Analysis of Toxic Effects of Fluoride on Ovine Follicular Granulosa Cells Using RNA-Seq
Abstract
1. Introduction
2. Materials and Methods
2.1. Ovary Collection
2.2. In Vitro Cultivation of Primary GCs
2.3. Cellular Identification and Cell Viability Assay
2.4. Screening of NaF Working Concentration
2.5. RNA Extraction, Library Construction, and Sequencing
2.6. Immunofluorescence Detection of Cellular ROS Levels
2.7. Analysis of RNA-Seq Data and qRT-PCR
2.8. Data Analysis
3. Results
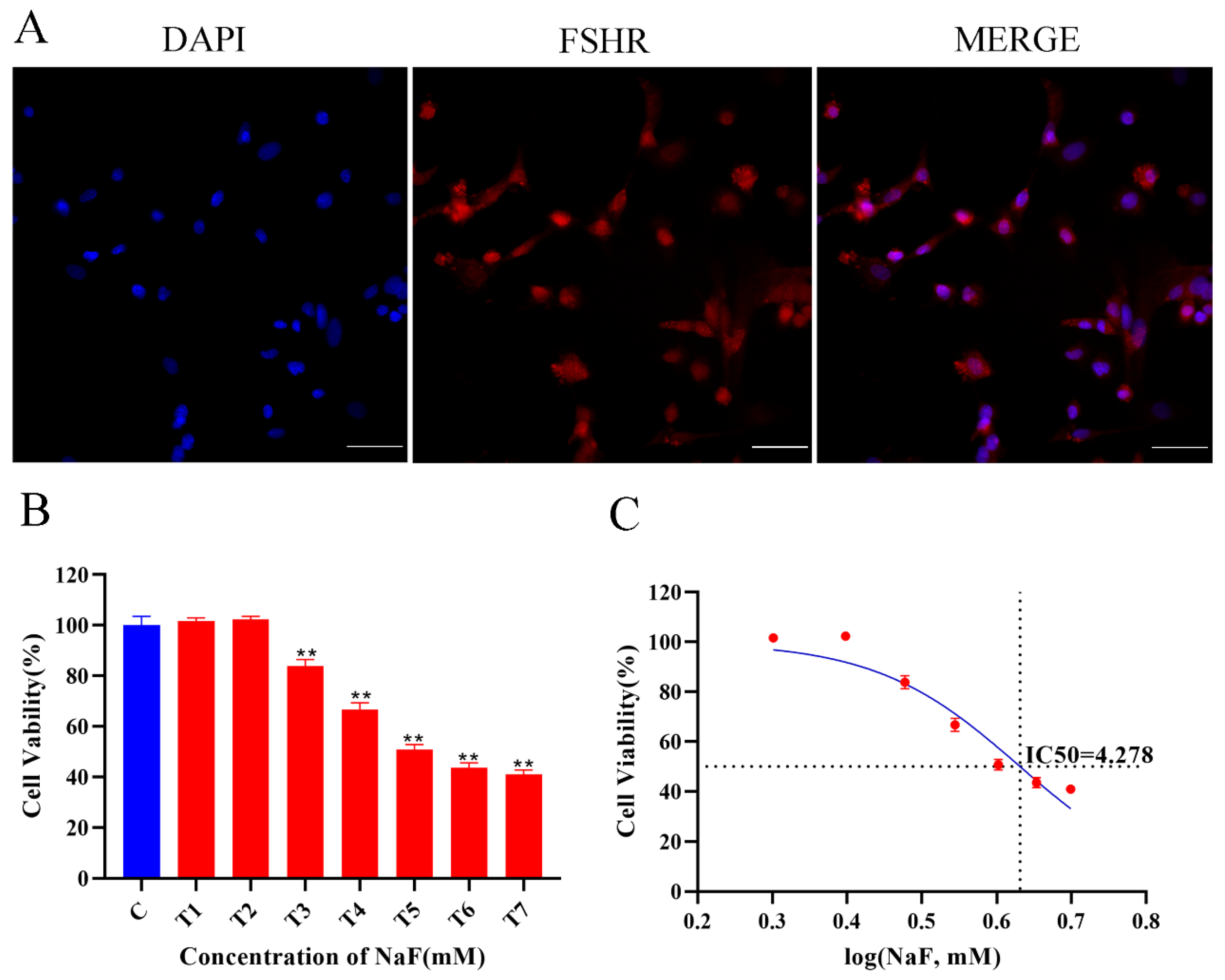
3.1. Cellular Identification and Cell Viability Assay
3.2. NaF Final Concentration Screening
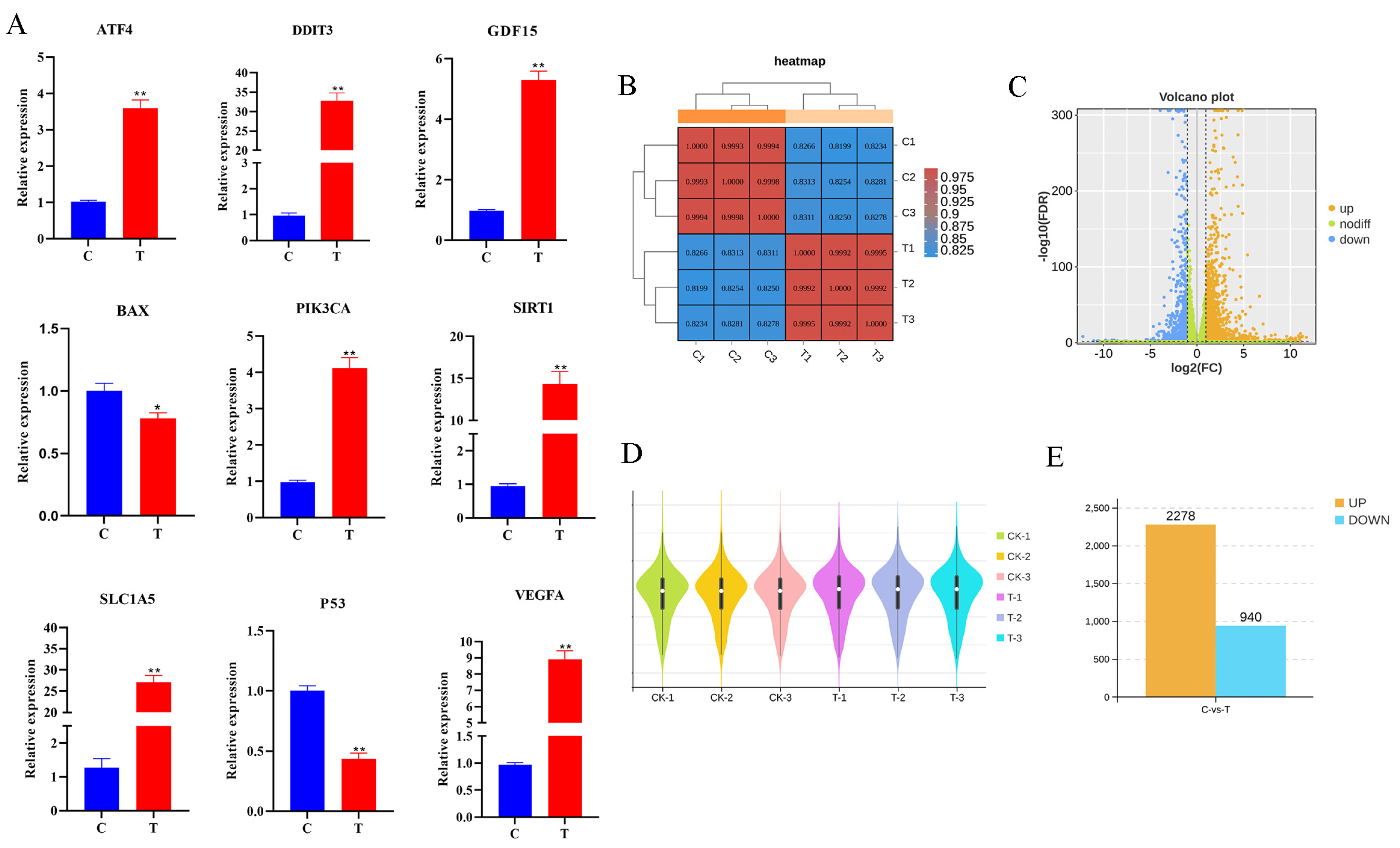
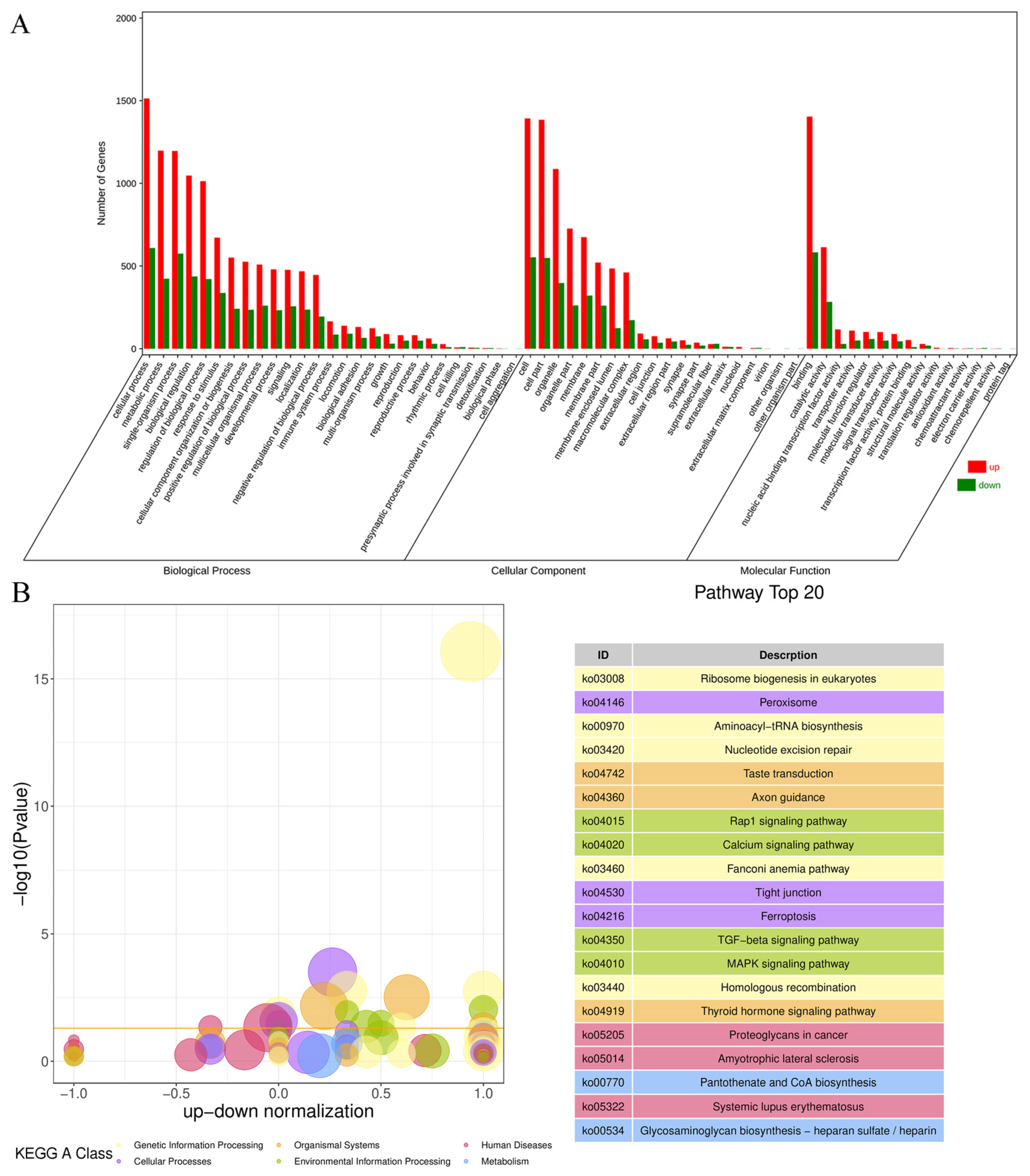
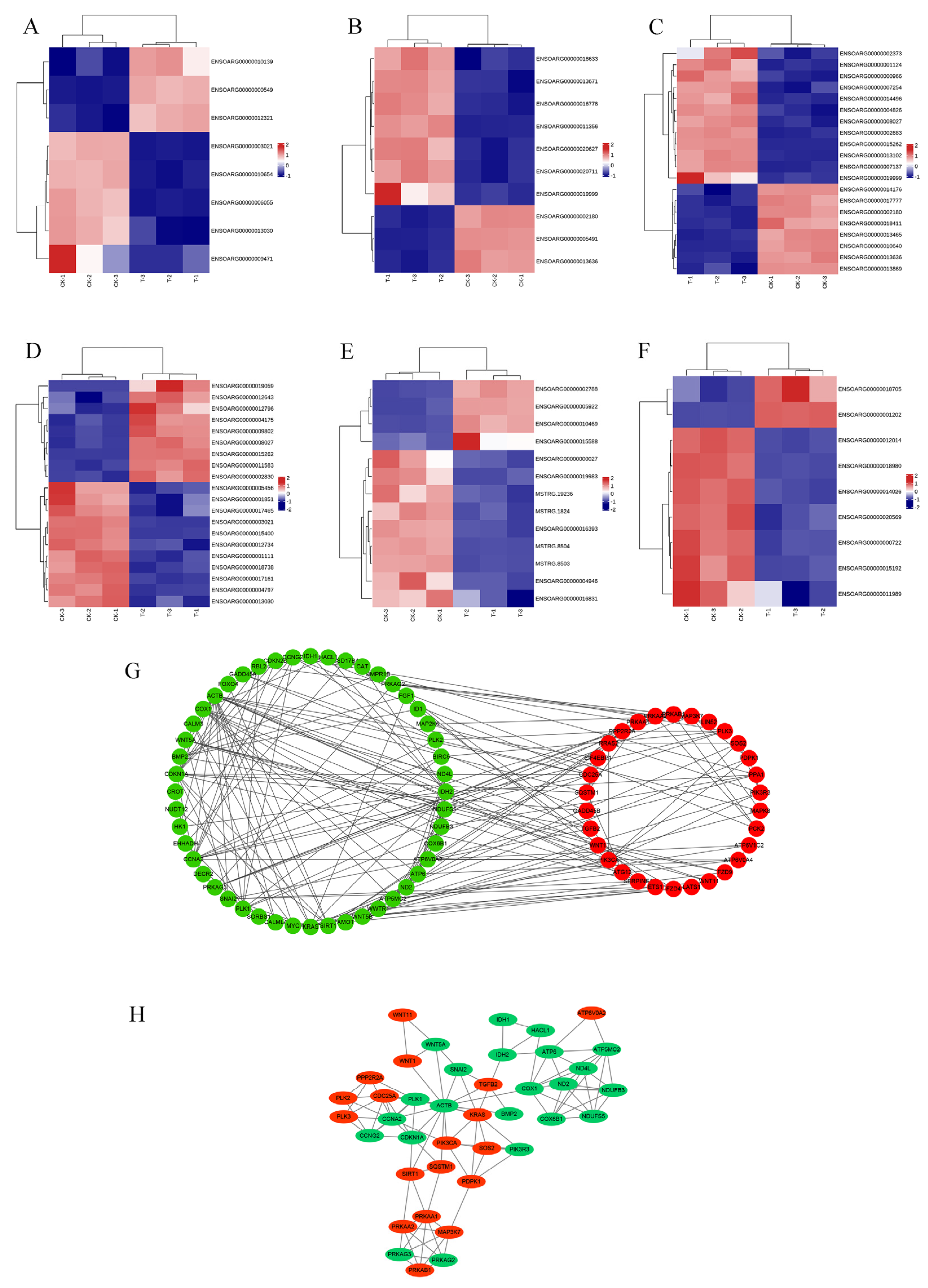
3.3. Analysis of RNA-Seq Data and qRT-PCR Validation
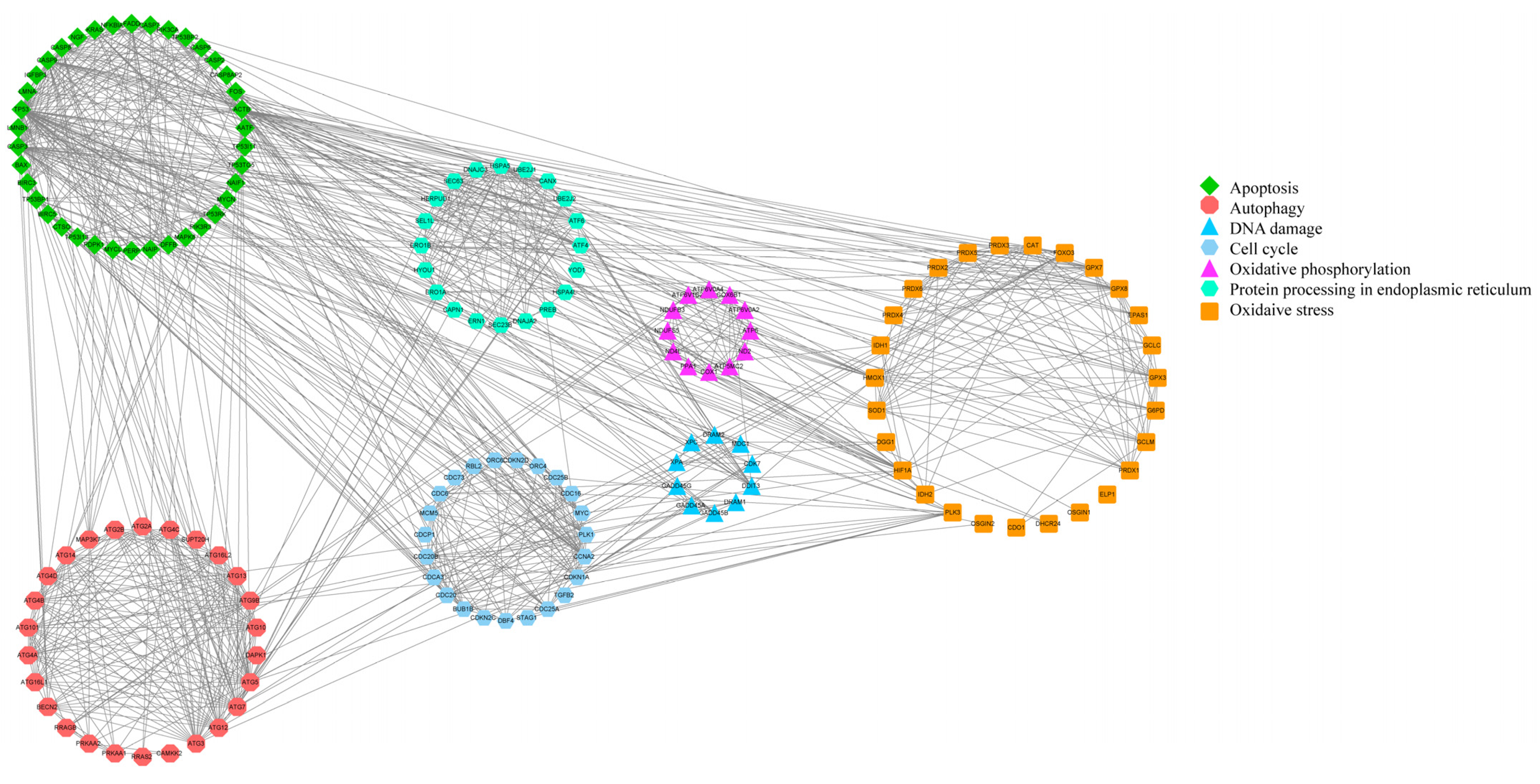
3.4. Inference Diagram of the Mechanism of Fluoride on Ovine Follicular GCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chakraborti, D.; Rahman, M.M.; Chatterjee, A.; Das, D.; Das, B.; Nayak, B.; Pal, A.; Chowdhury, U.K.; Ahmed, S.; Biswas, B.K.; et al. Fate of over 480 million inhabitants living in arsenic and fluoride endemic Indian districts: Magnitude, health, socio-economic effects and mitigation approaches. J. Trace Elem. Med. Biol. 2016, 38, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Kimambo, V.; Bhattacharya, P.; Mtalo, F.; Mtamba, J.; Ahmad, A. Fluoride occurrence in groundwater systems at global scale and status of defluoridation—State of the art. Groundw. Sustain. Dev. 2019, 9, 100223. [Google Scholar] [CrossRef]
- Rahim, A.; Essamadi, A.; El Amiri, B. A comprehensive review on endemic and experimental fluorosis in sheep: Its diverse effects and prevention. Toxicology 2022, 465, 153025. [Google Scholar] [CrossRef] [PubMed]
