How Bacteria Cope with Oxidative Stress Induced by Cadmium: Volatile Communication Is Differentially Perceived among Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures
2.2. Experimental Conditions
2.3. Biochemical Analysis
2.3.1. Extraction
2.3.2. Protein Content
2.3.3. Protein Carbonylation
2.3.4. Lipid Peroxidation
2.3.5. Superoxide Dismutase
2.3.6. Catalase
2.3.7. Glutathione S-Transferases
2.3.8. Electron Transport System
2.4. Volatiles Released by Rhizobium sp. E20-8
2.5. Data Analysis
3. Results
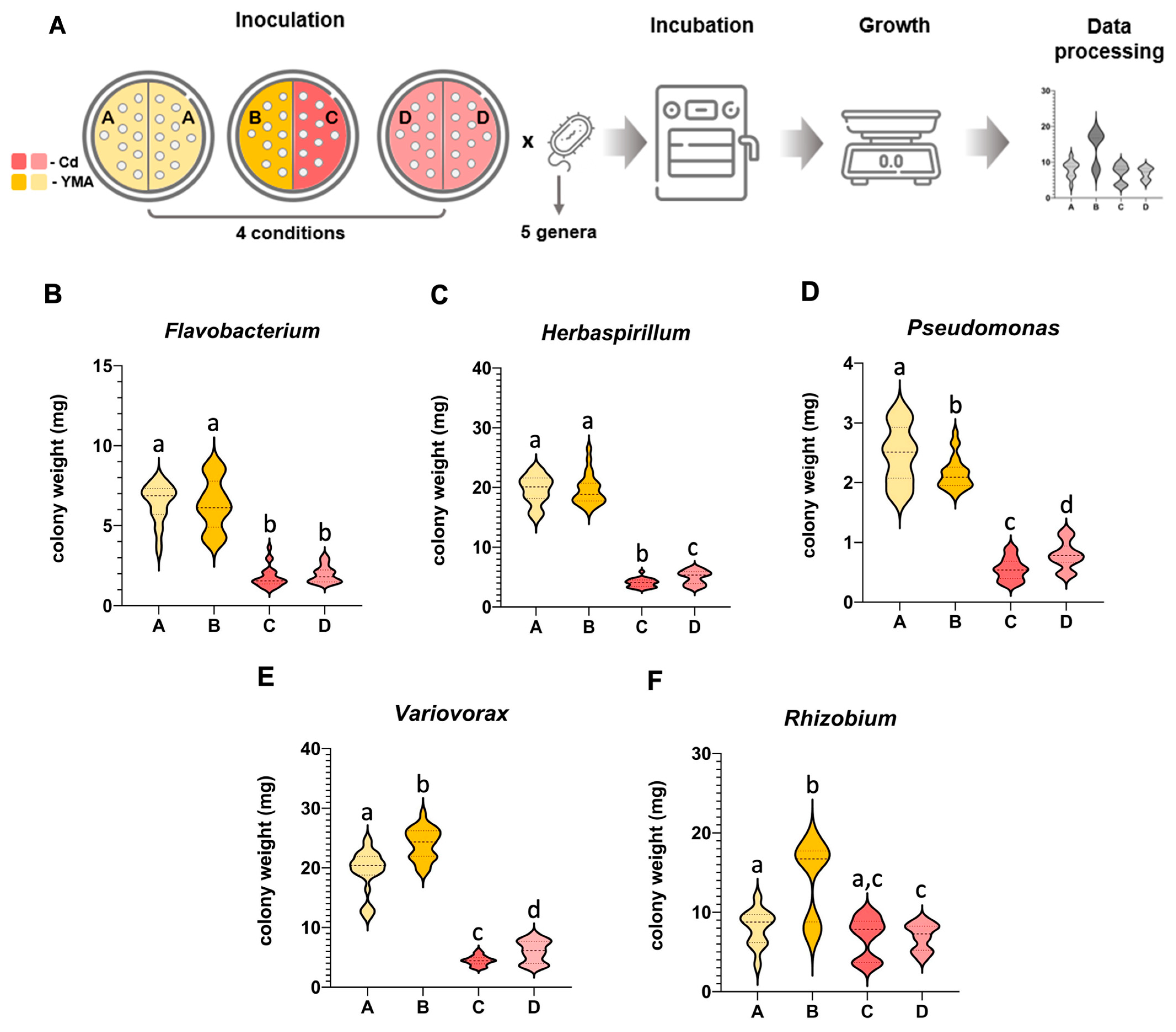
3.1. Mutual Airborne Influence of a Bacterial Strain Exposed to Different Conditions Varied among Bacterial Genera
3.2. The Volatiles Produced by Rhizobium Grown in Cd Influenced Distinctly the Growth of 13 Bacterial Strains
3.3. The Antioxidant and Biotransformation Mechanisms Seem to Be Induced in Strains That Grew Less and to Be Reduced in Those That Grew More in the Presence of Rhizobium Volatiles (Rz VOCs)
3.4. Changes in Cell Metabolism Seem to Be Inversely Related to the Influence of Rz VOCs on Growth
3.5. Strains with Lower Growth Evidenced Higher Membrane and Protein Damage
3.6. Most Biochemical Changes Were Highly Correlated with Strains That Grew Less under the Influence of Rz VOCs
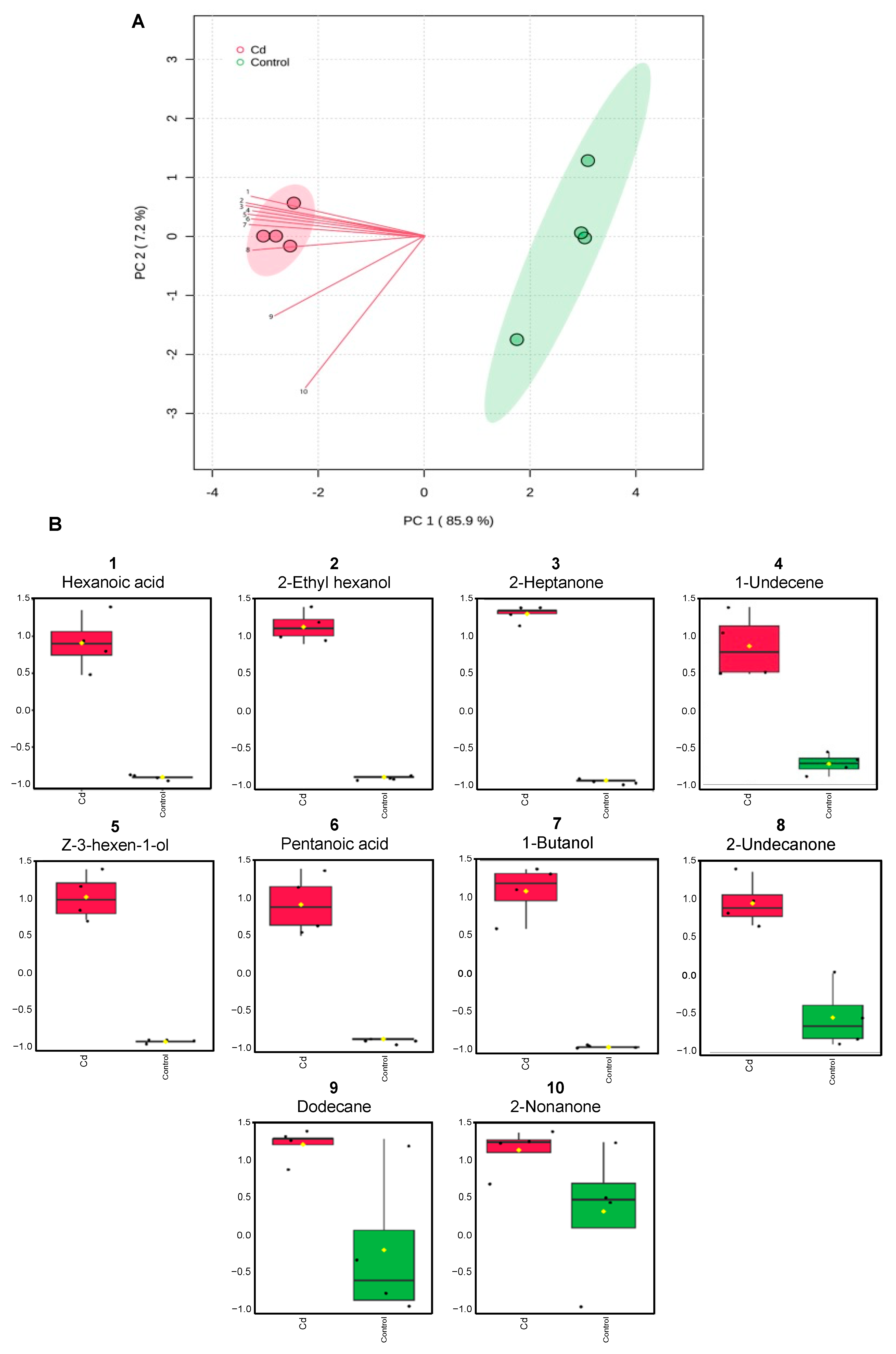
3.7. Exposure to Cd Alters Rhizobium Volatilome
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, M.D. An Ecological Perspective on Extreme Climatic Events: A Synthetic Definition and Framework to Guide Future Research. J. Ecol. 2011, 99, 656–663. [Google Scholar] [CrossRef]
- Reichstein, M.; Bahn, M.; Ciais, P.; Frank, D.; Mahecha, M.D.; Seneviratne, S.I.; Zscheischler, J.; Beer, C.; Buchmann, N.; Frank, D.C.; et al. Climate Extremes and the Carbon Cycle. Nature 2013, 500, 287–295. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of Soil Bacterial and Fungal Communities to Extreme Desiccation and Rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Evans, S.E.; Wallenstein, M.D. Climate Change Alters Ecological Strategies of Soil Bacteria. Ecol. Lett. 2014, 17, 155–164. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial Persister Cell Formation and Dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Trastoy, R.; Manso, T.; Fernández-García, L.; Blasco, L.; Ambroa, A.; Pérez del Molino, M.L.; Bou, G.; García-Contreras, R.; Wood, T.K.; Tomás, M. Mechanisms of Bacterial Tolerance and Persistence in the Gastrointestinal and Respiratory Environments. Clin. Microbiol. Rev. 2018, 31. [Google Scholar] [CrossRef]
