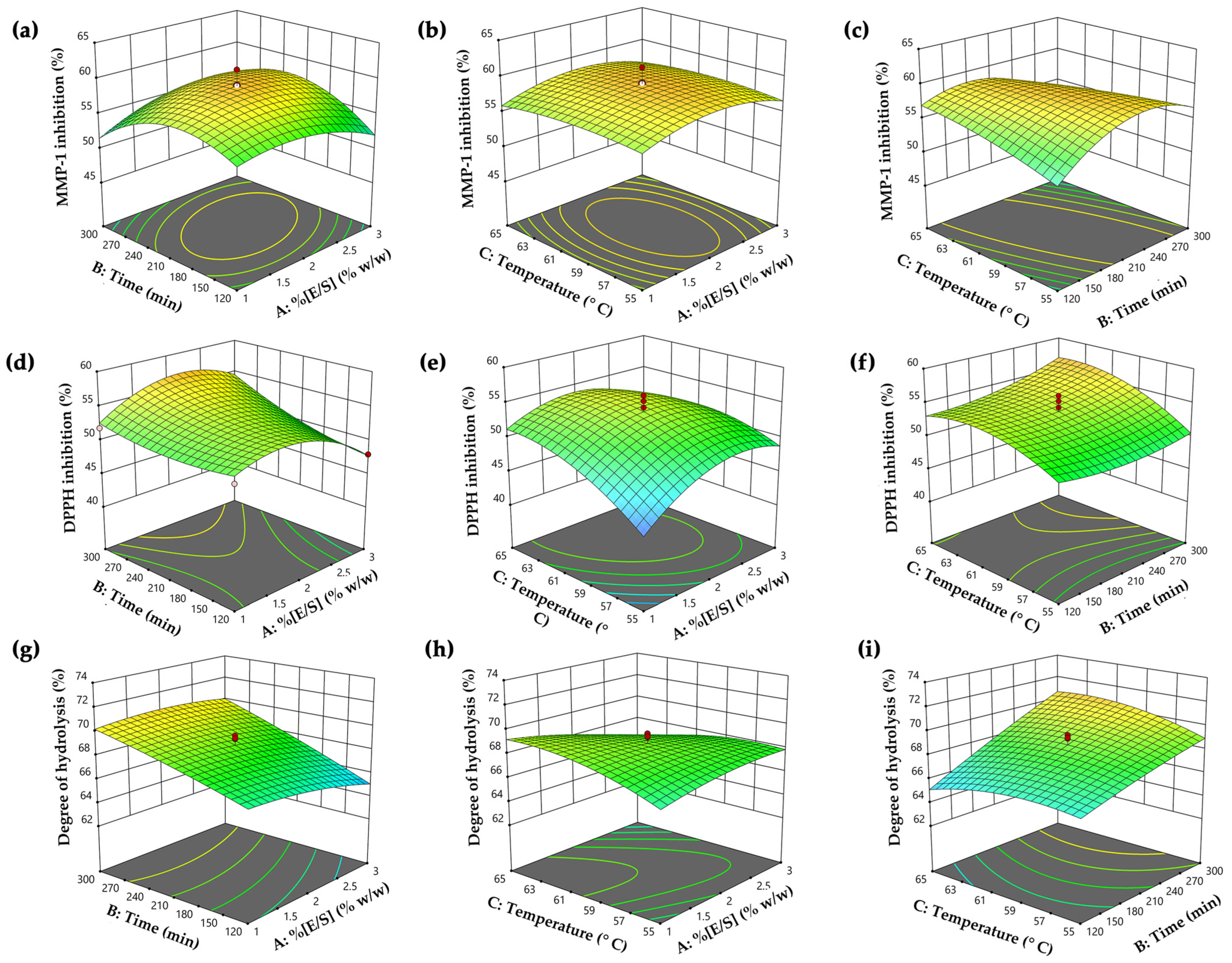
3.1. Optimized Hydrolysis Conditions for PH Generation
3.1.1. MMP-1 Inhibitory Activity
3.1.2. DPPH● Scavenging Activity
3.1.3. Degree of Hydrolysis
3.2. Predictive Optimal Hydrolysis Conditions and Verification
3.3. Chemical Composition of CE, PC, and PH
3.3.1. Total Protein Content and MW Distribution
3.3.2. Amino Acid Composition
3.4. Anti-Skin Aging Activity
3.5. Anti-Inflammatory Activity
3.6. Anti-Skin Aging Bioactive Peptides Derived from PH
3.6.1. Anti-Skin Aging Fractions from PH
3.6.2. Anti-Skin Aging Peptides Isolated from PH-I Fraction
3.6.3. Amino Acid Sequences of Anti-Skin Aging Peptides from PH
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviations | Definitions |
| AA | Ascorbic acid |
| AAE | Ascorbic acid equivalent |
| ALC | Average local confidence |
| Ala | Alanine |
| AP-1 | Activator protein 1 |
| Arg | Arginine |
| Asn | Asparagine |
| Asp | Aspartic acid |
| CCD | Central composite design |
| CE | Crude extract |
| Cys | Cysteine |
| DEX | Dexamethasone |
| DH | Degree of hydrolysis |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| E/S | Enzyme-substrate concentration |
| ECM | Extracellular matrix |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| Glu | Glutamic acid |
| Gln | Glutamine |
| Gly | Glycine |
| HaCaT | Immortalized human keratinocyte |
| His | Histidine |
| IC20 | Concentration reducing cell viability by 20% |
| IC50 | Half-maximal inhibitory concentration |
| Ile | Isoluecine |
| kDa | Kilodaltan |
| Lue | Luesine |
| Lys | Lysine |
| NF-κB | Nuclear factor kappa B |
| MAPKs | Mitogen-activated protein kinases |
| Met | Methionine |
| MMPs | Matrix metalloproteinases |
| MW | Molecular weight |
| OA | Oleanolic acid |
| OAE | Oleanolic acid equivalent |
| PC | Protein concentration |
| PH | Protein hydrolysate |
| Phe | Phenylalanine |
| Pro | Proline |
| RSM | Response surface methodology |
| Ser | Serine |
| Smad | Suppressor of mothers against decapentaplegic |
| TE | Trolox |
| TGF | Transforming growth factor |
| Thr | Threonine |
| Trp | Trptophan |
| Tyr | Tyrosine |
| UV | Ultraviolet |
| Val | Valine |
| VC | Vehicle control |
References
- Chen, C.; Ding, S.; Wang, J. Digital health for aging populations. Nat. Med. 2023, 29, 1623–1630. [Google Scholar] [CrossRef]
- Lee, H.; Hong, Y.; Kim, M. Structural and functional changes and possible molecular mechanisms in aged skin. Int. J. Mol. Sci. 2021, 22, 12489. [Google Scholar] [CrossRef] [PubMed]
- Bocheva, G.; Slominski, R.M.; Janjetovic, Z.; Kim, T.K.; Böhm, M.; Steinbrink, K.; Reiter, R.J.; Kleszczyński, K.; Slominski, A.T. Protective role of melatonin and its metabolites in skin aging. Int. J. Mol. Sci. 2022, 23, 1238. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; Travers, J.B.; Spandau, D.F. A new paradigm for the role of aging in the development of skin cancer. J. Investig. Dermatol. 2009, 129, 787. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.A.; Gupta, A.K. Photodamaged skin and quality of life: Reasons for therapy. J. Dermatol. Treat. 1996, 7, 261–264. [Google Scholar] [CrossRef]
- Puizina-Ivic, N.J.A.D.A. Skin aging. Acta Dermatovenerol. Alp. Panon. Adriat. 2008, 17, 47. [Google Scholar]
- Chen, J.; Liu, Y.; Zhao, Z.; Qiu, J. Oxidative stress in the skin: Impact and related protection. Int. J. Cosmet. Sci. 2021, 43, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, C.W.; Kim, E.K.; Lee, S.J.; Park, N.H.; Kim, H.S.; Kim, H.K.; Char, K.; Jang, Y.P.; Kim, J.W. Inhibition effect of Gynura procumbens extract on UV-B-induced matrix-metalloproteinase expression in human dermal fibroblasts. J. Ethnopharmacol. 2011, 137, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.M.; Chen, H.C.; Chiu, H.H.; Chen, C.W.; Wang, S.M.; Wen, K.C. Neonauclea reticulata (Havil.) Merr stimulates skin regeneration after UVB exposure via ROS scavenging and modulation of the MAPK/MMPs/collagen pathway. eCAM 2013, 2013, 324864. [Google Scholar] [CrossRef]