- Vitoria, I.; Maraver, F.; Almerich-Silla, J.M. Flúor en aguas de consumo público españolas y prevención de la caries dental. Gac. Sanit. 2014, 28, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Pollick, H.F. Water Fluoridation and the Environment: Current Perspective in the United States. Int. J. Occup. Environ. Health 2013, 10, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhu, Y.; Zhang, D.; Yan, Z.; Zhao, Y.; Manthari, R.K.; Cheng, X.; Wang, J.; Wang, J. Effects of Different Doses of Calcium on the Mitochondrial Apoptotic Pathway and Rho/ROCK Signaling Pathway in the Bone of Fluorosis Rats. Biol. Trace Elem. Res. 2020, 199, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Malin, A.J.; Lesseur, C.; Busgang, S.A.; Curtin, P.; Wright, R.O.; Sanders, A.P. Fluoride exposure and kidney and liver function among adolescents in the United States: NHANES, 2013–2016. Environ. Int. 2019, 132, 105012. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, J.; Li, Y.; Liu, H.; Hou, C.; Zeng, Q.; Cui, Y.; Zhao, L.; Li, P.; Zhou, Z.; et al. Threshold effects of moderately excessive fluoride exposure on children’s health: A potential association between dental fluorosis and loss of excellent intelligence. Environ. Int. 2018, 118, 116–124. [Google Scholar] [CrossRef]
- Wang, M.; Liu, L.; Li, H.; Li, Y.; Liu, H.; Hou, C.; Zeng, Q.; Li, P.; Zhao, Q.; Dong, L.; et al. Thyroid function, intelligence, and low-moderate fluoride exposure among Chinese school-age children. Environ. Int. 2020, 134, 105229. [Google Scholar] [CrossRef]
- Niu, Q.; Chen, J.; Xia, T.; Li, P.; Zhou, G.; Xu, C.; Zhao, Q.; Dong, L.; Zhang, S.; Wang, A. Excessive ER stress and the resulting autophagic flux dysfunction contribute to fluoride-induced neurotoxicity. Environ. Pollut. 2018, 233, 889–899. [Google Scholar] [CrossRef]