- Fernández-Raga, M.; Palencia, C.; Keesstra, S.; Jordán, A.; Fraile, R.; Angulo-Martínez, M.; Cerdà, A. Splash Erosion: A Review with Unanswered Questions. Earth-Sci. Rev. 2017, 171, 463–477. [Google Scholar] [CrossRef]
- Pagiola, S.; Ramirez, E.; Gobbi, J.; De haan, C.; Ibrahim, M.; Murgueitio, E.; Ruíz, J. Paying for the Environmental Services of Silvopastoral Practices in Nicaragua. Ecol. Econ. 2007, 64, 374–385. [Google Scholar] [CrossRef]
- Jie, C.; Jing-zhang, C.; Man-zhi, T.; Zi-tong, G. Soil Degradation: A Global Problem Endangering Sustainable Development. J. Geogr. Sci. 2002, 12, 243–252. [Google Scholar] [CrossRef]
- Gilland, B. World Population and Food Supply: Can Food Production Keep Pace with Population Growth in the next Half-Century? Food Policy 2002, 27, 47–63. [Google Scholar] [CrossRef]
- Aktar, W.; Sengupta, D.; Chowdhury, A. Impact of Pesticides Use in Agriculture: Their Benefits and Hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Megha, P.; Sreedev, P. Review Article. Organochlorine Pesticides, Their Toxic Effects on Living Organisms and Their Fate in the Environment. Interdiscip. Toxicol. 2016, 9, 90–100. [Google Scholar] [CrossRef]
- Roberts, T.L. Cadmium and Phosphorous Fertilizers: The Issues and the Science. Procedia Eng. 2014, 83, 52–59. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, H.; He, J.; Lyu, D.; Li, H. Integration of Cadmium Accumulation, Subcellular Distribution, and Physiological Responses to Understand Cadmium Tolerance in Apple Rootstocks. Front. Plant Sci. 2017, 8, 261167. [Google Scholar] [CrossRef]
- Asgher, M.; Khan, M.I.; Anjum, N.A.; Khan, N.A. Minimising Toxicity of Cadmium in Plants—Role of Plant Growth Regulators. Protoplasma 2015, 252, 399–413. [Google Scholar] [CrossRef]
- Lebeau, T.; Braud, A.; Jézéquel, K. Performance of Bioaugmentation-Assisted Phytoextraction Applied to Metal Contaminated Soils: A Review. Environ. Pollut. Barking Essex 1987 2008, 153, 497–522. [Google Scholar] [CrossRef]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The Role of Plant-Associated Bacteria in the Mobilization and Phytoextraction of Trace Elements in Contaminated Soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.H.; Anderson, C.W.N.; Dickinson, N.M. Phytoextraction: Where’s the Action? J. Geochem. Explor. 2015, 151, 34–40. [Google Scholar] [CrossRef]