- Vicentini, F.T.; He, T.; Shao, Y.; Fonseca, M.J.; Verri, W.A., Jr.; Fisher, G.J.; Xu, Y. Quercetin inhibits UV irradiation-induced inflammatory cytokine production in primary human keratinocytes by suppressing NF-κB pathway. J. Dermatol. Sci. 2011, 61, 162–168. [Google Scholar] [CrossRef]
- Fasciglione, G.F.; Gioia, M.; Tsukada, H.; Liang, J.; Iundusi, R.; Tarantino, U.; Coletta, M.; Pourmotabbed, T.; Marini, S. The collagenolytic action of MMP-1 is regulated by the interaction between the catalytic domain and the hinge region. JBIC 2012, 17, 663–672. [Google Scholar] [CrossRef]
- Pardo, A.; Selman, M. MMP-1: The elder of the family. Int. J. Biochem. Cell Biol. 2005, 37, 283–288. [Google Scholar] [CrossRef]
- Pittayapruek, P.; Meephansan, J.; Prapapan, O.; Komine, M.; Ohtsuki, M. Role of matrix metalloproteinases in photoaging and photocarcinogenesis. Int. J. Mol. Sci. 2016, 17, 868. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Roth, M.; Karakiulakis, G. Hyaluronic acid: A key molecule in skin aging. Derm.-Endocrinol. 2012, 4, 253–258. [Google Scholar] [CrossRef]
- Essendoubi, M.; Gobinet, C.; Reynaud, R.; Angiboust, J.F.; Manfait, M.; Piot, O. Human skin penetration of hyaluronic acid of different molecular weights as probed by Raman spectroscopy. Ski. Res. Technol. 2016, 22, 55–62. [Google Scholar] [CrossRef]
- Girish, K.S.; Kemparaju, K. The magic glue hyaluronan and its eraser hyaluronidase: A biological overview. Life Sci. 2007, 80, 1921–1943. [Google Scholar] [CrossRef]
- Friedberg, F.; Winnick, T.; Greenberg, D.M. Peptide synthesis in vivo. J. Biol. Chem. 1947, 169, 763–764. [Google Scholar] [CrossRef]
- Singh, B.P.; Vij, S.; Hati, S. Functional significance of bioactive peptides derived from soybean. Peptides 2014, 54, 171–179. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Lee, B.H.; Oh, D.H. Current trends and perspectives of bioactive peptides. Crit. Rev. Food Sci. Nutr. 2018, 58, 2273–2284. [Google Scholar] [CrossRef]
- Tadesse, S.A.; Emire, S.A. Production and processing of antioxidant bioactive peptides: A driving force for the functional food market. Heliyon 2020, 6, e04765. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Vidal-Limon, A.; Liceaga, A.M. Nutricosmetics: A new frontier in bioactive peptides’ research toward skin aging. In Advances in Food and Nutrition, 1st ed.; Fidel Toldrá, Academic Press: Cambridge, MA, USA, 2023; Volume 104, pp. 205–228. [Google Scholar]
- Cheng, J.H.; Zhang, X.Y.; Wang, Z.; Zhang, X.; Liu, S.C.; Song, X.Y.; Zhang, Y.Z.; Ding, J.M.; Chen, X.L.; Xu, F. Potential of thermolysin-like protease A69 in preparation of bovine collagen peptides with moisture-retention ability and antioxidative activity. Mar. Drugs 2021, 19, 676. [Google Scholar] [CrossRef]
- Iosageanu, A.; Ilie, D.; Craciunescu, O.; Seciu-Grama, A.M.; Oancea, A.; Zarnescu, O.; Moraru, I.; Oancea, F. Effect of fish bone bioactive peptides on oxidative, inflammatory and pigmentation processes triggered by UVB irradiation in skin cells. Molecules 2021, 26, 2691. [Google Scholar] [CrossRef]
- Demidova-Rice, T.N.; Geevarghese, A.; Herman, I.M. Bioactive peptides derived from vascular endothelial cell extracellular matrices promote microvascular morphogenesis and wound healing in vitro. Wound Repair Regen. 2011, 19, 59–70. [Google Scholar] [CrossRef]
- Beltrán-Barrientos, L.M.; García, H.S.; Torres-Llanez, M.J.; González-Córdova, A.F.; Hernández-Mendoza, A.; Vallejo-Cordoba, B. Safety of milk-derived bioactive peptides. Int. J. Dairy Technol. 2017, 70, 16–22. [Google Scholar] [CrossRef]
- Schaafsma, G. Safety of protein hydrolysates, fractions thereof and bioactive peptides in human nutrition. Eur. J. Clin. Nutr. 2009, 63, 1161–1168. [Google Scholar] [CrossRef]
- Hall, F.; Johnson, P.E.; Liceaga, A. Effect of enzymatic hydrolysis on bioactive properties and allergenicity of cricket (Gryllodes sigillatus) protein. Food Chem. 2018, 262, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive peptides: Synthesis, sources, applications, and proposed mechanisms of action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef] [PubMed]