- Li, X.; Zhang, J.; Niu, R.; Manthari, R.K.; Yang, K.; Wang, J. Effect of fluoride exposure on anxiety- and depression-like behavior in mouse. Chemosphere 2019, 215, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Tang, S.; Yang, L.; Niu, Q.; Chen, J.; Xia, T.; Wang, S.; Wang, M.; Zhao, Q.; Liu, L.; et al. Effects of long-term fluoride exposure on cognitive ability and the underlying mechanisms: Role of autophagy and its association with apoptosis. Toxicol. Appl. Pharmacol. 2019, 378, 114608. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Dong, N.; Hao, X.; Xing, Y.; Tian, X.; Feng, J.; Xie, J.; Lv, Y.; Wei, C.; Gao, Y.; et al. Comparative Transcriptomics Reveals the Role of the Toll-Like Receptor Signaling Pathway in Fluoride-Induced Cardiotoxicity. J. Agric. Food Chem. 2019, 67, 5033–5042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.-P.; Wang, H.-W.; Liu, J.; Zhang, Z.-H.; Zhu, S.-Q.; Zhou, B.-H. Mitochondrial respiratory chain complex abnormal expressions and fusion disorder are involved in fluoride-induced mitochondrial dysfunction in ovarian granulosa cells. Chemosphere 2019, 215, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, H.; Ba, Y.; Cheng, X.-M.; Cui, L.-X. Effect of oxidative stress on fluoride-induced apoptosis in primary cultured Sertoli cells of rats. Int. J. Environ. Health Res. 2014, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Qiu, Y.; Liu, X.; Chen, X.; Ding, Y.; Liu, S.; Zhao, Y.; Gao, R.; Wang, Y.; He, J. Sodium fluoride activates ERK and JNK via induction of oxidative stress to promote apoptosis and impairs ovarian function in rats. J. Hazard. Mater. 2014, 272, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiang, C.; Liu, H.; Guan, Z.; Zeng, Q.; Zhang, C.; Lei, R.; Xia, T.; Gao, H.; Yang, L.; et al. Fluoride-elicited developmental testicular toxicity in rats: Roles of endoplasmic reticulum stress and inflammatory response. Toxicol. Appl. Pharmacol. 2013, 271, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Luo, G.; Sun, Z.; Wang, S.; Wang, J. Chronic fluoride exposure-induced testicular toxicity is associated with inflammatory response in mice. Chemosphere 2016, 153, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Nie, Q.; Zhang, L.; Niu, R.; Wang, J.; Wang, S. Fluoride reduced the immune privileged function of mouse Sertoli cells via the regulation of Fas/FasL system. Chemosphere 2017, 168, 318–325. [Google Scholar] [CrossRef]