- Krämer, U. Phytoremediation: Novel Approaches to Cleaning up Polluted Soils. Curr. Opin. Biotechnol. 2005, 16, 133–141. [Google Scholar] [CrossRef]
- Somero, G.N. The Physiology of Climate Change: How Potentials for Acclimatization and Genetic Adaptation Will Determine ‘Winners’ and ‘Losers’. J. Exp. Biol. 2010, 213, 912–920. [Google Scholar] [CrossRef]
- Or, D.; Smets, B.F.; Wraith, J.M.; Dechesne, A.; Friedman, S.P. Physical Constraints Affecting Bacterial Habitats and Activity in Unsaturated Porous Media—A Review. Adv. Water Resour. 2007, 30, 1505–1527. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J.S. Bacterial Volatiles: The Smell of Small Organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.K.; King, A.J.; Meier, C.L.; Bowman, W.D.; Farrer, E.C.; Suding, K.N.; Nemergut, D.R. Plant–Microbe Interactions at Multiple Scales across a High-Elevation Landscape. Plant Ecol. Divers. 2015, 8, 703–712. [Google Scholar] [CrossRef]
- Kai, M.; Haustein, M.; Molina, F.; Petri, A.; Scholz, B.; Piechulla, B. Bacterial Volatiles and Their Action Potential. Appl. Microbiol. Biotechnol. 2009, 81, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.S.; Crippen, T.L.; Hofstetter, R.W.; Tomberlin, J.K. Microbial Volatile Emissions as Insect Semiochemicals. J. Chem. Ecol. 2013, 39, 840–859. [Google Scholar] [CrossRef]
- Sá, C.; Matos, D.; Cardoso, P.; Figueira, E. Do Volatiles Affect Bacteria and Plants in the Same Way? Growth and Biochemical Response of Non-Stressed and Cd-Stressed Arabidopsis Thaliana and Rhizobium E20-8. Antioxidants 2022, 11, 2303. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Surette, M.G. Communication in Bacteria: An Ecological and Evolutionary Perspective. Nat. Rev. Microbiol. 2006, 4, 249–258. [Google Scholar] [CrossRef]