- Sarker, A. A review on the application of bioactive peptides as preservatives and functional ingredients in food model systems. J. Food Process. Preserv. 2022, 46, e16800. [Google Scholar] [CrossRef]
- Cruz-Casas, D.E.; Aguilar, C.N.; Ascacio-Valdés, J.A.; Rodríguez-Herrera, R.; Chávez-González, M.L.; Flores-Gallegos, A.C. Enzymatic hydrolysis and microbial fermentation: The most favorable biotechnological methods for the release of bioactive peptides. Food Chem. Mol. Sci. 2021, 3, 100047. [Google Scholar] [CrossRef]
- Mora, L.; Toldrá, F. Advanced enzymatic hydrolysis of food proteins for the production of bioactive peptides. Curr. Opin. Food Sci. 2023, 49, 100973. [Google Scholar] [CrossRef]
- da Silva Lucas, A.J.; de Oliveira, L.M.; Da Rocha, M.; Prentice, C. Edible insects: An alternative of nutritional, functional and bioactive compounds. Food Chem. 2020, 311, 126022. [Google Scholar] [CrossRef]
- Fernandez-Cassi, X.; Supeanu, A.; Vaga, M.; Jansson, A.; Boqvist, S.; Vagsholm, I. The house cricket (Acheta domesticus) as a novel food: A risk profile. J. Insects Food Feed. 2019, 5, 137–157. [Google Scholar] [CrossRef]
- Udomsil, N.; Imsoonthornruksa, S.; Gosalawit, C.; Ketudat-Cairns, M. Nutritional values and functional properties of house cricket (Acheta domesticus) and field cricket (Gryllus bimaculatus). Food Sci. Technol. Res. 2019, 25, 597–605. [Google Scholar] [CrossRef]
- Lucas-González, R.; Fernández-López, J.; Pérez-Álvarez, J.A.; Viuda-Martos, M. Effect of drying processes in the chemical, physico-chemical, techno-functional and antioxidant properties of flours obtained from house cricket (Acheta domesticus). Eur. Food Res. Technol. 2019, 245, 1451–1458. [Google Scholar] [CrossRef]
- Teixeira, C.S.; Villa, C.; Sousa, S.F.; Costa, J.; Ferreira, I.M.; Mafra, I. An in silico approach to unveil peptides from Acheta domesticus with potential bioactivity against hypertension, diabetes, cardiac and pulmonary fibrosis. Food Res. Int. 2023, 169, 112847. [Google Scholar] [CrossRef] [PubMed]
- de Matos, F.M.; de Lacerda, J.T.J.G.; Zanetti, G.; de Castro, R.J.S. Production of black cricket protein hydrolysates with α-amylase, α-glucosidase and angiotensin I-converting enzyme inhibitory activities using a mixture of proteases. Biocatal. Agric. Biotechnol. 2022, 39, 102276. [Google Scholar] [CrossRef]
- Quinteros, M.F.; Martínez, J.; Barrionuevo, A.; Rojas, M.; Carrillo, W. Functional, antioxidant, and anti-inflammatory properties of cricket protein concentrate (Gryllus assimilis). Biology 2022, 11, 776. [Google Scholar] [CrossRef]
- De Marchi, L.; Mainente, F.; Leonardi, M.; Scheurer, S.; Wangorsch, A.; Mahler, V.; Pilolli, R.; Sorio, D.; Zoccatelli, G. Allergenicity assessment of the edible cricket Acheta domesticus in terms of thermal and gastrointestinal processing and IgE cross-reactivity with shrimp. Food Chem. 2021, 359, 129878. [Google Scholar] [CrossRef]
- Yeerong, K.; Sriyab, S.; Somwongin, S.; Punyoyai, C.; Chantawannakul, P.; Anuchapreeda, S.; Prommaban, A.; Chaiyana, W. Skin irritation and potential antioxidant, anti-collagenase, and anti-elastase activities of edible insect extracts. Sci. Rep. 2021, 11, 22954. [Google Scholar] [CrossRef]
- Trinh, B.T.; Supawong, S. Enzymatic hydrolysis of cricket (Gryllodes sigillatus) protein: Influence of Alcalase and Neutrase enzyme on functional properties of recovered protein. TSTJ 2021, 10, 342–353. [Google Scholar]
- Thring, T.S.; Hili, P.; Naughton, D.P. Anti-collagenase, anti-elastase and anti-oxidant activities of extracts from 21 plants. BMC Complement Altern. Med. 2009, 9, 27. [Google Scholar] [CrossRef]
- Jiamphun, S.; Chaiyana, W. Enhanced antioxidant, hyaluronidase, and collagenase inhibitory activities of glutinous rice husk extract by aqueous enzymatic extraction. Molecules 2022, 27, 3317. [Google Scholar] [CrossRef]