- Sun, Z.; Xue, X.; Zhang, Y.; Niu, R.; Wang, J. Effect of sodium fluoride on the sperm mitochondrial DNA in mice. Biochem. Biophys. Res. Commun. 2017, 492, 295–299. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, W.; Li, S.; Xue, X.; Niu, R.; Shi, L.; Li, B.; Wang, X.; Wang, J. Altered miRNAs expression profiling in sperm of mice induced by fluoride. Chemosphere 2016, 155, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Liang, C.; Manthari, R.K.; Wang, C.; Zhang, J. Effects of Fluoride on Autophagy in Mouse Sertoli Cells. Biol. Trace Elem. Res. 2018, 187, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-C.; Yin, S.; Song, C.; Wu, H.; Chen, X.; Zhang, Y. Adverse Effects of High Concentrations of Fluoride on Characteristics of the Ovary and Mature Oocyte of Mouse. PLoS ONE 2015, 10, e0129594. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Zhao, W.-P.; Tan, P.-P.; Liu, J.; Zhao, J.; Zhou, B.-H. The MMP-9/TIMP-1 System is Involved in Fluoride-Induced Reproductive Dysfunctions in Female Mice. Biol. Trace Elem. Res. 2017, 178, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.P.; Li, L.L.; Zhu, M.K.; Dong, X.Y.; Elwan, H.A.M.; Zou, X.T. Excess dietary fluoride affects laying performance, egg quality, tissue retention, serum biochemical indices, and reproductive hormones of laying hens. Poult. Sci. 2019, 98, 6873–6879. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Irving-Rodgers, H.F.; Hummitzsch, K.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genom. 2014, 15, 24. [Google Scholar] [CrossRef]