- Stephens, K.; Pozo, M.; Tsao, C.-Y.; Hauk, P.; Bentley, W.E. Bacterial Co-Culture with Cell Signaling Translator and Growth Controller Modules for Autonomously Regulated Culture Composition. Nat. Commun. 2019, 10, 4129. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, A.; Perazzolli, M.; Pertot, I.; Puopolo, G. The Perception of Rhizosphere Bacterial Communication Signals Leads to Transcriptome Reprogramming in Lysobacter Capsici AZ78, a Plant Beneficial Bacterium. Front. Microbiol. 2021, 12, 725403. [Google Scholar] [CrossRef]
- Cardoso, P.; Alves, A.; Silveira, P.; Sá, C.; Fidalgo, C.; Freitas, R.; Figueira, E. Bacteria from Nodules of Wild Legume Species: Phylogenetic Diversity, Plant Growth Promotion Abilities and Osmotolerance. Sci. Total Environ. 2018, 645, 1094–1102. [Google Scholar] [CrossRef]
- Farag, M.A.; Song, G.C.; Park, Y.-S.; Audrain, B.; Lee, S.; Ghigo, J.-M.; Kloepper, J.W.; Ryu, C.-M. Biological and Chemical Strategies for Exploring Inter- and Intra-Kingdom Communication Mediated via Bacterial Volatile Signals. Nat. Protoc. 2017, 12, 1359–1377. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Lee, S.; Ryu, C.M. Interspecific Bacterial Sensing through Airborne Signals Modulates Locomotion and Drug Resistance. Nat. Commun. 2013, 4, 1809–1812. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.W.; Hogden, C.G. The Biuret Reaction in the Determination of Serum Proteins. J. Biol. Chem. 1940, 135, 707–725. [Google Scholar] [CrossRef]
- Mesquita, C.S.; Oliveira, R.; Bento, F.; Geraldo, D.; Rodrigues, J.V.; Marcos, J.C. Simplified 2,4-Dinitrophenylhydrazine Spectrophotometric Assay for Quantification of Carbonyls in Oxidized Proteins. Anal. Biochem. 2014, 458, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Buege, J.A.; Aust, S.D. Microsomal Lipid Peroxidation. Methods Enzymol. 1978, 52, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, C.; Fridovich, I. Superoxide Dismutase: Improved Assays and an Assay Applicable to Acrylamide Gels. Anal. Biochem. 1971, 287, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.H.; Borg, L.A. A Spectrophotometric Method for Determination of Catalase Activity in Small Tissue Samples. Anal. Biochem. 1988, 174, 331–336. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-Transferases. The First Enzymatic Step in Mercapturic Acid Formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- De Coen, W.M.; Janssen, C.R. The Use of Biomarkers in Daphnia Magna Toxicity Testing. IV. Cellular Energy Allocation: A New Methodology to Assess the Energy Budget of Toxicant-Stressed Daphnia Populations. J. Aquat. Ecosyst. Stress Recovery 1997, 6, 43–55. [Google Scholar] [CrossRef]