- Somwongin, S.; Sirilun, S.; Chantawannakul, P.; Anuchapreeda, S.; Yawootti, A.; Chaiyana, W. Ultrasound-assisted green extraction methods: An approach for cosmeceutical compounds isolation from Macadamia integrifolia pericarp. Ultrason. Sonochem. 2023, 92, 106266. [Google Scholar] [CrossRef]
- Montowska, M.; Kowalczewski, P.Ł.; Rybicka, I.; Fornal, E. Nutritional value, protein and peptide composition of edible cricket powders. Food Chem. 2019, 289, 130–138. [Google Scholar] [CrossRef]
- Yin, F.; Zhang, Z.; Huang, J.; Yin, Y. Digestion rate of dietary starch affects systemic circulation of amino acids in weaned pigs. Br. J. Nutr. 2010, 103, 1404–1412. [Google Scholar] [CrossRef]
- Panyajai, P.; Viriyaadhammaa, N.; Tima, S.; Chiampanichayakul, S.; Dejkriengkraikul, P.; Okonogi, S.; Anuchapreeda, S. Anticancer activity of Curcuma aeroginosa essential oil and its nano-formulations: Cytotoxicity, apoptosis and cell migration effects. BMC Complement. Med. Ther. 2024, 24, 16. [Google Scholar] [CrossRef]
- Neimkhum, W.; Anuchapreeda, S.; Lin, W.C.; Lue, S.C.; Lee, K.H.; Chaiyana, W. Effects of Carissa carandas Linn. Fruit, pulp, leaf, and seed on oxidation, inflammation, tyrosinase, matrix metalloproteinase, elastase, and hyaluronidase inhibition. Antioxidants 2021, 10, 1345. [Google Scholar] [CrossRef]
- Doungapai, C.; Siriwoharn, T.; Malila, Y.; Autsavapromporn, N.; Makkhun, S.; Yarnpakdee, S.; Jantanasakulwong, K.; Regenstein, J.M.; Wangtueai, S. UV-B Protective and antioxidant activities of protein hydrolysate from sea cucumber (Holothuria scabra) using enzymatic hydrolysis. Front. Mar. Sci. 2022, 9, 892255. [Google Scholar] [CrossRef]
- Krobthong, S.; Yingchutrakul, Y. Identification and enhancement of antioxidant P1-peptide isolated from Ganoderma lucidum hydrolysate. Food Biotechnol. 2020, 34, 338–351. [Google Scholar] [CrossRef]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007, 102, 1317–1327. [Google Scholar] [CrossRef]
- Anh, P.T.N.; Le, B.; Yang, S.H. Anti-aging skin and antioxidant assays of protein hydrolysates obtained from salted shrimp fermented with Salinivibrio cibaria BAO-01. J. Appl. Biol. Chem. 2020, 63, 203–209. [Google Scholar] [CrossRef]
- Zhang, R.; Wei, Y.; Zhang, J.; Cai, M.; Lu, L.; Fang, L.; Qin, X.; Gu, R. Protection effects of rice protein hydrolysate on UVB-irradiated photodamage in Hartley guinea pigs skin and human skin fibroblasts. J. Funct. Foods 2021, 82, 104504. [Google Scholar] [CrossRef]
- Kang, Y.A.; Kim, Y.J.; Jin, S.K.; Choi, H.J. Antioxidant, collagenase inhibitory, and antibacterial effects of bioactive peptides derived from enzymatic hydrolysate of Ulva australis. Mar. Drugs 2023, 21, 469. [Google Scholar] [CrossRef]
- Ding, J.; Liang, R.; Yang, Y.; Sun, N.; Lin, S. Optimization of pea protein hydrolysate preparation and purification of antioxidant peptides based on an in silico analytical approach. LWT-Food Sci. Technol. 2020, 123, 109126. [Google Scholar] [CrossRef]
- de Castro, R.J.S.; Cason, V.G.; Sato, H.H. Binary mixture of proteases increases the antioxidant properties of white bean (Phaseolus vulgaris L.) protein-derived peptides obtained by enzymatic hydrolysis. Biocatal. Agric. Biotechnol. 2017, 10, 291–297. [Google Scholar] [CrossRef]
- Zhang, S.B.; Wang, Z.; Xu, S.Y. Optimization of the aqueous enzymatic extraction of rapeseed oil and protein hydrolysates. J. Am. Oil Chem. Soc. 2007, 84, 97–105. [Google Scholar] [CrossRef]
- Kang, P.Y.; Ishak, N.H.; Sarbon, N.M. Optimization of enzymatic hydrolysis of shortfin scad (Decapterus macrosoma) myofibrillar protein with antioxidant effect using alcalase. Int. Food Res. J. 2018, 25, 1808–1817. [Google Scholar]
- Jino, T.; Surawang, S. Functional properties of hydrolysate protein from house cricket (Acheta domestica) extracted by Alcalase. YRU J. Sci. Technol. 2022, 7, 1–11. [Google Scholar]
- Grossmann, K.K.; Merz, M.; Appel, D.; De Araujo, M.M.; Fischer, L. New insights into the flavoring potential of cricket (Acheta domesticus) and mealworm (Tenebrio molitor) protein hydrolysates and their Maillard products. Food Chem. 2021, 364, 130336. [Google Scholar] [CrossRef] [PubMed]