- Richards, J.S.; Pangas, S.A. The ovary: Basic biology and clinical implications. J. Clin. Investig. 2010, 120, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, Z.; Xu, Z.; Chen, C.; Wang, J.; Zhu, J.; Dong, Z. miR-378d is Involved in the Regulation of Apoptosis and Autophagy of and E2 Secretion from Cultured Ovarian Granular Cells Treated by Sodium Fluoride. Biol. Trace Elem. Res. 2021, 199, 4119–4128. [Google Scholar] [CrossRef]
- Cavanagh, C.R.; Jonas, E.; Hobbs, M.; Thomson, P.C.; Tammen, I.; Raadsma, H.W. Mapping Quantitative Trait Loci (QTL) in sheep. III. QTL for carcass composition traits derived from CT scans and aligned with a meta-assembly for sheep and cattle carcass QTL. Genet. Sel. Evol. 2010, 42, 36. [Google Scholar] [CrossRef]
- Alberto, F.J.; Boyer, F.; Orozco-terWengel, P.; Streeter, I.; Servin, B.; de Villemereuil, P.; Benjelloun, B.; Librado, P.; Biscarini, F.; Colli, L.; et al. Convergent genomic signatures of domestication in sheep and goats. Nat. Commun. 2018, 9, 813. [Google Scholar] [CrossRef]
- Alapala Demirhan, S. Sheep farming business in Uşak city of Turkey: Economic structure, problems and solutions. Saudi J. Biol. Sci. 2019, 26, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.K.; Jain, A.; Kulkarni, V.S.; Govindaiah, M.G.; Aswathnarayan, T.; Sadana, D.K. Classification of four ovine breeds of southern peninsular zone of India: Morphometric study using classical discriminant function analysis. SpringerPlus 2013, 2, 29. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Qiu, D.; Zaky, M.Y.; Liu, J.; Xu, S.; Xiong, Y.; Fu, B.; Lin, X. Folic acid Ameliorates the Declining Quality of Sodium Fluoride-Exposed Mouse Oocytes through the Sirt1/Sod2 Pathway. Aging Dis. 2022, 13, 1471–1487. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ning, H.; Hua, L.; Ren, F.; Chen, L.; Ma, Z.; Li, R.; Ge, Y.; Yin, Z. Exposure to fluoride induces apoptosis in the liver, kidney, and heart of Xenopus laevis by regulating the Caspase-8/3 signaling pathway. Acta Histochem. 2023, 125, 151999. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Fu, B.; Zhang, J.; Zhao, J.; Yuan, M.; Peng, W.; Zhang, Y.; Wu, H. Sodium fluoride induces nephrotoxicity via oxidative stress-regulated mitochondrial SIRT3 signaling pathway. Sci. Rep. 2017, 7, 672. [Google Scholar] [CrossRef]