- Farag, M.A.; Ryu, C.-M.; Sumner, L.W.; Paré, P.W. GC-MS SPME Profiling of Rhizobacterial Volatiles Reveals Prospective Inducers of Growth Promotion and Induced Systemic Resistance in Plants. Phytochemistry 2006, 67, 2262–2268. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards More Transparent and Integrative Metabolomics Analysis. Nucl. Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Audrain, B.; Létoffé, S.; Ghigo, J.-M. Airborne Bacterial Interactions: Functions Out of Thin Air? Front. Microbiol. 2015, 6, 1476. [Google Scholar] [CrossRef]
- Tyc, O.; Zweers, H.; de Boer, W.; Garbeva, P. Volatiles in Inter-Specific Bacterial Interactions. Front. Microbiol. 2015, 6, 166507. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Keren-Paz, A.; Korenblum, E.; Oved, R.; Malitsky, S.; Kolodkin-Gal, I. Weaponizing Volatiles to Inhibit Competitor Biofilms from a Distance. Npj Biofilms Microbiomes 2021, 7, 2. [Google Scholar] [CrossRef]
- Groenhagen, U.; Baumgartner, R.; Bailly, A.; Gardiner, A.; Eberl, L.; Schulz, S.; Weisskopf, L. Production of Bioactive Volatiles by Different Burkholderia Ambifaria Strains. J. Chem. Ecol. 2013, 39, 892–906. [Google Scholar] [CrossRef]
- Kehe, J.; Ortiz, A.; Kulesa, A.; Gore, J.; Blainey, P.C.; Friedman, J. Positive Interactions Are Common among Culturable Bacteria. Sci. Adv. 2021, 7, eabi7159. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Walker, D.M.; Harshey, R.M. Dead Cells Release a ‘Necrosignal’ That Activates Antibiotic Survival Pathways in Bacterial Swarms. Nat. Commun. 2020, 11, 4157. [Google Scholar] [CrossRef] [PubMed]
- Audrain, B.; Farag, M.A.; Ryu, C.-M.; Ghigo, J.-M. Role of Bacterial Volatile Compounds in Bacterial Biology. FEMS Microbiol. Rev. 2015, 39, 222–233. [Google Scholar] [CrossRef]
- Corre, M.-H.; Mercier, A.; Bouteiller, M.; Khalil, A.; Ginevra, C.; Depayras, S.; Dupont, C.; Rouxel, M.; Gallique, M.; Grac, L.; et al. Bacterial Long-Range Warfare: Aerial Killing of Legionella Pneumophila by Pseudomonas Fluorescens. Microbiol. Spectr. 2021, 9, 10–1128. [Google Scholar] [CrossRef]