- Luna, G.C.; Martin-Gonzalez, F.S.; Mauer, L.J.; Liceaga, A.M. Cricket (Acheta domesticus) protein hydrolysates’ impact on the physicochemical, structural and sensory properties of tortillas and tortilla chips. J. Insects Food Feed 2021, 7, 109–120. [Google Scholar] [CrossRef]
- Rutherfurd, S.M. Methodology for determining degree of hydrolysis of proteins in hydrolysates: A review. J. AOAC Int. 2010, 93, 1515–1522. [Google Scholar] [CrossRef]
- Meshginfar, N.; Mahoonak, A.S.; Hosseinian, F.; Tsopmo, A. Physicochemical, antioxidant, calcium binding, and angiotensin converting enzyme inhibitory properties of hydrolyzed tomato seed proteins. J. Food Biochem. 2019, 43, e12721. [Google Scholar] [CrossRef]
- Walker, J.M. (Ed.) The bicinchoninic acid (BCA) assay for protein quantitation. In Basic Protein and Peptide Protocols, 1st ed.; Humana Press: Totowa, NJ, USA, 1994; Volume 32, pp. 5–8. [Google Scholar]
- Sosa-Flores, M.L.; García-Hernández, D.G.; Amaya-Guerra, C.A.; Bautista-Villarreal, M.; González-Luna, A.R. Obtención de aislados e hidrolizados proteicos de grillo (Acheta domesticus) y evaluación de su actividad antioxidante. Investig. Desarro. Cienc. Tecnol. Aliment. 2023, 8, 608–618. [Google Scholar] [CrossRef]
- Yi, L.; Lakemond, C.M.; Sagis, L.M.; Eisner-Schadler, V.; van Huis, A.; van Boekel, M.A. Extraction and characterisation of protein fractions from five insect species. Food Chem. 2013, 141, 3341–3348. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Boys, E.L.; Yan, M.; Bryant, K.; Cameron, B.; Desai, A.; Thomas, P.S.; Tedia, N.T. Hypersensitivity pneumonitis caused by house cricket, Acheta domesticus. J Clin. Cell Immunol. 2014, 5, 2. [Google Scholar] [CrossRef]
- Brogan, E.N.; Park, Y.L.; Matak, K.E.; Jaczynski, J. Characterization of protein in cricket (Acheta domesticus), locust (Locusta migratoria), and silk worm pupae (Bombyx mori) insect powders. LWT-Food Sci. Technol. 2021, 152, 112314. [Google Scholar] [CrossRef]
- Li, X.X.; Han, L.J.; Chen, L.J. In vitro antioxidant activity of protein hydrolysates prepared from corn gluten meal. J. Sci. Food Agric. 2008, 88, 1660–1666. [Google Scholar] [CrossRef]
- Nimalaratne, C.; Lopes-Lutz, D.; Schieber, A.; Wu, J. Free aromatic amino acids in egg yolk show antioxidant properties. Food Chem. 2011, 129, 155–161. [Google Scholar] [CrossRef]
- Krane, S.M. The importance of proline residues in the structure, stability, and susceptibility to proteolytic degradation of collagens. Amino Acids 2008, 35, 703–710. [Google Scholar] [CrossRef]
- Mirzaee, H.; Ahmadi Gavlighi, H.; Nikoo, M.; Udenigwe, C.C.; Khodaiyan, F. Relation of amino acid composition, hydrophobicity, and molecular weight with antidiabetic, antihypertensive, and antioxidant properties of mixtures of corn gluten and soy protein hydrolysates. Food Sci. Nutr. 2023, 11, 1257–1271. [Google Scholar] [CrossRef]
- Nehete, J.Y.; Bhambar, R.S.; Narkhede, M.R.; Gawali, S.R. Natural proteins: Sources, isolation, characterization and applications. Pharmacogn. Rev. 2013, 7, 107. [Google Scholar] [CrossRef]
- Fountoulakis, M.; Lahm, H.W. Hydrolysis and amino acid composition analysis of proteins. J. Chromatogr. A 1998, 826, 109–134. [Google Scholar] [CrossRef]
- Lone, A.B.; Bhat, H.F.; Aït-Kaddour, A.; Hassoun, A.; Aadil, R.M.; Dar, B.N.; Bhat, Z.F. Cricket protein hydrolysates pre-processed with ultrasonication and microwave improved storage stability of goat meat emulsion. Innov. Food Sci. Emerg. Technol. 2023, 86, 103364. [Google Scholar] [CrossRef]
- Kim, D.; Kim, H.J.; Chae, H.S.; Park, N.G.; Kim, Y.B.; Jang, A. Anti-oxidation and anti-wrinkling effects of Jeju horse leg bone hydrolysates. Korean J. Food Sci. Anim. Resour. 2014, 34, 844. [Google Scholar] [CrossRef]
- Wang, K.; Siddanakoppalu, P.N.; Ahmed, I.; Pavase, T.R.; Lin, H.; Li, Z. Purification and identification of anti-allergic peptide from Atlantic Salmon (Salmo salar) byproduct enzymatic hydrolysates. J. Funct. Foods 2020, 72, 104084. [Google Scholar] [CrossRef]