- Flora, S.J.S.; Mittal, M.; Mishra, D. Co-exposure to arsenic and fluoride on oxidative stress, glutathione linked enzymes, biogenic amines and DNA damage in mouse brain. J. Neurol. Sci. 2009, 285, 198–205. [Google Scholar] [CrossRef]
- Ridley, W.; Matsuoka, M. Fluoride-induced cyclooxygenase-2 expression and prostaglandin E2 production in A549 human pulmonary epithelial cells. Toxicol. Lett. 2009, 188, 180–185. [Google Scholar] [CrossRef]
- Sun, Z.; Niu, R.; Su, K.; Wang, B.; Wang, J.; Zhang, J.; Wang, J. Effects of sodium fluoride on hyperactivation and Ca2+ signaling pathway in sperm from mice: An in vivo study. Arch. Toxicol. 2010, 84, 353–361. [Google Scholar] [CrossRef]
- Guth, S.; Hüser, S.; Roth, A.; Degen, G.; Diel, P.; Edlund, K.; Eisenbrand, G.; Engel, K.-H.; Epe, B.; Grune, T.; et al. Toxicity of fluoride: Critical evaluation of evidence for human developmental neurotoxicity in epidemiological studies, animal experiments and in vitro analyses. Arch. Toxicol. 2020, 94, 1375–1415. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, A.; He, W.; He, P.; Xu, B.; Xia, T.; Chen, X.; Yang, K. Effects of fluoride on the expression of NCAM, oxidative stress, and apoptosis in primary cultured hippocampal neurons. Toxicology 2007, 236, 208–216. [Google Scholar] [CrossRef]
- Gao, Q.; Liu, Y.-J.; Guan, Z.-Z. Oxidative stress might be a mechanism connected with the decreased α7 nicotinic receptor influenced by high-concentration of fluoride in SH-SY5Y neuroblastoma cells. Toxicol. Vitr. 2008, 22, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Carwile, J.L.; Ahrens, K.A.; Seshasayee, S.M.; Lanphear, B.; Fleisch, A.F. Predictors of Plasma Fluoride Concentrations in Children and Adolescents. Int. J. Environ. Res. Public Health 2020, 17, 9205. [Google Scholar] [CrossRef] [PubMed]