- Lemire, J.; Alhasawi, A.; Appanna, V.P.; Tharmalingam, S.; Appanna, V.D. Metabolic Defence against Oxidative Stress: The Road Less Travelled so Far. J. Appl. Microbiol. 2017, 123, 798–809. [Google Scholar] [CrossRef]
- Garbeva, P.; Hordijk, C.; Gerards, S.; De Boer, W. Volatile-Mediated Interactions between Phylogenetically Different Soil Bacteria. Front. Microbiol. 2014, 5, 289. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.G.; Dong, S.; Catalano, C.; Moore, R.; Liang, X.; Mekalanos, J.J. Generation of Reactive Oxygen Species by Lethal Attacks from Competing Microbes. Proc. Natl. Acad. Sci. USA 2015, 112, 2181–2186. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, P.; Santos, M.; Freitas, R.; Rocha, S.M.; Figueira, E. Response of Rhizobium to Cd Exposure: A Volatile Perspective. Environ. Pollut. 2017, 231, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S.; Maier, R.M.; Pepper, I.L. Microbial Communication. In Environmental Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 461–481. [Google Scholar] [CrossRef]
- Abis, L.; Loubet, B.; Ciuraru, R.; Lafouge, F.; Houot, S.; Nowak, V.; Tripied, J.; Dequiedt, S.; Maron, P.A.; Sadet-Bourgeteau, S. Reduced Microbial Diversity Induces Larger Volatile Organic Compound Emissions from Soils. Sci. Rep. 2020, 10, 6104. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardoso, P.; Pinto, R.; Lopes, T.; Figueira, E. How Bacteria Cope with Oxidative Stress Induced by Cadmium: Volatile Communication Is Differentially Perceived among Strains. Antioxidants 2024, 13, 565. https://doi.org/10.3390/antiox13050565
Cardoso P, Pinto R, Lopes T, Figueira E. How Bacteria Cope with Oxidative Stress Induced by Cadmium: Volatile Communication Is Differentially Perceived among Strains. Antioxidants. 2024; 13(5):565. https://doi.org/10.3390/antiox13050565
Chicago/Turabian StyleCardoso, Paulo, Ricardo Pinto, Tiago Lopes, and Etelvina Figueira. 2024. "How Bacteria Cope with Oxidative Stress Induced by Cadmium: Volatile Communication Is Differentially Perceived among Strains" Antioxidants 13, no. 5: 565. https://doi.org/10.3390/antiox13050565
APA StyleCardoso, P., Pinto, R., Lopes, T., & Figueira, E. (2024). How Bacteria Cope with Oxidative Stress Induced by Cadmium: Volatile Communication Is Differentially Perceived among Strains. Antioxidants, 13(5), 565. https://doi.org/10.3390/antiox13050565