- Chen, H.J.; Dai, F.J.; Fan, S.L.; Huang, Y.C.; Chau, C.F.; Lin, Y.S.; Chen, C.S. Kinetics of hyaluronidase inhibition by rice (Oryza sativa L.) protein hydrolysate. Appl. Sci. 2020, 10, 9087. [Google Scholar] [CrossRef]
- Tilstra, J.S.; Clauson, C.L.; Niedernhofer, L.J.; Robbins, P.D. NF-κB in aging and disease. Aging Dis. 2011, 2, 449. [Google Scholar] [PubMed]
- Borg, M.; Brincat, S.; Camilleri, G.; Schembri-Wismayer, P.; Brincat, M.; Calleja-Agius, J. The role of cytokines in skin aging. Climacteric 2013, 16, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Asamitsu, K.; Uranishi, H.; Iddamalgoda, A.; Ito, K.; Kojima, H.; Okamoto, T. Protecting skin photoaging by NF-κB inhibitor. Curr. Drug Metab. 2010, 11, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Wong, N.A.; Chae, M.; Auh, J.H. Comparative characterization of protein hydrolysates from three edible insects: Mealworm larvae, adult crickets, and silkworm pupae. Foods 2019, 8, 563. [Google Scholar] [CrossRef] [PubMed]
- Hall, F.; Reddivari, L.; Liceaga, A.M. Identification and characterization of edible cricket peptides on hypertensive and glycemic in vitro inhibition and their anti-inflammatory activity on RAW 264.7 macrophage cells. Nutrients 2020, 12, 3588. [Google Scholar] [CrossRef]
- Zielińska, E.; Baraniak, B.; Karaś, M. Antioxidant and anti-inflammatory activities of hydrolysates and peptide fractions obtained by enzymatic hydrolysis of selected heat-treated edible insects. Nutrients 2017, 9, 970. [Google Scholar] [CrossRef]
- Won, H.R.; Lee, P.; Oh, S.R.; Kim, Y.M. Epigallocatechin-3-gallate suppresses the expression of TNF-α-induced MMP-1 via MAPK/ERK signaling pathways in human dermal fibroblasts. Biol. Pharm. Bull. 2021, 44, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hwang, E.; Lin, P.; Gao, W.; Ngo, H.T.; Yi, T.H. Prunella vulgaris L. exerts a protective effect against extrinsic aging through NF-κB, MAPKs, AP-1, and TGF-β/Smad signaling pathways in UVB-aged normal human dermal fibroblasts. Rejuvenation Res. 2018, 21, 313–322. [Google Scholar] [CrossRef]
- Han, A.R.; Nam, M.H.; Lee, K.W. Plantamajoside Inhibits UVB and Advanced Glycation End Products-Induced MMP-1 Expression by Suppressing the MAPK and NF-κB Pathways in HaCaT Cells. Photochem. Photobiol. 2016, 92, 708–719. [Google Scholar] [CrossRef]
- Hong, G.P.; Min, S.G.; Jo, Y.J. Anti-oxidative and anti-aging activities of porcine by-product collagen hydrolysates produced by commercial proteases: Effect of hydrolysis and ultrafiltration. Molecules 2019, 24, 1104. [Google Scholar] [CrossRef]
- Fashakin, O.O.; Tangjaidee, P.; Unban, K.; Klangpetch, W.; Khumsap, T.; Sringarm, K.; Rawdkuen, S.; Phongthai, S. Isolation and identification of antioxidant peptides derived from cricket (Gryllus bimaculatus) protein fractions. Insects 2023, 14, 674. [Google Scholar] [CrossRef] [PubMed]
- Ishak, N.H.; Sarbon, N.M. A review of protein hydrolysates and bioactive peptides deriving from wastes generated by fish processing. Food Bioproc Tech. 2018, 11, 2–16. [Google Scholar] [CrossRef]
- Stellwagen, E. Gel filtration. In Methods in Enzymology, 1st ed.; Deutscher, M.P., Ed.; Academic Press: Cambridge, MA, USA, 1990; Volume 182, pp. 317–328. [Google Scholar]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.H.; Tavano, O.; Berenguer-Murcia, A.; Fernandez-Lafuente, R. Use of Alcalase in the production of bioactive peptides: A review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef]
- Mazorra-Manzano, M.A.; Ramírez-Suarez, J.C.; Yada, R.Y. Plant proteases for bioactive peptides release: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2147–2163. [Google Scholar] [CrossRef]
- Yeo, I.; Lee, Y.J.; Song, K.; Jin, H.S.; Lee, J.E.; Kim, D.; Lee, D.W.; Kang, N.J. Low-molecular weight keratins with anti-skin aging activity produced by anaerobic digestion of poultry feathers with Fervidobacterium islandicum AW-1. J. Biotech. 2018, 271, 17–25. [Google Scholar] [CrossRef]