- Márton, M.; Bánhegyi, G.; Gyöngyösi, N.; Kálmán, E.É.; Pettkó-Szandtner, A.; Káldi, K.; Kapuy, O. A systems biological analysis of the ATF4-GADD34-CHOP regulatory triangle upon endoplasmic reticulum stress. FEBS Open Bio 2022, 12, 2065–2082. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Ren, L.; Liu, J.; Li, W.; Zheng, X.; Wang, J.; Du, G. Withaferin A triggers G2/M arrest and intrinsic apoptosis in glioblastoma cells via ATF4-ATF3-CHOP axis. Cell Prolif. 2019, 53, e12706. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Zhao, J.; Bu, S.; Teng, H.; Yang, J.; Zhang, X.; Li, X.; Dong, L. KLF6 Induces Apoptosis in Human Lens Epithelial Cells Through the ATF4-ATF3-CHOP Axis. Drug Des. Dev. Ther. 2020, 14, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Yang, W.-L.; Wang, P. Endoplasmic Reticulum Stress in Sepsis. Shock 2015, 44, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.D.; Harding, H.P.; Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004, 167, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Li, Y.; Wang, C.; Gao, S.; Zhao, Y.; Yang, Z.; Wang, H.; Gao, Z.; Jiang, Y.; He, Y.; et al. Diterpenoid Vinigrol specifically activates ATF4/DDIT3-mediated PERK arm of unfolded protein response to drive non-apoptotic death of breast cancer cells. Pharmacol. Res. 2022, 182, 106285. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]




| Primers | Primer Sequences | NCBI Reference Sequence | Product Length (bp) |
|---|---|---|---|
| CAT | CCAGCCCTGACAAAATGCTT | XM_060400055.1 | 242 |
| AAAGCGGGTCCTATGTTCCA | |||
| SOD1 | GGCAATGTGAAGGCTGACAA | NM_001145185.2 | 130 |
| TGCCCAAGTCATCTGGTCTT | |||
| SOD2 | GGACAAATCTGAGCCCCAAC | NM_001280703.1 | 180 |
| CAATCTGTAAGCGTCCCTGC | |||
| GPX1 | CAGTTTGGGCATCAGGAAAAC | XM_004018462.5 | 100 |
| CGAAGAGCATGAAATTGGGC | |||
| CASP3 | ACGGAAGCAAATCAGTGGAC | XM_060406953.1 | 167 |
| GGTTTCCCTGAGGTTTGCTG | |||