- Jin, H.S.; Song, K.; Baek, J.H.; Lee, J.E.; Kim, D.J.; Nam, G.W.; Kang, N.J.; Lee, D.W. Identification of matrix metalloproteinase-1-suppressive peptides in feather keratin hydrolysate. J. Agric. Food Chem. 2018, 66, 12719–12729. [Google Scholar] [CrossRef] [PubMed]
- Zaky, A.A.; Simal-Gandara, J.; Eun, J.B.; Shim, J.H.; Abd El-Aty, A.M. Bioactivities, applications, safety, and health benefits of bioactive peptides from food and by-products: A review. Front. Nutr. 2022, 8, 815640. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Ning, J.; Li, M.; Chen, W.; Yang, M.; Chen, J.; Luo, X.; Yue, X. Characterization and biological function analysis of endogenous peptides derived from donkey colostrum proteins. Food Funct. 2023, 14, 8261–8275. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Hernández-Mendoza, A.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Liceaga, A.M. Potential role of natural bioactive peptides for development of cosmeceutical skin products. Peptides 2019, 122, 170170. [Google Scholar] [CrossRef]
- Peng, Z.; Gao, J.; Su, W.; Cao, W.; Zhu, G.; Qin, X.; Zhang, C.; Qi, Y. Purification and identification of peptides from oyster (Crassostrea hongkongensis) protein enzymatic hydrolysates and their anti-skin photoaging effects on UVB-irradiated HaCaT cells. Mar. Drugs 2022, 20, 749. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Toalá, J.E.; Liceaga, A.M. Identification of chia seed (Salvia hispanica L.) peptides with enzyme inhibition activity towards skin-aging enzymes. Amino Acids 2020, 52, 1149–1159. [Google Scholar] [CrossRef]
- Brown, P.D. Matrix metalloproteinase inhibitors. Angiogenesis 1997, 1, 142–154. [Google Scholar] [CrossRef]
- Castro-Jácome, T.P.; Alcántara-Quintana, L.E.; Montalvo-González, E.; Chacón-López, A.; Kalixto-Sánchez, M.A.; del Pilar Rivera, M.; López-García, U.M.; Tovar-Pérez, E.G. Skin-protective properties of peptide extracts produced from white sorghum grain kafirins. Ind. Crops Prod. 2021, 167, 113551. [Google Scholar] [CrossRef]
- Huang, J.J.; Li, H.L.; Xiong, G.Q.; Cai, J.; Liao, T.; Zu, X.Y. Extraction, identification and anti-photoaging activity evaluation of collagen peptides from silver carp (Hypophthalmichthys molitrix) skin. LWT-Food Sci. Technol. 2023, 173, 114384. [Google Scholar] [CrossRef]
- Lu, J.; Hou, H.; Fan, Y.; Yang, T.; Li, B. Identification of MMP-1 inhibitory peptides from cod skin gelatin hydrolysates and the inhibition mechanism by MAPK signaling pathway. J. Funct. Foods 2017, 33, 251–260. [Google Scholar] [CrossRef]
- Liang, R.; Xu, L.; Fan, C.; Cao, L.; Guo, X. Structural characteristics and antioxidant mechanism of donkey-hide gelatin peptides by molecular dynamics simulation. Molecules 2023, 28, 7975. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.W.; Li, B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: Key sequence positions and their amino acid properties. J. Theor. Biol. 2013, 318, 29–43. [Google Scholar] [CrossRef]
- Matsui, R.; Honda, R.; Kanome, M.; Hagiwara, A.; Matsuda, Y.; Togitani, T.; Ikemoto, N.; Terashima, M. Designing antioxidant peptides based on the antioxidant properties of the amino acid side-chains. Food Chem. 2018, 245, 750–755. [Google Scholar] [CrossRef]
- Korkmaz, K.; Tokur, B. Optimization of hydrolysis conditions for the production of protein hydrolysates from fish wastes using response surface methodology. Food Biosci. 2022, 45, 101312. [Google Scholar] [CrossRef]

| Run | Variable Factors | ||
|---|---|---|---|
| X1 (E/S: % w/w) | X2 (Time: min) | X3 (Temp: °C) | |
| 1 | 1.0 | 300.0 | 55.0 |
| 2 | 1.0 | 300.0 | 65.0 |
| 3 | 3.0 | 300.0 | 55.0 |
| 4 | 2.0 | 58.6 | 60.0 |
| 5 | 2.0 | 210.0 | 60.0 |
| 6 | 2.0 | 210.0 | 68.4 |
| 7 | 2.0 | 210.0 | 60.0 |
| 8 | 2.0 | 210.0 | 60.0 |
| 9 | 2.0 | 210.0 | 51.6 |
| 10 | 2.0 | 210.0 | 60.0 |
| 11 | 3.0 | 120.0 | 55.0 |
| 12 | 3.7 | 320.0 | 60.0 |
| 13 | 1.0 | 120.0 | 65.0 |
| 14 | 3.0 | 120.0 | 65.0 |
| 15 | 2.0 | 210.0 | 60.0 |
| 16 | 0.3 | 210.0 | 60.0 |
| 17 | 3.0 | 300.0 | 65.0 |
| 18 | 2.0 | 361.4 | 60.0 |
| 19 | 2.0 | 210.0 | 60.0 |
| 20 | 1.0 | 120.0 | 55.0 |
| Run | Experimental Responses | ||
|---|---|---|---|
| Y1 (% MMP-1 Inhibition) | Y2 (% DPPH● Inhibition) | Y3 (% DH) | |
| 1 | 53.6 | 42.2 | 66.9 |
| 2 | 48.3 | 51.9 | 71.2 |
| 3 | 58.2 | 51.6 | 69.1 |
| 4 | 48.2 | 54.6 | 63.8 |
| 5 | 61.3 | 55.3 | 69.5 |
| 6 | 57.1 | 53.2 | 66.0 |
| 7 | 57.8 | 52.7 | 68.2 |
| 8 | 59.1 | 51.9 | 68.3 |
| 9 | 55.8 | 43.9 | 66.9 |
| 10 | 58.6 | 56.1 | 69.2 |
| 11 | 50.0 | 48.1 | 66.2 |
| 12 | 52.5 | 46.7 | 66.1 |
| 13 | 55.8 | 50.8 | 67.3 |
| 14 | 54.0 | 48.0 | 63.6 |
| 15 | 59.0 | 52.0 | 69.2 |
| 16 | 53.3 | 43.6 | 68.7 |
| 17 | 52.9 | 53.0 | 69.4 |
| 18 | 58.0 | 59.7 | 72.3 |
| 19 | 59.0 | 54.3 | 67.5 |
| 20 | 52.4 | 46.7 | 65.6 |
| MMP-1 Inhibitory Activity (%) | DPPH● Scavenging Activity (%) | DH (%) | |
|---|---|---|---|
| Predicted values | 58.8 | 54.5 | 68.9 |
| Experimental values | 59.3 ± 1.3 | 55.4 ± 2.6 | 65.7 ± 6.8 |
| Amino Acid Composition of PH | Amount Based on a Dry Basis of PH (% w/w) |
|---|---|
| Essential amino acid | |
| Leucine | 24.4 |
| Valine | 15.5 |
| Lysine | 14.7 |
| Phenylalanine | 10.7 |
| Isoleucine | 10.5 |
| Threonine | 8.7 |
| Histidine | 4.4 |
| Methionine | 3.7 |
| Non-essential amino acid | |
| Glutamic acid and its derivative glutamine | 41.0 |
| Aspartic acid and its derivative asparagine | 38.0 |
| Alanine | 17.8 |
| Serine | 17.4 |
| Glycine | 11.5 |
| Arginine | 11.4 |
| Tyrosine | 7.3 |
| Proline | 7.3 |
| Cysteine | 0.5 |
| Total essential amino acids (%) | 37.8 |
| Total hydrophobic amino acids (%) | 36.7 |
| Samples | IC50 (µg/mL) | |||
|---|---|---|---|---|
| MMP-1 Inhibition | Hyaluronidase Inhibition | DPPH● Inhibition | Lipid Peroxidation Inhibition | |
| CE | 31.3 ± 0.8 a | 180.7 ± 2.0 a | 251.3 ± 6.2 a | 172.8 ± 2.2 a |
| PC | 19.2 ± 0.1 b | 89.8 ± 6.9 b | 143.7 ± 13.3 b | 157.6 ± 4.0 b |
| PH | 15.5 ± 1.8 c | 36.7 ± 3.5 c | 91.0 ± 6.2 c | 121.6 ± 7.6 c |
| OA | 2.7 ± 0.8 d | 28.7 ± 0.4 c | - | - |
| AA | - | - | 4.1 ± 0.0 d | - |
| TE | - | - | - | 4.8 ± 0.3 d |
| MW (kDa) | Fraction | MMP-1 Inhibition (µg OAE/mg Protein) | DPPH● Inhibition (µg AAE/mg Protein) |
|---|---|---|---|
| <1 kDa | PH-I | 6.2 ± 0.1 a | 3.5 ± 0.1 a |
| 1–3 kDa | PH-II | 3.4 ± 0.2 b | 2.3 ± 0.0 b |
| 3–10 kDa | PH-III | 1.3 ± 0.6 c | 1.8 ± 0.0 c |
| >10 kDa | PH-IV | 0 ± 0.9 d | 1.2 ± 0.1 d |
| Fraction no. | Amino Acid Sequence | Length | Mass | ALC Score |
|---|---|---|---|---|
| 99 | AVTKADPYTDQ | 11 | 1208 | 98 |
| AENQRVSFD | 9 | 1064 | 97 | |
| YLGGEGHNLQEH | 12 | 1353 | 96 | |
| SPLPKY | 6 | 703 | 96 | |
| EAKAAASAPVALHKAK | 16 | 1562 | 96 | |
| NGEPVYHP | 8 | 911 | 95 | |
| 106 | TVMELNDLVKAF | 12 | 1379 | 98 |
| FGGEAKDYSQ | 10 | 1100 | 97 | |
| WAPDLPGL | 8 | 867 | 97 | |
| FGSQDLSK | 8 | 880 | 97 | |
| AENQRVSFD | 9 | 1064 | 96 | |
| FGGEAKDY | 8 | 885 | 95 | |
| TGTVVSDKMD | 10 | 1051 | 95 | |
| AAAPAAPAAD | 10 | 824 | 95 | |
| 113 | VPLLGPW | 7 | 780 | 99 |
| VGTLGHVD | 8 | 796 | 97 | |
| YVAGAEGPQ | 9 | 890 | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeerong, K.; Chantawannakul, P.; Anuchapreeda, S.; Wangtueai, S.; Chaiyana, W. Optimization of Hydrolysis Conditions, Isolation, and Identification of Biologically Active Peptides Derived from Acheta domesticus for Antioxidant and Collagenase Inhibition. Antioxidants 2024, 13, 367. https://doi.org/10.3390/antiox13030367
Yeerong K, Chantawannakul P, Anuchapreeda S, Wangtueai S, Chaiyana W. Optimization of Hydrolysis Conditions, Isolation, and Identification of Biologically Active Peptides Derived from Acheta domesticus for Antioxidant and Collagenase Inhibition. Antioxidants. 2024; 13(3):367. https://doi.org/10.3390/antiox13030367
Chicago/Turabian StyleYeerong, Kankanit, Panuwan Chantawannakul, Songyot Anuchapreeda, Sutee Wangtueai, and Wantida Chaiyana. 2024. "Optimization of Hydrolysis Conditions, Isolation, and Identification of Biologically Active Peptides Derived from Acheta domesticus for Antioxidant and Collagenase Inhibition" Antioxidants 13, no. 3: 367. https://doi.org/10.3390/antiox13030367