| CASP8 | AGTGAGTTGCAGACATCCGA | XM_060410232.1 | 172 |
| AGGTCTTGTCCAAAGCCTCT | |||
| P53 | TCTTCAGATCCGTGGGCGTA | NM_001009403.1 | 158 |
| TTTTATGGCAGGAGGGAGAAGG | |||
| BAX | TTCCGACGGCAACTTCAACT | XM_027978594.3 | 211 |
| CCATGTGGGTGTCCCAAAGT | |||
| LC3 | ACGCCTCTCAGGAGACTTTTG | XM_004014953.4 | 121 |
| ACCTCAGTTGGTAACATCCCT | |||
| PIK3CA | GAGGAGCCCCGAGCATTTCT | NM_006218.4 | 134 |
| AAGTGGATGCCCCACAGTTC | |||
| ATF4 | AGGAGGATGCCCACTCAGAT | XM_012158819.3 | 172 |
| TCTCCAGGAGGGTCGTAAGG | |||
| DDIT3 | GCTGCCCTTCCCTTTTTGACTA | XM_060412904.1 | 111 |
| CTCAGTAAGCCAAGCCAGAGA | |||
| SIRT1 | GAGGCGGTTGAAAGATGGCG | XM_015104377.4 | 76 |
| TCAGCCGCCACTACCGAG | |||
| VEGFA | ACCAAAGCCAGCACATAGGA | NM_001025370.3 | 105 |
| GCCTCGGCTTGTCACATTTTTC | |||
| GDF15 | TGCTCATGTTCTCCTGGCTG | XM_004008419.5 | 179 |
| GTCTTCCCAGGTCTGGTTCG | |||
| SLC1A5 | CTTGATCTTGGCCGTGGACT | XM_027978525.2 | 99 |
| TCCAGGTAACTCGGGAGGAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, T.; Liu, W.; Jiang, D.; Zhang, G.; Zhao, X.; Zhang, Y.; Li, Z. Analysis of Toxic Effects of Fluoride on Ovine Follicular Granulosa Cells Using RNA-Seq. Antioxidants 2024, 13, 506. https://doi.org/10.3390/antiox13050506
Ma T, Liu W, Jiang D, Zhang G, Zhao X, Zhang Y, Li Z. Analysis of Toxic Effects of Fluoride on Ovine Follicular Granulosa Cells Using RNA-Seq. Antioxidants. 2024; 13(5):506. https://doi.org/10.3390/antiox13050506
Chicago/Turabian StyleMa, Tian, Wanruo Liu, Didi Jiang, Guolin Zhang, Xingxu Zhao, Yong Zhang, and Zongshuai Li. 2024. "Analysis of Toxic Effects of Fluoride on Ovine Follicular Granulosa Cells Using RNA-Seq" Antioxidants 13, no. 5: 506. https://doi.org/10.3390/antiox13050506
APA StyleMa, T., Liu, W., Jiang, D., Zhang, G., Zhao, X., Zhang, Y., & Li, Z. (2024). Analysis of Toxic Effects of Fluoride on Ovine Follicular Granulosa Cells Using RNA-Seq. Antioxidants, 13(5), 506. https://doi.org/10.3390/antiox13050506






